Abstract
Sialoadhesin is a macrophage-restricted cellular interaction molecule and a prototypic member of the Siglec family of sialic acid binding immunoglobulin (Ig)-like lectins. So far, it has only been characterized in rodents. Here, we report the molecular cloning, binding properties, and expression pattern of human sialoadhesin. The predicted protein sequences of human and mouse sialoadhesin are about 72% identical, with the greatest similarity in the extracellular region, which comprises 17 Ig domains in both species. A recombinant protein consisting of the first 4 N-terminal domains of human sialoadhesin fused to the Fc region of human IgG1 mediated sialic acid–dependent binding with a specificity similar to its mouse counterpart, preferring sialic acid in the α2,3 glycosidic linkage over the α2,6 linkage. By flow cytometry with peripheral blood leukocytes, recombinant sialoadhesin bound strongly to granulocytes with intermediate binding to monocytes, natural killer cells, B cells, and a subset of CD8 T cells. Using antibodies raised to the recombinant protein, sialoadhesin was immunoprecipitated from the THP-1 human monocytic cell line as an approximate 200-kd glycoprotein. The expression pattern of human sialoadhesin was found to be similar to that of the mouse receptor, being absent from monocytes and other peripheral blood leukocytes, but expressed strongly by tissue macrophages in the spleen, lymph node, bone marrow, liver, colon, and lungs. High expression was also found on inflammatory macrophages present in affected tissues from patients with rheumatoid arthritis.
Introduction
Macrophages constitute a heterogeneous population of bone marrow–derived cells, present throughout the body, where they perform myriad functions, both during steady-state conditions and in pathology.1 This is reflected in the variable cell surface phenotypes exhibited by macrophage subsets. Cell surface receptors initiate and orchestrate many of the activities of macrophages, such as growth, differentiation, adhesion, phagocytosis, activation, chemotaxis, and apoptosis. Most such receptors are expressed on other leukocytes, with only a restricted few being exclusive to macrophages and hence implicated in macrophage-specific functions.
Sialoadhesin (Sn) is one such receptor. It was originally characterized in the mouse on isolated resident bone marrow macrophages as a nonphagocytic, sialic acid–dependent sheep erythrocyte receptor.2 In situ, these macrophages bind avidly to developing myeloid and erythroid cells and are therefore well positioned to dispose of apoptotic cells and extruded erythroblast nuclei that are generated during hemopoiesis3,4 and B lymphopoiesis.5 Sialylated ligands for Sn are displayed on the surface of the attached hemopoietic cells, and the receptor is clustered at the contact points between macrophages and developing myeloid cells.6 Sn is therefore likely to be involved in macrophage-hemopoietic cell interactions in the bone marrow. In addition to a scavenging function, resident macrophages may contribute to the trophic microenvironment of the bone marrow, for example, through recycling of heme-derived iron required for sustained erythropoiesis.7
Immunocytochemical staining of tissue sections showed that Sn is also expressed at high levels on discrete subsets of tissue macrophages, especially those in secondary lymphoid organs.8 For example, in the spleen of rodents, Sn is expressed mainly on macrophages in the marginal zone, which have been implicated in specialized functions in innate and acquired immunity.9Lower levels of Sn were seen on many other tissue macrophage populations, such as in the liver, gut, and lung, and certain macrophage populations, notably the resident brain microglia, expressed undetectable levels of the receptor.10 Studies on the ED3 antigen in the rat, which has a similar macrophage-restricted distribution, showed that this molecule is the likely rat ortholog of Sn.11 12
Mature, circulating neutrophils have been shown to express high levels of Sn ligands, whereas other cells such as thymocytes and resting T cells have relatively low levels.13 Because Sn is not a phagocytic receptor,2 it is unlikely to be involved directly in scavenging functions, but it could cooperate with phagocytic receptors to increase the efficiency of recognition and uptake as well as being involved in other types of cell-cell interactions. For example, recent findings in a murine model of allogeneic tumor rejection have shown that transferred, activated CD4 and CD8 T cells can cluster in vivo with Sn+ macrophages, an association that may be important for T-cell effector functions.14 Because potential sialylated ligands are also present on molecules like laminin in the extracellular matrix,15 Sn also may be involved in macrophage-matrix interactions.
Sn is the prototypic member of the Siglec (sialic acid binding Ig-like lectin) family of cellular interaction molecules and was designated Siglec-1.16Siglecs possess a homologous N-terminal V-set domain that contains the sialic acid binding site followed by varying numbers of C2-set domains. Sn contains the unusually large number of 16 C2-set domains, a feature that may be important in its ability to mediate macrophage adhesive functions.17 Apart from Sn, members of the Siglec family include CD22 (Siglec-2) on B cells,18 CD33 (Siglec-3) on immature myeloid cells and monocytes,19 myelin-associated glycoprotein (MAG) (Siglec-4A) on Schwann cells and oligodendrocytes,20 and Siglecs-5, -6, -7, -8, and -9 expressed on various hemopoietic subsets.21-26 The restricted expression patterns of different Siglecs implies that they mediate distinct functions, some of which are likely to involve sialic acid recognition. Each protein displays a binding preference for sialic acid in either α2,3 or α2,6 glycosidic linkage, and naturally occurring modifications of sialic acid can have a profound effect on protein recognition for all Siglecs studied.20,27 28
The molecular basis for carbohydrate binding by Sn has been determined recently by X-ray crystallography29 in conjunction with site-directed mutagenesis30 and nuclear magnetic resonance analysis.31 In the crystal structure of the Sn V-set domain complexed to 3′-sialyllactose, a highly conserved arginine residue (Arg97 in mouse Sn) forms a bidentate salt bridge with the carboxylate group of sialic acid, and 2 well-conserved aromatic groups (both tryptophan for Sn) make hydrophobic interactions with theN-acetyl and glycerol moieties ofN-acetylneuraminic acid. The crucial importance of the arginine has been demonstrated by the finding that even a conservative substitution with lysine leads to an approximate 10-fold loss in binding affinity of Sn for sialic acid.31
Despite detailed knowledge of the expression pattern of Sn and the molecular basis for its recognition of sialic acid, the biologic functions of this receptor are relatively poorly understood. Insights into the potential functions of a given protein can be obtained by comparing the molecular properties and expression patterns of different species' orthologs. In addition, for proteins expressed by cells of the immune system, an understanding of their expression during disease states can give valuable insight into potential roles in pathologic situations. So far, Sn has only been characterized in rodents. The purpose of the present study was to identify a human ortholog of Sn in order to compare its structure and binding properties. The production of specific antibodies then allowed us to investigate the expression pattern in normal and diseased situations.
Materials and methods
Materials
All reagents were obtained from Sigma Chemical Co (Poole, United Kingdom) unless otherwise specified. Restriction and modifying enzymes were purchased from Amersham Pharmacia Biotech (St Albans, United Kingdom) or Life Technologies Ltd (Paisley, Scotland). Also, [α-32P]-dCTP and [α-35S]-dATP were purchased from Amersham Pharmacia Biotech. UCHM1, anti-CD14 was obtained from the Imperial Cancer Research Fund (ICRF) (London, United Kingdom), and mouse monoclonal antibodies (mAbs) anti-CD3, anti-CD4, and phycoerythrin (PE)-conjugated anti-CD16 were purchased from Dako (High Wycombe, United Kingdom). Anti-CD8–PE, anti-CD19–PE, and anti-CD69–PE were from Serotec (Kidlington, United Kingdom). Human interferon-γ and human tumor necrosis factor (TNF)-α were purchased from Peprotech (London, United Kingdom).
Cells
All cell lines were provided by the ICRF Cell Production Service (South Mimms, United Kingdom). COS-1 cells were cultured in Dulbecco's modified Eagle's medium containing 5% fetal calf serum and antibiotics. Other cell lines were cultured in RPMI 1640 containing 10% fetal calf serum and antibiotics. Human red blood cells (RBCs) and peripheral blood leukocytes were obtained from healthy donors.
Preparation and screening of cDNA libraries
A complementary DNA (cDNA) library in λZAPII (Stratagene, Cambridge, United Kingdom) was prepared as described32using human monocytic THP-1 cells cultured for 24 hours with 100 ng/mL phorbol myristate acetate (PMA) as a source of messenger RNA (mRNA). A total of 6 × 105 plaque-forming units were screened with a 1.35-kilobase (kb) BamHI/EcoRI fragment of mouse Sn cDNA (nucleotides 4337-568732) labeled with [α-32P]-dCTP by random priming as described.32 One positive plaque was identified on duplicate filters and the cDNA insert excised as a pBluescript SK− phagemid. The nucleotide sequence of the 2.9-kb cDNA insert (pTH1) was determined by the dideoxynucleotide chain termination method using double-stranded DNA templates with Sequenase Version 2.0 (United States Biochemical Corp, Cleveland, OH). A cDNA library was also prepared from human spleen poly(A) RNA as described above but with random priming.
Isolation of a full-length genomic clone
A human genomic P1-derived artificial chromosome (PAC) library33 was used to isolate the Sn gene by polymerase chain reaction (PCR) using primers derived from the cDNA sequence of pTH1.34 A single clone was identified that, on Southern blotting, hybridized with a 5′ EcoRI fragment of pTH1. When the Southern blot was hybridized with a 0.3-kb BamHI fragment of mouse Sn cDNA (encoding most of domain 1), 6.5-kbBamHI and 2.2-kb PstI fragments were identified. Hybridization with a 0.9-kb BamHI mouse cDNA probe (encoding domains 3-6) identified a fragment of 12 kb. These 3 restriction fragments were subcloned into pBluescript and sequenced using AmpliTaq FS and dye-labeled terminators (Perkin Elmer, Warrington, United Kingdom) followed by analysis on an ABI Prism 377 DNA Sequencer (Perkin Elmer). End-sequencing revealed that the 2.2-kb PstI fragment spanned from 5′ of the gene to the N-terminal immunoglobulin (Ig)-like domain (domain 1); the 6.5-kb BamHI fragment spanned from 5′ of the gene to domain 2; and the 12-kb BamHI fragment spanned from the intron between domains 3 and 4 to aBamHI site approximately 1 kb from the poly(A) tail. The nucleotide sequence of the leader peptide, domain 1, and most of domain 2 was obtained from the PstI and 6.5-kb BamHI genomic subclones. Repeated attempts to subclone a fragment encoding the 3′ end of domain 2 and all of domain 3 were unsuccessful, and therefore this sequence was obtained from 3 independent PCR products spanning this region, using reverse-transcribed THP-1 mRNA as the template. The region encoding domains 4 to 13 was amplified by PCR from human spleen cDNA and sequenced. The sequence was confirmed using the 12-kb BamHI subclone as a genomic template.
Computer analysis
Nucleotide and amino acid comparisons were performed using Blast Version 2.0 (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD). Protein motif searches were carried out using the Prosite database (Swiss Institute for Bioinformatics, Geneva, Switzerland). The amino acid sequences of human and mouse Sn were aligned using the ClustalW 1.7 software available on the Baylor College of Medicine server (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html).
Northern blot analysis
Poly(A) RNA was purified from various cell lines and from human spleen and electrophoresed in a 1% (wt/vol) agarose formaldehyde gel. The RNA was transferred to Hybond N+ nylon membrane (Amersham Phamacia Biotech) and hybridized with a 1.2-kbEcoRI fragment of pTH1 or human actin cDNA (Clontech, Palo Alto, CA) labeled with [α-32P]-dCTP by random priming. Hybridization was performed using 50% formamide in the hybridization buffer and washing at 55°C with 0.1 × SSC.
Fc protein production
A soluble, truncated form of human Sn was produced consisting of the first 4 N-terminal domains (d1-4) of the receptor fused to the Fc portion of human IgG. Human Sn d1-4 was generated by the PCR from spleen cDNA using the following primers: forward, 5′-AGGAATTCGCTATGGGCTTCTTGCCCAAGCTTCTC-3′; reverse, 5′-AGGAATTCACTTACCTGTGTTGACTACCACGCTGACAGG-3′; and cloned into the pIG1 vector provided by Dr D. L. Simmons.35 Recombinant Fc protein was produced in transiently transfected COS cells and purified on protein A–Sepharose (Amersham Pharmacia Biotech) as previously described.35 The protein was dialysed against 20 mM Tris (pH 8.0), concentrated, and filter-sterilized. Control Fc proteins, CD22-Fc, MAG-Fc, and neural cell adhesion molecule (NCAM)-Fc were prepared as described.20
Binding assays to RBCs and polyacrylamide glycoconjugates
RBC binding assays were performed as detailed elsewhere.20 For polyacrylamide (PAA) assays, various concentrations of biotinylated PAA glycoconjugates carrying either NeuAcα2,3Galβ1,4Glc (2,3-PAA) or NeuAcα2,6Galβ1,4Glc (2,6-PAA) (Syntesome, Munich, Germany) were added to wells that had been coated with Fc proteins as described above and incubated on ice for 1 hour. After washing, horse radish peroxidase (HRP)-conjugated streptavidin was incubated in the wells for a further hour, and theno-phenylenediamine dihydrochloride substrate was added and the optical density measured at 450 nm.
Fc protein binding to leukocytes
Mononuclear cells and granulocytes were separated from peripheral blood by sedimentation of RBCs on Dextran T-500 (Amersham Pharmacia Biotech) followed by centrifugation on Lymphoprep (Nycomed Pharma, Oslo, Norway). The leukocytes were labeled immediately or following incubation at 37°C for 2 hours in RPMI 1640 with or without 0.1 U/mL Vibrio cholerae sialidase. All subsequent steps were performed at 4°C. Cells were incubated with 10 μg/mL Fc protein and mAbs for 30 minutes, followed by fluorescein isothiocynate (FITC)-conjugated goat antihuman IgG and PE-conjugated F(ab′)2 goat antimouse Ig (Dako), to detect CD3 and CD4. In the case of CD8, CD19, CD16, and CD69, labeling was carried out directly using PE-conjugated mAbs. The cells were analyzed immediately on a FACScan (Becton Dickinson, Oxford, United Kingdom). The monocytes and lymphocyte/natural killer (NK) cell fractions were distinguished by their characteristic forward and side scatter properties. Data were routinely collected for 20 000 to 50 000 mononuclear cells and 20 000 granulocytes.
Monoclonal antibody production
Mice were immunized with 5 injections of 10 μg purified hSn(d1-4)Fc protein and fusions carried out 4 days after the last immunization. Hybridoma supernatants were screened for reactivity with hSn(d1-4)Fc but not with an irrelevant Fc protein, MAG(d1-3)Fc. Two positive hybridomas, clones 7D2 and 8H2, were identified from 2 separate fusions and the mAbs, both IgG1, designated HSN1 and HSN2, respectively.
Immunoprecipitation
THP-1 cells (4 × 107) were surface-biotinylated for 10 minutes at 37°C with 0.5 mM sulfo-NHS-biotin (Pierce and Warriner, Chester, United Kingdom). Immunoprecipitations of Nonidet P-40 lysates were carried out as described8 and samples run out under reducing and nonreducing conditions on 6.5% PAA gels. Following transfer to nitrocellulose, immunoprecipitated proteins were detected with HRP-conjugated streptavidin and enhanced chemiluminescence (ECL, Amersham Pharmacia Biotech).
Monocyte-derived macrophages
Monocytes were purified from granulocyte-depleted blood leukocytes by adhesion to plastic Petri dishes as described.36 Cells were cultured in RPMI plus 10% autologous serum alone for 6 days or with interferon-γ (500 U/mL) and TNF-α (10 ng/mL) for 2 days. Cells were lifted with phosphate-buffered saline containing 5 mM ethylenediaminetetraacetic acid immediately prior to immunostaining and fluorescence-activated cell sorter (FACS) analysis.
FACS analysis
A total of 100 μL whole blood or cell suspensions at 5 × 106/mL were incubated with 10 μL mAb culture supernatant or purified mAb at 5 μg/mL for 30 minutes on ice, washed, and incubated for a further 30 minutes with FITC-conjugated F(ab′)2 goat antimouse Ig (Dako). Samples were then treated with Becton Dickinson FACS lysing solution prior to analysis on a FACScan. Data were collected for 1000 THP-1 cells and cultured macrophages or 2500 whole leukocytes and cell populations gated according to their characteristic forward and side scatter profiles.
Immunohistochemistry
Acetone-fixed, 6 μm frozen sections of various human tissues were incubated with HSN1 or HSN2 culture supernatant or control antibodies diluted in phosphate-buffered saline containing 10% normal human serum. Binding of mAbs was detected by incubating sections with biotinylated goat antimouse IgG (Vector Labs, Burlingame, CA), followed by biotin-avidin complexes (ABC, Vector Labs) conjugated to either peroxidase or alkaline phosphatase and then diaminobenzidine or Fast Red substrates, respectively. Samples of synovial tissue from patients with seropositive rheumatoid arthritis were obtained during routine surgical operations and biopsies performed at the Nuffield Orthopaedic Centre, Oxford Radcliffe Trust Hospital, United Kingdom. Cryostat sections measuring 8 μm were stained with the following primary mAbs at 10 μg/mL: HSN1 or Y182A anti-CD68,37 followed by peroxidase-conjugated secondary antibodies. Omission of the primary antibody was used as a negative control. All sections were counterstained with hematoxylin prior to mounting.
Results
Molecular cloning of human Sn
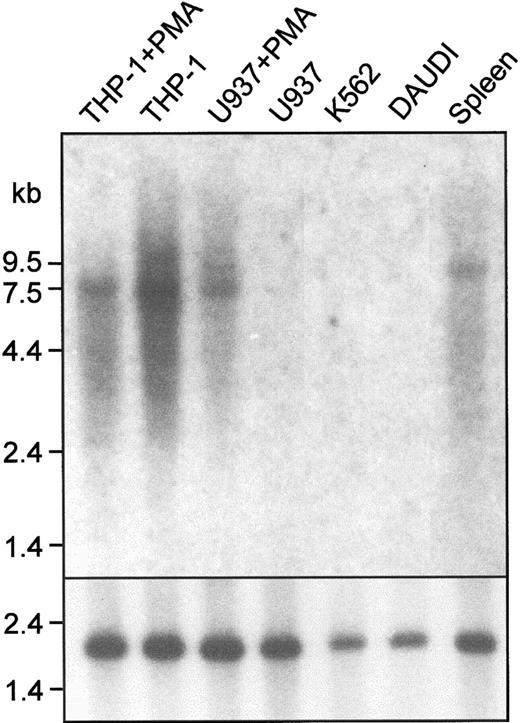
Clone pTH1 was isolated from a cDNA library prepared from PMA-treated THP-1 cells by low-stringency hybridization with a murine Sn probe. The 2.9-kb insert from pTH1 had significant homology to mouse Sn and corresponded to a region between domain 14 and the 3′ untranslated sequence. However, further clones isolated from either the THP-1 library or a human spleen cDNA library did not extend beyond the 5′ end of pTH1. As an alternative strategy, a clone containing the entire human Sn gene was isolated from a PAC library.33Sequencing revealed an open reading frame of 5127 base pairs (bp) encoding a protein of 1709 amino acids (Figure1) and a 3′ untranslated region of 1593 bp. Northern blot analysis showed a single transcript of 7.5 kb in the spleen and certain cell lines (Figure2), suggesting that the 5′ untranslated region is about 780 bp. Northern analysis was consistent with Sn gene expression being restricted to macrophages. Signals were detected with THP-1 cells (monocytic), both unstimulated and after culture with PMA to promote macrophage differentiation. In comparision, U937 cells (promonocytic) failed to express detectable Sn mRNA unless they were induced to differentiate with PMA (Figure 2). The Daudi B-cell line and the K562 myeloerythroid cell line did not express detectable Sn mRNA (Figure 2).
The predicted amino acid sequence of human Sn shows a high degree of similarity to mouse Sn.
The hydrophobic leader peptide (amino acids 1-19) and transmembrane region (1642-1662) are underlined. Potential N-linked glycosylation sites are boxed in black. The conserved cysteine residues in domains 1 and 2 characteristic of Siglecs are boxed in gray. Residues in domain 1 that interact with sialic acid are boxed in white. The Ig domain boundaries, deduced from the exon-intron boundaries of the mouse Sn gene,63 are indicated. Residues that are identical with the mouse ortholog are indicated by stars; residues that are similar are indicated by dashes; and residues that are not represented in mouse Sn are indicated by open circles. The cDNA sequence of human Sn has been assigned the GenBank accession number AF230073.
The predicted amino acid sequence of human Sn shows a high degree of similarity to mouse Sn.
The hydrophobic leader peptide (amino acids 1-19) and transmembrane region (1642-1662) are underlined. Potential N-linked glycosylation sites are boxed in black. The conserved cysteine residues in domains 1 and 2 characteristic of Siglecs are boxed in gray. Residues in domain 1 that interact with sialic acid are boxed in white. The Ig domain boundaries, deduced from the exon-intron boundaries of the mouse Sn gene,63 are indicated. Residues that are identical with the mouse ortholog are indicated by stars; residues that are similar are indicated by dashes; and residues that are not represented in mouse Sn are indicated by open circles. The cDNA sequence of human Sn has been assigned the GenBank accession number AF230073.
Northern blot analysis shows that human Sn mRNA is expressed in macrophage-like cell lines and in the spleen.
A total of 10 μg mRNA from the indicated sources was loaded in each lane. The blot was probed with a 1.2-kb EcoRI fragment of pTH1 (top panel) or with a human actin cDNA to control for loading (bottom panel). A single transcript of 7.5 kb is present in THP-1 cells, PMA-treated U937 cells, and in human spleen. Cell lines are as follows: THP-1, monocytic; U937, promonocytic; Daudi, B-cell; K562, myeloerythroid.
Northern blot analysis shows that human Sn mRNA is expressed in macrophage-like cell lines and in the spleen.
A total of 10 μg mRNA from the indicated sources was loaded in each lane. The blot was probed with a 1.2-kb EcoRI fragment of pTH1 (top panel) or with a human actin cDNA to control for loading (bottom panel). A single transcript of 7.5 kb is present in THP-1 cells, PMA-treated U937 cells, and in human spleen. Cell lines are as follows: THP-1, monocytic; U937, promonocytic; Daudi, B-cell; K562, myeloerythroid.
Sequence comparisons with mouse Sn
The long open reading frame encodes a type 1 transmembrane glycoprotein that is 72% identical to mouse Sn. It consists of a leader peptide of 19 amino acids, a large extracellular domain of 1622 amino acids, and a short cytoplasmic tail of 47 amino acids (Figure 1, Table 1). Like the mouse protein,32 the extracellular region of human Sn consists of 17 Ig-like domains, comprising an N-terminal V-set domain and 16 C2-set domains that show an alternating pattern of long and short domains (Table 1). The size of each Ig-like domain is well conserved between the human and murine receptors, with only 4 of the 17 domains differing in amino acid length. The Ig-like domains are 60% to 80% identical between species, the N-terminal domain being the most highly conserved (Table 1). There are 14 potential N-linked glycosylation sites, 12 of which are present in the mouse (Figure 1). The cytoplasmic tail is poorly conserved, being 12 amino acids longer than its mouse counterpart and only 30% identical over the full length (Table 1). There are 2 potential phosphorylation sites, one for protein kinase C at position 1664 and one for casein kinase-2 at position 1674, only the former being conserved between species.
Comparison of the predicted protein structure of human and mouse sialoadhesin
| Domain . | Length HSn (mSn) . | Identity, % . | Similarity, % . |
|---|---|---|---|
| Leader | 19 (19) | 57.9 | 57.9 |
| 1 (V) | 117 (117) | 78.6 | 89.7 |
| 2 (C2) | 99 (100) | 60.6 | 71.7 |
| 3 (C2) | 89 (89) | 67.4 | 82.0 |
| 4 (C2) | 85 (85) | 74.1 | 87.1 |
| 5 (C2) | 100 (100) | 76.0 | 86.0 |
| 6 (C2) | 86 (86) | 79.1 | 84.9 |
| 7 (C2) | 112 (107) | 65.2 | 77.7 |
| 8 (C2) | 86 (86) | 76.7 | 86.0 |
| 9 (C2) | 101 (101) | 79.2 | 89.1 |
| 10 (C2) | 87 (86) | 75.8 | 87.4 |
| 11 (C2) | 104 (104) | 69.2 | 77.9 |
| 12 (C2) | 84 (84) | 72.6 | 79.8 |
| 13 (C2) | 90 (90) | 78.9 | 87.8 |
| 14 (C2) | 84 (84) | 70.2 | 79.8 |
| 15 (C2) | 100 (102) | 76.0 | 82.0 |
| 16 (C2) | 87 (87) | 73.6 | 79.3 |
| 17 (C2) | 111 (111) | 73.0 | 83.8 |
| TD | 24 (21) | 71.4 | 71.4 |
| CT | 44 (35) | 29.8 | 40.4 |
| Total | 1709 (1694) | 71.9 | 81.5 |
| Domain . | Length HSn (mSn) . | Identity, % . | Similarity, % . |
|---|---|---|---|
| Leader | 19 (19) | 57.9 | 57.9 |
| 1 (V) | 117 (117) | 78.6 | 89.7 |
| 2 (C2) | 99 (100) | 60.6 | 71.7 |
| 3 (C2) | 89 (89) | 67.4 | 82.0 |
| 4 (C2) | 85 (85) | 74.1 | 87.1 |
| 5 (C2) | 100 (100) | 76.0 | 86.0 |
| 6 (C2) | 86 (86) | 79.1 | 84.9 |
| 7 (C2) | 112 (107) | 65.2 | 77.7 |
| 8 (C2) | 86 (86) | 76.7 | 86.0 |
| 9 (C2) | 101 (101) | 79.2 | 89.1 |
| 10 (C2) | 87 (86) | 75.8 | 87.4 |
| 11 (C2) | 104 (104) | 69.2 | 77.9 |
| 12 (C2) | 84 (84) | 72.6 | 79.8 |
| 13 (C2) | 90 (90) | 78.9 | 87.8 |
| 14 (C2) | 84 (84) | 70.2 | 79.8 |
| 15 (C2) | 100 (102) | 76.0 | 82.0 |
| 16 (C2) | 87 (87) | 73.6 | 79.3 |
| 17 (C2) | 111 (111) | 73.0 | 83.8 |
| TD | 24 (21) | 71.4 | 71.4 |
| CT | 44 (35) | 29.8 | 40.4 |
| Total | 1709 (1694) | 71.9 | 81.5 |
HSn indicates human sialoadhesin; mSn, mouse sialoadhesin; V, V-set Ig-like domain; C2, C2-set Ig-like domain; TD, transmembrane domain; CT, cytoplasmic tail.
Alignment of the N-terminal region of human and mouse Sn showed that all of the characteristic structural features of Siglecs are present (Figure 1). In particular, amino acids important for sialic acid binding29 are identical in human and mouse Sn. These include an invariant arginine on the F strand at position 116 and 2 aromatic residues (both tryptophan in Sn) at positions 21 and 125 (Figure 1). In addition, there is conservation of the unusual pattern of cysteine residues that is characteristic of the Siglec family and thought to give rise to an intrasheet disulfide bond within domain 1 and a disulfide bond between domains 1 and 2.29
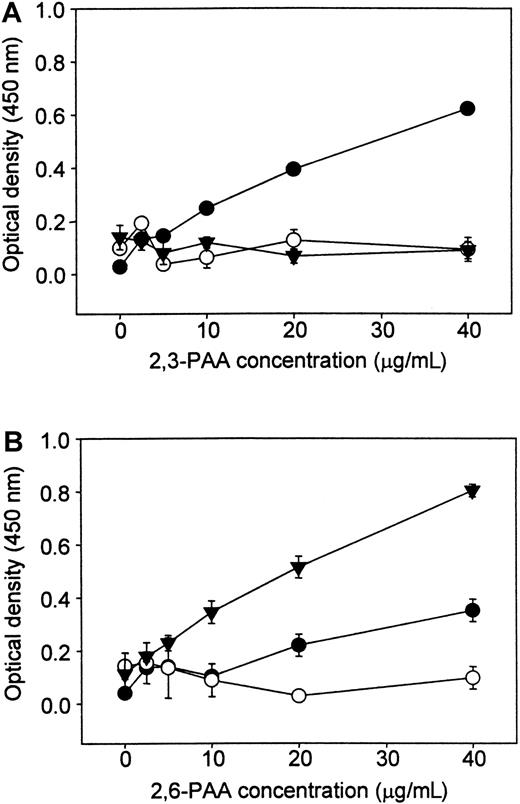
Sialic acid–dependent binding properties of recombinant human Sn
To study the binding properties of human Sn, a soluble, truncated form of the receptor consisting of the first 4 N-terminal domains (d1-4) fused to the Fc portion of human IgG (hSn[d1-4]Fc) was produced in COS cells. Human RBCs, which have abundant cell surface sialic acid, were used as indicator cells to investigate whether human Sn could mediate sialic acid–dependent binding to cells similar to that seen with murine Sn.13 Untreated, but not sialidase-treated, RBCs bound avidly to hSn(d1-4)Fc immobilized on microtiter wells (data not shown).
To determine the sialic acid linkage preference of human Sn, binding assays were carried out with biotinylated PAA carrying either 3′- or 6′-sialyllactose (2,3-PAA or 2,6-PAA). The hSn(d1-4)Fc exhibited dose-dependent binding of 2,3-PAA (Figure3A) but, under the same conditions, only low levels of binding were seen with 2,6-PAA (Figure 3B). CD22-Fc was also included as a control and, as expected, bound strongly to 2,6-PAA but not to 2,3-PAA (Figure 3). No binding was seen with NCAM-Fc used as a nonsialic acid binding control protein. Therefore, human Sn has a specificity similar to that of murine Sn, preferring α2,3-linked sialic acid over the α2,6 linkage.
Human Sn(d1-4)Fc binds preferentially to PAA glycoconjugates carrying 3′-sialyllactose.
Fc proteins were coated onto plastic microwells, and biotinylated PAA glycoconjugates linked either to 3′-sialyllactose (2,3-PAA, panel A) or 6′-sialyllactose (2,6-PAA, panel B) were added at the indicated concentrations. Unbound conjugate was washed off and binding detected with streptavidin-peroxidase and o-phenylenediamine dihydrochloride as substrate. ● indicates hSn(d1-4)Fc, and comparisons were made with CD22-Fc (▾) that binds specifically to 2,6-PAA and NCAM-Fc (○), a control protein that does not mediate sialic acid binding. Data show mean absorbance values ± range of duplicates and are representative of 2 experiments performed.
Human Sn(d1-4)Fc binds preferentially to PAA glycoconjugates carrying 3′-sialyllactose.
Fc proteins were coated onto plastic microwells, and biotinylated PAA glycoconjugates linked either to 3′-sialyllactose (2,3-PAA, panel A) or 6′-sialyllactose (2,6-PAA, panel B) were added at the indicated concentrations. Unbound conjugate was washed off and binding detected with streptavidin-peroxidase and o-phenylenediamine dihydrochloride as substrate. ● indicates hSn(d1-4)Fc, and comparisons were made with CD22-Fc (▾) that binds specifically to 2,6-PAA and NCAM-Fc (○), a control protein that does not mediate sialic acid binding. Data show mean absorbance values ± range of duplicates and are representative of 2 experiments performed.
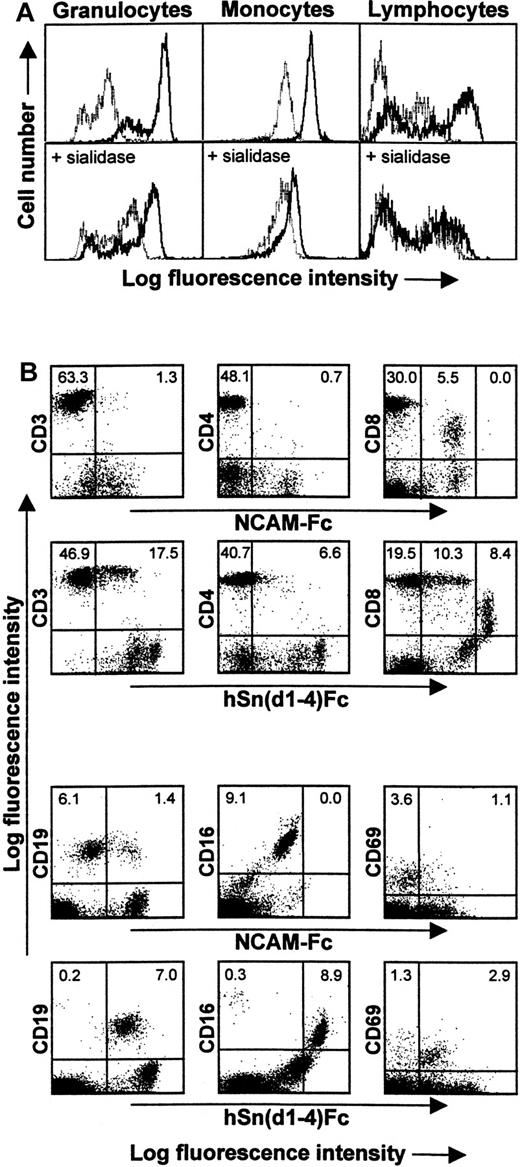
Binding of recombinant Sn to human peripheral blood leukocytes
In flow cytometric assays with human peripheral blood leukocytes, hSn(d1-4)Fc showed strong binding to granulocytes and intermediate levels of binding to monocytes compared with the control protein, NCAM-Fc. Within the lymphocyte/NK cell fraction, a subset of cells showed background binding to NCAM-Fc, but binding was clearly stronger with hSn(d1-4)Fc (Figure 4A). Sn-specific binding to all 3 populations was sialic acid–dependent because sialidase pretreatment of the leukocytes reduced hSn(d1-4)Fc binding to a level similar to that seen with the control protein, NCAM-Fc (Figure4). To characterize the lymphocyte/NK cell–reactive subset in more detail, double labeling was carried out by combining hSn(d1-4)Fc labeling with staining for CD3 (pan T cell), CD4 (helper T cells), CD8 (cytotoxic T cells plus NK cell subset), CD19 (B cells), and CD16 (NK cells) (Figure 4B). This showed that hSn(d1-4)Fc bound specifically to most CD19+ B cells and CD16+ NK cells and about 30% of the CD3+ T cells, most of which belonged to the CD8+ subset (Figure 4B). Most CD4+ T cells were unlabeled. To investigate the possibility that hSn(d1-4)Fc interacts with activated cells, double labeling was carried out for CD69, an early activation marker of lymphocytes. This showed that although only about 4% of lymphocytes expressed CD69, most of these activated cells were specifically recognized by hSn(d1-4)Fc (Figure 4B).
Human Sn(d1-4)Fc binds specifically to granulocytes, monocytes, and a subset of lymphocytes/NK cells.
(A) Blood leukocytes were either untreated (top panels) or treated with sialidase (bottom panels) and then labeled with Fc proteins followed by FITC-antihuman IgG to detect Fc protein binding. FACS histograms show staining of granulocytes, monocytes, and lymphocytes/NK cells with either hSn(d1-4)Fc (solid lines) or with the control protein, NCAM-Fc (dotted lines). The background labeling with NCAM-Fc is likely to be due to nonspecific Fc receptor binding. The population of granulocytes with lower fluorescence following staining with hSn(d1-4)Fc corresponds to eosinophils, as determined by double labeling for CD16 found at high levels on neutrophils and low levels on eosinophils (not shown). Following sialidase treatment, binding of hSn(d1-4)Fc is reduced to the background levels seen with NCAM-Fc for all cell populations. (B) Double labeling of the lymphocyte/NK cell population with NCAM-Fc or hSn(d1-4)Fc together with the indicated mAbs. The percentages of cells in the upper quadrants are indicated. This analysis shows that Sn stains a subset of CD3+ T cells, most of which are CD8high T cells. The CD8dim cells correspond to NK cells that are labeled nonspecifically by NCAM-Fc but show additional staining with hSn(d1-4)Fc. Most CD19+ B cells and most CD16+ NK cells were also labeled. The majority of recently activated CD69+ lymphocytes were also stained by hSn(d1-4)Fc. Similar results were obtained in 3 independent experiments.
Human Sn(d1-4)Fc binds specifically to granulocytes, monocytes, and a subset of lymphocytes/NK cells.
(A) Blood leukocytes were either untreated (top panels) or treated with sialidase (bottom panels) and then labeled with Fc proteins followed by FITC-antihuman IgG to detect Fc protein binding. FACS histograms show staining of granulocytes, monocytes, and lymphocytes/NK cells with either hSn(d1-4)Fc (solid lines) or with the control protein, NCAM-Fc (dotted lines). The background labeling with NCAM-Fc is likely to be due to nonspecific Fc receptor binding. The population of granulocytes with lower fluorescence following staining with hSn(d1-4)Fc corresponds to eosinophils, as determined by double labeling for CD16 found at high levels on neutrophils and low levels on eosinophils (not shown). Following sialidase treatment, binding of hSn(d1-4)Fc is reduced to the background levels seen with NCAM-Fc for all cell populations. (B) Double labeling of the lymphocyte/NK cell population with NCAM-Fc or hSn(d1-4)Fc together with the indicated mAbs. The percentages of cells in the upper quadrants are indicated. This analysis shows that Sn stains a subset of CD3+ T cells, most of which are CD8high T cells. The CD8dim cells correspond to NK cells that are labeled nonspecifically by NCAM-Fc but show additional staining with hSn(d1-4)Fc. Most CD19+ B cells and most CD16+ NK cells were also labeled. The majority of recently activated CD69+ lymphocytes were also stained by hSn(d1-4)Fc. Similar results were obtained in 3 independent experiments.
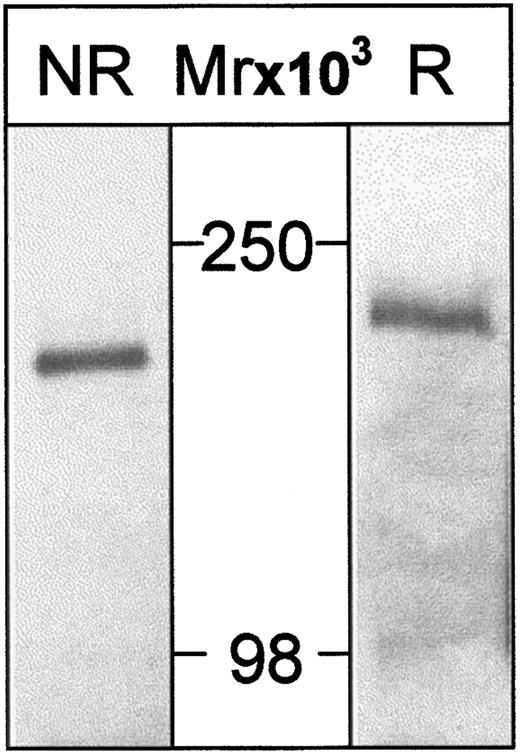
Antibody production and immunoprecipitation
Two mouse mAbs, HSN1 and HSN2, were raised to hSn(d1-4)Fc. Using lysates from surface-biotinylated THP-1 cells, HSN1 immunoprecipitated a single protein corresponding to human Sn, which migrated at about 200 kd on sodium dodecyl sulfate–polyacrylamide gel electrophoresis under reducing conditions and about 180 kd under nonreducing conditions (Figure 5). This is similar to the mouse protein8 and is consistent with the presence of multiple intramolecular disulfide bonds.
Human Sn is immunoprecipitated from THP-1 cells as a single species of about 200 kd.
THP-1 cells were surface-labeled with biotin and lysates subjected to immunoprecipitation with HSN1 mAb. Samples were run either nonreduced (NR) or reduced (R) on 6.5% PAA gels, transferred to nitrocellulose, and proteins on Western blots detected with streptavidin-HRP followed by enhanced chemiluminescence. A single species is detected that migrates at about 180 kd (nonreduced) or about 200 kd (reduced).
Human Sn is immunoprecipitated from THP-1 cells as a single species of about 200 kd.
THP-1 cells were surface-labeled with biotin and lysates subjected to immunoprecipitation with HSN1 mAb. Samples were run either nonreduced (NR) or reduced (R) on 6.5% PAA gels, transferred to nitrocellulose, and proteins on Western blots detected with streptavidin-HRP followed by enhanced chemiluminescence. A single species is detected that migrates at about 180 kd (nonreduced) or about 200 kd (reduced).
Immunostaining of blood leukocytes and cultured monocytes
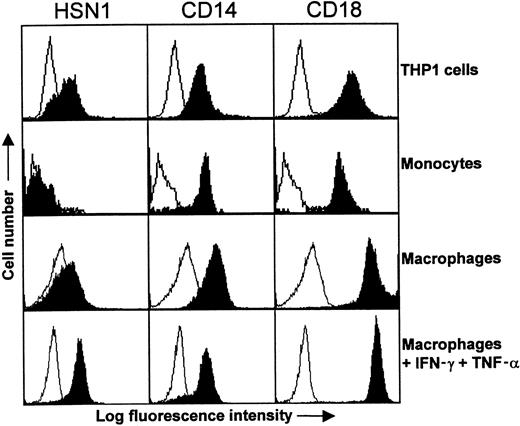
Northern blot analysis (Figure 2) suggested that expression of human Sn was likely to be restricted to macrophages, as is the case in mouse8 and rat.38 To investigate this further, FACS analysis was initially performed with peripheral blood leukocytes and THP-1 cells. Using either HSN1 or HSN2, Sn was found to be expressed weakly by THP-1 cells but not by blood monocytes, although CD14 and CD18 expression was comparable on both cell types (Figure6). Neutrophils and lymphocytes were also found to be negative for Sn expression (data not shown).
Expression of Sn on THP-1 cells and monocyte-derived macrophages.
Either whole blood, THP-1 cells, or monocyte-derived macrophages were labeled with the indicated mAbs followed by FITC-conjugated F(ab′)2 goat antimouse IgG. Monocytes were gated according to their characteristic forward and side scatter profiles following lysis of RBCs. Sn was expressed by THP-1 cells but was undetectable on the surface of monocytes. In comparison, the myeloid antigens CD14 and CD18 are expressed at high levels on both cell populations. Monocytes cultured for 6 days with autologous serum expressed only low levels of Sn, whereas addition of IFN-γ and TNF-α for 2 days of culture led to significant Sn expression. The data are representative of 2 to 3 experiments.
Expression of Sn on THP-1 cells and monocyte-derived macrophages.
Either whole blood, THP-1 cells, or monocyte-derived macrophages were labeled with the indicated mAbs followed by FITC-conjugated F(ab′)2 goat antimouse IgG. Monocytes were gated according to their characteristic forward and side scatter profiles following lysis of RBCs. Sn was expressed by THP-1 cells but was undetectable on the surface of monocytes. In comparison, the myeloid antigens CD14 and CD18 are expressed at high levels on both cell populations. Monocytes cultured for 6 days with autologous serum expressed only low levels of Sn, whereas addition of IFN-γ and TNF-α for 2 days of culture led to significant Sn expression. The data are representative of 2 to 3 experiments.
Previously, it was found that cultivation of murine monocytes and macrophages in autologous serum can lead to induction of Sn expression.39 To determine whether human monocyte-derived macrophages behaved similarly, human monocytes were cultured for 6 days in autologous serum and FACS analysis performed. Despite repeated attempts, Sn could only be detected at the cell surface at very low levels under these conditions (Figure 6). Interestingly, however, addition of interferon-γ or TNF-α for 2 days of culture resulted in induction of Sn on macrophages, and the effect was additive when both cytokines were combined, resulting in levels of Sn that were similar to those of CD14 used as a control (Figure 6).
Expression of Sn in normal human tissues
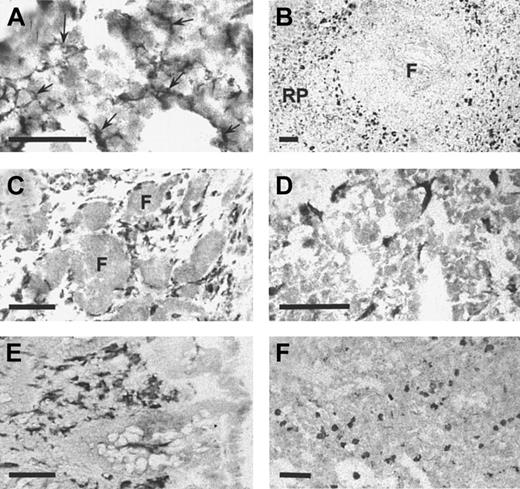
In the mouse, Sn is expressed at high levels on distinctive populations of macrophages in spleen and lymph node as well as stromal macrophages in the bone marrow.8 Lower levels are found on many other tissue macrophages although certain macrophages, notably resident brain microglia, express undetectable levels of Sn.10 When immunohistochemistry was performed on frozen sections of human bone marrow using HSN1, a reticular staining pattern of resident macrophages was observed (Figure7A) that was strikingly similar to the previous description in the mouse.8 In the spleen, a subset of macrophages was intensely stained (Figure 7B), in a region defined as the perifollicular zone,40 whereas red pulp macrophages were stained only weakly. Lymph node perifollicular sinusoidal macrophages stained strongly with mAb HSN1 (Figure 7C). In addition, other resident macrophages in various human tissues were found to be Sn+, including liver Kupffer cells (Figure 7D), macrophages in the lamina propria of the colon (Figure 7E), and alveolar and interstitial macrophages in the lung (Figure 7F). In human brain sections, perivascular macrophages were intensely positive, but the resident microglia did not express detectable levels of Sn (not shown). In conclusion, expression of human Sn is restricted to stromal tissue macrophage subsets in a remarkably similar way to the mouse protein.
Sn is expressed specifically on resident macrophages in various human tissues.
Cryostat sections measuring 6 μm from bone marrow (A), spleen (B), reactive lymph node (C), liver (D), colon (E), and lung (F) were stained immunocytochemically with HSN1 mAb. Macrophage populations were stained specifically in all tissues. In bone marrow (A), the stellate processes of resident bone marrow macrophages (arrows) are evident. In the spleen (B), strong staining is present on perifollicular macrophages with weaker staining in the red pulp (RP). Tingible body macrophages within the follicle (F) are not labeled. In reactive lymph node (C), sinusoidal perifollicular macrophages are intensely stained, but no labeling is seen in the follicles (F). In the liver (D), the sinus lining Kupffer cells are labeled. In the colon (E), macrophages in the lamina propria are intensely labeled. In the lung (F), alveolar and interstitial macrophages are positive for Sn. Bars correspond to 50 μm (A) or 100 μm (B-F).
Sn is expressed specifically on resident macrophages in various human tissues.
Cryostat sections measuring 6 μm from bone marrow (A), spleen (B), reactive lymph node (C), liver (D), colon (E), and lung (F) were stained immunocytochemically with HSN1 mAb. Macrophage populations were stained specifically in all tissues. In bone marrow (A), the stellate processes of resident bone marrow macrophages (arrows) are evident. In the spleen (B), strong staining is present on perifollicular macrophages with weaker staining in the red pulp (RP). Tingible body macrophages within the follicle (F) are not labeled. In reactive lymph node (C), sinusoidal perifollicular macrophages are intensely stained, but no labeling is seen in the follicles (F). In the liver (D), the sinus lining Kupffer cells are labeled. In the colon (E), macrophages in the lamina propria are intensely labeled. In the lung (F), alveolar and interstitial macrophages are positive for Sn. Bars correspond to 50 μm (A) or 100 μm (B-F).
Expression of Sn in rheumatoid arthritis
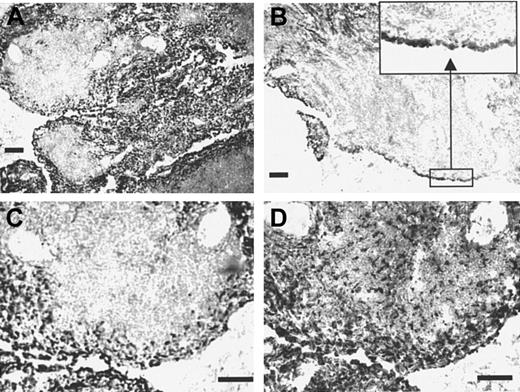
To investigate the expression of Sn under inflammatory conditions, immunohistochemistry was carried out using tissue sections from patients with rheumatoid arthritis, a disease in which inflammatory macrophages are believed to play key etiologic roles.41Parallel staining with HSN1 and the panmacrophage marker CD68 revealed both resident macrophages within the synovial membrane and infiltrating macrophages present in the subintima as strongly positive for Sn. However, macrophages within the T- and B-cell–rich extralymphoid follicles that were positive for CD68 were largely negative for Sn (Figure 8). Interestingly, the follicular macrophages also expressed several other monocyte/macrophage markers, including CD14, the high-affinity IgG Fc receptor CD64, and CD163 (data not shown). This differential staining pattern within an area of active lymphoid proliferation indicates that Sn is expressed by a functionally distinct macrophage subpopulation in inflammatory tissue.
Sn is expressed on inflammatory macrophages in rheumatoid synovial membrane.
Cryostat sections of rheumatoid synovial membrane measuring 8 μm were stained immunocytochemically with the mAb HSN1 (A-C) or with the antimacrophage marker CD68 (D). Most cells expressing Sn are located in the intima (inset to panel B), which contain a large proportion of CD68+ macrophage-like synoviocytes. In contrast, little if any Sn staining is seen within the T- and B-cell–rich extralymphoid follicles, despite the presence of large numbers of CD68+macrophages pervading the tissue (D). Bars correspond to 100 μm.
Sn is expressed on inflammatory macrophages in rheumatoid synovial membrane.
Cryostat sections of rheumatoid synovial membrane measuring 8 μm were stained immunocytochemically with the mAb HSN1 (A-C) or with the antimacrophage marker CD68 (D). Most cells expressing Sn are located in the intima (inset to panel B), which contain a large proportion of CD68+ macrophage-like synoviocytes. In contrast, little if any Sn staining is seen within the T- and B-cell–rich extralymphoid follicles, despite the presence of large numbers of CD68+macrophages pervading the tissue (D). Bars correspond to 100 μm.
Discussion
The most striking feature of the data presented here is the remarkable degree to which Sn is conserved between mouse and man. This is apparent at all levels examined, namely sequence similarity, domain organization, binding properties, and expression patterns. Interestingly, the highest degree of sequence similarity was seen in the extracellular region, particularly within the V-set domain that contains the sialic acid binding site. In comparison, the cytoplasmic tail was poorly conserved. Taken together, these observations indicate that Sn has evolved primarily to mediate extracellular functions, namely cell-cell or cell-matrix interactions. The interspecies conservation of all 17 Ig domains is relevant in this regard, because it has been proposed that this large number of domains could be important in extending the binding site of the molecule away from the plasma membrane, which then favors cell-cell interactions.17 Similar conclusions have been made regarding the importance of length for the adhesion molecule, P-selectin,42 which has 9 complement control protein domains in addition to an EGF-like domain and the ligand binding C-type lectin domain.42
Although the ligand binding studies of human Sn are not as extensive as those carried out previously for the mouse protein, the results presented here show that human Sn has very similar properties. It binds strongly to human RBCs in a sialic acid–dependent manner, prefers Neu5Ac in α2,3 glycosidic linkage, and binds preferentially to granulocytes when compared with other blood leukocyte populations. The strong binding of human Sn to autologous RBCs is surprising, because previous studies with mouse Sn showed that it bound weakly to autologous RBCs. This was shown to be due to the presence of the 9-O-acetylated form of Neu5Ac (Neu5,9Ac2) on mouse (but not human) RBCs, which prevents recognition by Sn.27It could be argued that masking of Sn ligands on autologous RBCs is important for preventing inappropriate interactions between circulating RBCs and Sn+ macrophages, especially in tissues like liver where resident macrophages in sinuses are in direct contact with blood. Because human RBCs are strongly recognized by human Sn, the question arises as to how RBCs avoid being trapped by Sn+macrophages exposed to the circulation, such as Kupffer cells. One possibility is that the binding activity of human Sn is regulated by interactions with sialic acids presented on the macrophage surface.19,43,44 Unlike all other mammals studied, humans do not express N-glycolylneuraminic acid (Neu5Gc) due to a partial deletion of the CMP-NeuAc hydroxylase gene required for its synthesis.45,46 Neu5Gc is not recognized by either mouse27 or human47 Sn. Therefore, if mouse macrophages preferentially expressed this form of sialic acid, the Sn binding site would not be masked and the receptor would be able to mediate adhesive functions. In contrast, the increased levels of Neu5Ac on human macrophages could partially mask the binding site of Sn, resulting in lowered adhesive activity of the receptor. In this way, the higher levels of Sn ligands on human versus mouse RBCs would be compensated for by the reduced adhesive activity of Sn on human versus mouse Kupffer cells. Further studies are needed to explore these possibilities.
Despite the marked differences in binding to autologous RBCs, human and mouse Sn exhibit similar strong, sialic acid–dependent binding to autologous neutrophils. Although the physiologic significance of this is unknown, one possibility is that Sn contributes to the clearance of these cells following apoptosis or senescence in tissues like liver, spleen, and lymph nodes, for example, in conjunction with defined phagocytic receptors like CD36 and the vitronectin receptor, αVβ3.48 In addition to granulocytes, we also observed clear binding to B cells, NK cells, and a subset of CD8 T cells. The finding that most of the CD69+ cells were recognized by Sn suggests that lymphocyte activation may lead to higher levels of Sn ligand expression and promote interactions with Sn+macrophages. This would be consistent with previous findings in the mouse that activated lymphocytes cluster selectively with Sn+ macrophages in vivo.14 Lymphocyte activation is known to result in altered cell surface sialylation, at least in part due to increased expression of the core 2 enzyme, which is needed for synthesis of branched O-glycans on mucinlike molecules such as CD43.49 Further studies are needed to determine whether such alterations in glycosylation lead to increased recognition by Sn.
The generation of specific mAbs to Sn has allowed us to investigate the expression pattern of the human protein. The immunocytochemical and FACS studies show that Sn is not detectable on monocytes but is expressed on a wide variety of tissue macrophages in both healthy and diseased tissues. As in the mouse and rat, human Sn appears to be exquisitely macrophage-restricted but, importantly, the expression is heterogeneous and not all macrophages express the receptor. As in the mouse, subsets of stromal macrophages in lymphoid and hemopoietic tissues express high levels, whereas Sn is undetectable on the resident microglia in the brain. It has been shown previously in the mouse that a factor(s) in plasma is important in regulating Sn expression in vitro.39 This could provide a partial explanation why macrophages not exposed to plasma proteins, such as those behind the blood-brain barrier, do not express Sn. Other factors such as glucocorticosteroids and interferon-β have also been shown to have Sn-inducing activity in vitro,11 but their in vivo significance is unclear. Certain cytokines have been found to inhibit Sn expression, for example, interferon-gamma,39interleukin-4,50 and interleukin-13.51 In the present study using human monocytes, we did not observe significant expression of Sn after culture in autologous serum, although addition of interferon-γ and TNF-α led to clear induction. The potential regulatory role of such cytokines could therefore be particularly relevant in inflammatory and immune reactions.
Here, we examined expression of Sn in rheumatoid arthritis, a commonly occurring inflammatory disorder in which macrophages are thought to be important.52 We found that most macrophages expressed high levels of Sn along with other markers like CD68 and CD163. These findings raise the possibility that Sn contributes to the cell-cell and cell-matrix interactions of macrophages during inflammatory reactions. In rheumatoid arthritis, macrophages, together with T cells, are the predominant cell types within the hyperplastic synovial intima. It is thought that macrophages are recruited from the circulation rather than produced locally. The synovial lining, which normally consists of a bilayer, is increased in thickness by 3- to 4-fold, mostly by activated macrophages. Synovial membrane and rheumatoid pannus macrophages are implicated in a number of key events, including production of interleukin-1 and TNF-α as part of the proinflammatory cytokine network.53 Macrophages are also important sources of matrix metalloproteinases like MMP-1 and MMP-3 that promote cartilage and proteoglycan degradation.54
From a pragmatic perspective, the antihuman Sn mAbs described here are useful reagents for detecting human tissue macrophages under a variety of different conditions. In mice, several macrophage-restricted markers have been described in addition to Sn, including F4/80 antigen,55 macrosialin,56MARCO,57 MOMA-1,58 MOMA-2,59 and ERTR9.60 However, in humans only a few macrophage-specific mAbs have been defined, such as those against CD6861 and CD163.62 Although CD68 is apparently expressed on all human macrophage populations, the antigen can also be detected on other myeloid cells and also on certain nonmyeloid cells.37 61In addition, CD68 is an intracellular antigen that, unlike Sn, does not allow the visualization of plasma membrane processes in tissue sections. Thus, the use of anti-Sn mAbs in conjunction with others like anti-CD163 and anti-CD68 mAbs may be a useful way to maximally visualize macrophages in normal and diseased human tissues.
Acknowledgments
We are grateful to Roger Cox for PAC library screening, Jonathon Fawcett for providing human spleen, Kevin Gatter for access to human tissue samples, and Jiquan Zhang for help with recombinant protein production.
Supported by the Imperial Cancer Research Fund and the Wellcome Trust.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Note added in proof
Human sialoadhesin has been designated CD169 at the recent CD antigen workshop (HLDA7) held at Harrogate, United Kingdom, June 20-24, 2000.
Author notes
P.R. Crocker, MSI/WTB Complex, Department of Biochemistry University of Dundee, Dow Street, Dundee DD1 5EH, Scotland; e-mail: p.r.crocker@dundee.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal