Abstract
It has recently been shown that monoclonal antibody (MoAb) 97A6 detects a surface antigen expressed on basophils and their CD34+ precursor cells, as well as the basophil cell line KU-812. In this report the partial amino acid sequence of affinity chromatography– and sodium dodecyl sulfate–polyacrylamide gel electrophoresis–separated 97A6 antigen(s) from KU-812 lysates was determined. Sequence alignment of high-performance liquid chromatography–selected tryptic peptides from the resulting 130- and 150-kd bands revealed a 100% identity with amino acids 393 to 405 of ectonucleotide pyrophosphatase/phosphodiesterase-3 (E-NPP3; CD203c) but not of the related ectoenzyme PC-1 (E-NPP1). Moreover, MoAb 97A6 selectively recognized 293 cells transfected with human E-NPP3, but did not react with cells transfected with PC-1 or parental 293 cells. In addition, E-NPP3 messenger RNA expression was detected in basophils but not other peripheral blood cells. Finally, MoAb 97A6 immunoprecipitated phosphodiesterase activity from KU-812 cells and peripheral blood basophils, but not from other cell populations. These data demonstrate that MoAb 97A6 recognizes the functionally active type II transmembrane ectoenzyme E-NPP3.
Introduction
In an attempt to generate antibodies against surface antigens expressed on rare hematopoietic cell populations, monoclonal antibody (MoAb) 97A6 was raised, which selectively recognizes basophils, mast cells, and their CD34+ precursor cells.1 Stimulation of the basophils from donors allergic for acarids allergen with either anti-IgE antiserum or allergen results in a dose-dependent increase of 97A6 antigen expression, suggesting that the detected antigen plays an important role in cell activation.1 Immunoprecipitation of the proteins recognized by MoAb 97A6 revealed 2 bands of 150 kd and 270 kd, respectively.1 However, the identity of the molecule remained unknown. In this report, we show that 97A6 affinity-purified lysates consist of ectonucleotide pyrophosphatase phosphodiesterase-3 (E-NPP3), also termed phosphodiesterase I/nucleotide pyrophosphatase-3 (PDNP3), an ectoenzyme previously found in uterus, prostate, and glioma.2-4
Study design
Affinity purification of 97A6 antigen
Coupling of purified MoAb 97A6 to an N-hydroxy succinimide (NHS)-activated Sepharose column (Pharmacia, Freiburg, Germany) was performed according to the manufacturer's recommendations. Lysates from 3 × 109 KU-812 cells were loaded on a previously prepared 97A6-Sepharose affinity column equilibrated with washing buffer (10 mM Tris/HCl, 150 mM NaCl, 0.025% NaN3, 0.5% Triton X-100, pH 8.0). Bound 97A6 antigen was eluted with 50 mM triethanolamine, 0.1% Triton X-100, 150 mM NaCl, pH 11.5 and collected in tubes containing 1M Tris/HCl, pH 6.7. The collected fractions were screened on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) for appearance of specific bands. In the next step, positive fractions were pooled and used for preparative separations on 7.5% SDS-PAGE. The proteins were visualized by silver staining and the resulting bands were cut for commercial microsequencing. This included tryptic digestion of the proteins, peptide mass fingerprint analysis (MALDI-MS), reverse phase–high-performance liquid chromatography (RP-HPLC), sequencing of the purified peptides, and search in the NCBI database (Toplab, Martinsried, Germany).
Generation of E-NPP3 transfectant cell line
The E-NPP3 complementary DNA (cDNA) was prepared and inserted into plasmids as described previously.2 The calcium phosphate method was used for transfection of 293 embryonic kidney cells with 15 μg plasmid.5 Transfected cells were grown in tissue culture flasks in the presence of 0.5 mg/mL Geneticin. Six days after transfection the cells were stained with MoAb 97A6–phycoerythrin (PE) and the 97A6+ fraction (9% of total cells) was sorted on a FACSVantage cell sorter (Becton Dickinson, Heidelberg, Germany). After 3 sorting rounds the cells stably expressed E-NPP3 on the cell surface. The transfectant cells were stained with MoAb 97A6–PE and analyzed on a FACSCalibur (Becton Dickinson).
Purification of peripheral blood basophils
Ficoll-Hypaque–selected buffy coat peripheral blood (PB) cells from healthy donors were stained with 97A6-PE plus anti-PE–MACS beads and selected by magnetic activated cell sorting (MACS) (Miltenyi, Bergisch Gladbach, Germany). Staining and separation were performed as described by the manufacturer (Miltenyi). Purity of 97A6+ cells was more than 98%. The 97A6− mononuclear cells that passed the column were further separated by FACS sorting. The resulting purity of these cells was more than 99.9%.
Analysis of E-NPP3 messenger RNA expression in KU-812 cells and basophils
Total RNA was extracted from 1.5 × 106KU-812 cells and Ficoll-separated neutrophils plus eosinophils, as well as MACS-selected 97A6+ and 97A6− mononuclear cells (interphase cells) using RNeasy column (Qiagen, Hilden, Germany). Reverse transcription–polymerase chain reaction (RT-PCR) for detection of E-NPP3 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) messenger RNA (mRNA) was performed essentially as described before.6 The sequences of E-NPP3 mRNA primers were as follows: forward primer: 5′-GTGGATCAACAGTGGCTG; reverse primer: 5′-TTCAGGACAGCTCTCCAC. The sequences of GAPDH primers were used as described.6 The primers were designed so that genomic DNA is not amplified. Amplification was achieved by performing 20 cycles, each cycle consisting of 94°C 1 minute, 60°C 1 minute, and 72°C 1 minute. The amplified product was run on a 1% agarose gel, transferred to a nylon membrane, and hybridized with each corresponding32P-labeled cDNA probe. The washed membrane was exposed to an x-ray film for 1.5 hours at −80°C.
Determination of phosphodiesterase-I activity
For assessment of phosphodiesterase-I activity, lysates from 1.5 × 105 KU-812 cells or 1.5 × 105 PB cells (97A6+, 97A6− cells, neutrophils plus eosinophils) were incubated for 5 hours at 37°C in 20 mM Tris/HCl, pH 9.6 containing 5 mM MgCl2 and 1 mM p-nitrophenyl thymidine-5′-l-monosphosphate (Sigma-Aldrich, St Louis, MO). The reaction was terminated by the addition of 0.1 N NaOH and the reaction product was quantified by reading the absorbance at 410 nm (A410 × 64 = nmol p-nitrophenol). For immunoprecipitation experiments, the lysates were incubated with either MoAb 97A6 or an isotype-matched control antibody (Coulter, Tokyo, Japan) for 30 minutes at room temperature. The antibody-labeled lysates were then coupled to avidin-agarose beads (Sigma-Aldrich). After washing the beads 4 times with phosphate-buffered saline, enzymatic activities were determined as described above.
Results and discussion
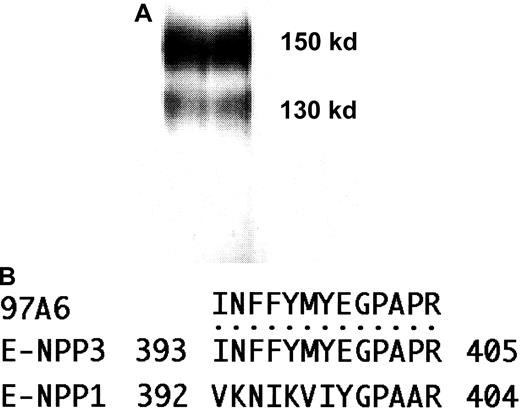
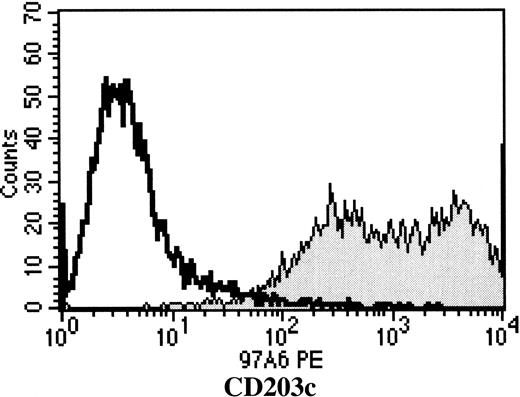
MoAb 97A6 was recently described to recognize 150- and 270-kd cell surface antigens expressed on basophils, mast cells, and their precursors.1 To identify the detected molecule(s) lysates from KU-812 cells were purified on 97A6-Sepharose affinity columns and the eluted proteins separated by SDS-PAGE. Using this approach 2 bands of 130 kd and 150 kd were obtained (Figure1A). In the next step the bands were cut and subjected to commercial microsequencing. Partial amino acid sequencing of selected, HPLC-purified tryptic peptides from both bands resulted in identical sequences. Comparisons with protein sequences from the NCBI database revealed 100% identity of the peptide with amino acids 393 to 405 from the recently cloned ectonucleotide pyrophosphatase phosphodiesterase 3 (E-NPP3: accession number: NM 005021.1).2,3 Figure 1B shows that the sequence of this peptide matches E-NPP3 sequences, but not the sequences of the most related membrane-bound phosphodiesterase I species, E-NPP1 (PC-1). E-NPP2 (autotaxin) that exists only in a soluble form did not show any similarity either (not shown). To confirm the specificity of MoAb 97A6, 293 embryonic kidney cells were transfected with full-length E-NPP3 cDNA and the resulting cells analyzed for reactivity with MoAb 97A6. Initially, 9% of the cells were 97A6+. This population was sorted by FACS and after 3 sorting and culture rounds a stable transfectant cell line was established that expressed high levels of E-NPP3. Figure2 shows that MoAb 97A6 selectively recognizes 293 cells transfected with human E-NPP3 (293/huE-NPP3) but not 293 cells transfected with a control plasmid. In addition, MoAb 97A6 does not cross-react with 293 cells transfected with the related protein E-NPP1 (not shown).2 During the last conference on human leukocyte differentiation antigens (HLDA) in Harrogate, United Kingdom (June 20-24, 2000), the specificity of MoAb 97A6 antibody for E-NPP3 was confirmed and clustered to CD203c.7
MoAb 97A6 detects 130- and 150-kd proteins that correspond to E-NPP3.
(A) Lysates of KU-812 cells were affinity purified on a 97A6-Sepharose column and separated on 7.5% SDS-PAGE. The silver-stained proteins had a molecular mass of 130 kd and 150 kd, respectively. (B) The sequences of E-NPP1 (PC-1) and E-NPP3 (PDNP3) are aligned with tryptic HPLC-purified 97A6 peptides derived from the cut bands shown in panel A. The sequence of the digested peptides (identical peptides from both bands) shows a complete match with amino acids 393 to 405 of E-NPP3. Although the primary structures of E-NPP family proteins share a significant similarity, the sequence of this region differs considerably between E-NPP1 and E-NPP3.
MoAb 97A6 detects 130- and 150-kd proteins that correspond to E-NPP3.
(A) Lysates of KU-812 cells were affinity purified on a 97A6-Sepharose column and separated on 7.5% SDS-PAGE. The silver-stained proteins had a molecular mass of 130 kd and 150 kd, respectively. (B) The sequences of E-NPP1 (PC-1) and E-NPP3 (PDNP3) are aligned with tryptic HPLC-purified 97A6 peptides derived from the cut bands shown in panel A. The sequence of the digested peptides (identical peptides from both bands) shows a complete match with amino acids 393 to 405 of E-NPP3. Although the primary structures of E-NPP family proteins share a significant similarity, the sequence of this region differs considerably between E-NPP1 and E-NPP3.
MoAb 97A6 recognizes E-NPP3.
HEK 293 cells were transfected with full-length E-NPP3 cDNA and the resulting cells were screened for reactivity with MoAb 97A6. The positive population was sorted by FACS and cultured in RPMI 1640 plus 10% fetal bovine serum. After 3 rounds of sorting, the cells were stained either with control IgG1-PE (dotted histogram) or 97A6-PE (filled histogram) and analyzed on a FACSCalibur flow cytometer. Nontransfected 293 cells were negative for MoAb 97A6 (not shown).
MoAb 97A6 recognizes E-NPP3.
HEK 293 cells were transfected with full-length E-NPP3 cDNA and the resulting cells were screened for reactivity with MoAb 97A6. The positive population was sorted by FACS and cultured in RPMI 1640 plus 10% fetal bovine serum. After 3 rounds of sorting, the cells were stained either with control IgG1-PE (dotted histogram) or 97A6-PE (filled histogram) and analyzed on a FACSCalibur flow cytometer. Nontransfected 293 cells were negative for MoAb 97A6 (not shown).
The calculated molecular mass of E-NPP3 is 100 089 D, but the 10 N-glycosylation sites suggested a higher molecular mass of this transmembrane protein.2 Indeed, the bands identified by SDS-PAGE revealed proteins of 130- and 150-kd apparent molecular masses. Our preliminary analyses indicate that the 150-kd protein is encoded by a new, not yet described, splice variant and not the result of differential glycosylation. The previously published 270-kd band1 probably represents a dimer of either 2 130-kd molecules or a complex of 130- and 150-kd molecules.
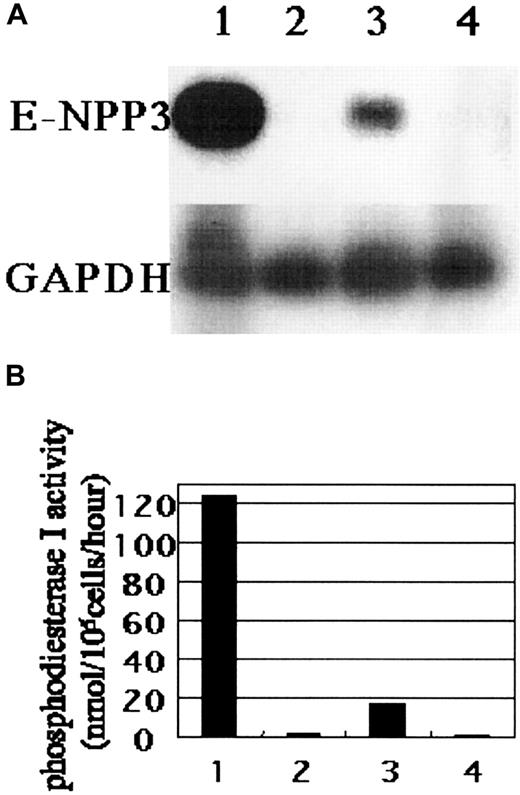
To further confirm that the molecule recognized by MoAb 97A6 on primary basophils is E-NPP3, the corresponding mRNA expression was analyzed on 97A6+ cells and compared with that of 97A6− PB cells. Figure 3A shows that basophils indeed express the E-NPP3 gene (lane 3) but neutrophils/eosinophils (lane 2) and 97A6− mononuclear cells (lane 4) do not express this gene to a measurable extent. High levels of E-NPP3 expression were also detected in KU-812 cells (lane 1). Next, we determined the phosphodiesterase-I enzymatic activity in these cells using 5′-l-thymidine monophosphate-p-nitrophenyl ester as a substrate. The specific activities of KU-812 cells, neutrophils/eosinophils, basophils, and 97A6− mononuclear cells were 124.0, 1.3, 17.2, and 0.8 nmol/105 cells/h, respectively (Figure 3B). Therefore, basophils contain the highest phosphodiesterase-I activity among the tested PB cell fractions. To determine whether MoAb 97A6 detects a functional phosphodiesterase I, lysates of these cell types were either precipitated with MoAb 97A6 or with an IgG1 control antibody, and subjected to the above-mentioned enzyme assay. This analysis revealed that MoAb 97A6 precipitates 99% of the enzymatic activity in KU-812 cells and 95% of that of basophils, whereas the control IgG did not precipitate any enzymatic activities (data not shown). Neither MoAb 97A6 nor control IgG precipitates enzymatic activities from neutrophils/eosinophils and 97A6− mononuclear cells. Thus, MoAb 97A6 detects functionally active E-NPP3 in both PB basophils and KU-812 cells, but not in other PB subpopulations. Most likely, the low level of phosphodiesterase-I activity detected in neutrophils/eosinophils and 97A6− mononuclear cells result from the activity of other E-NPP family molecules such as E-NPP1.
Expression of E-NPP3 mRNA in KU-812 cells and PB basophils.
(A) Total RNA extracted from KU-812 cells (lane 1), neutrophils/eosinophils (lane 2), 97A6+ basophils (lane 3), and 97A6− mononuclear cells (lane 4) were subjected to RT-PCR analysis as described in “Study design.” Expression of E-NPP3 mRNA was observed in KU-812 cells and basophils but not in other cell fractions. (B) Phosphodiesterase-I activity of lysates from KU-812 cells and PB subpopulations (lanes are described in panel A) was determined using p-nitrophenyl thymidine-5′-l-monophosphate as a substrate. Data represent the mean of triplicate assays.
Expression of E-NPP3 mRNA in KU-812 cells and PB basophils.
(A) Total RNA extracted from KU-812 cells (lane 1), neutrophils/eosinophils (lane 2), 97A6+ basophils (lane 3), and 97A6− mononuclear cells (lane 4) were subjected to RT-PCR analysis as described in “Study design.” Expression of E-NPP3 mRNA was observed in KU-812 cells and basophils but not in other cell fractions. (B) Phosphodiesterase-I activity of lysates from KU-812 cells and PB subpopulations (lanes are described in panel A) was determined using p-nitrophenyl thymidine-5′-l-monophosphate as a substrate. Data represent the mean of triplicate assays.
The E-NPPs comprise a family of ectonucleotidases consisting of E-NPP1 (PC-1), E-NPP2 (PD-Iα, autotaxin), and E-NPP3 (PD-Iβ, B10, gp130RB14-6).8-14 These type II transmembrane proteins (N-terminus inside) are highly homologous within their extracellular domains but differ in their transmembrane and cytosolic domains. E-NPP1 was identified as a plasma cell differentiation antigen.10 A naturally occurring mutation in the mouse E-NPP1 gene results in abnormal calcification in spinal ligaments, joints, and soft tissues, suggesting an involvement of E-NPP1 in bone mineralization.14 E-NPP2 was cloned from melanoma cells and shows strong motility stimulating activity toward a variety of tumor cells.11 E-NPP3 was cloned from rat embryonic neural cells12 and its overexpression in 3T3 fibroblasts results in the up-regulation of glial fibrillary acidic protein.13 In hematopoietic tissues, surface E-NPP3 is exclusively expressed on basophils, mast cells, and their progenitors.1 Interestingly, E-NPP3 expression on basophils is up-regulated after stimulation with allergen or cross-linking with IgE.1 These observations suggest that E-NPP3 is a basophil activation antigen. Studies are in progress to reveal the precise role of this ectoenzyme in basophil activation.
The authors thank Heike Letzkus for excellent technical assistance, Dr Selim Kuçi (Children's Hospital of Tübingen) for critical discussion, Dr Andreas van Agthoven (Immunotech SA, a Beckman-Coulter company, Marseilles, France) for providing 97A6-PE conjugate, and Dr James W. Goding (Monash University, Australia) for providing human PC-1 cDNA.
Supported by a grant from the Deutsche Forschungsgemeinschaft SFB 510, project A1 (H.-J.B. and C.G.) and by Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture, Japan (K.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hans-Jörg Bühring, Medizinische Klinik II, Otfried-Müller-Str 10, 72076 Tübingen, Germany; e-mail: hans-joerg.buehring@med.uni-tuebingen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal