Abstract
The human protein ABC7 belongs to the adenosine triphosphate-binding cassette transporter superfamily, and its yeast orthologue, Atm1p, plays a central role in the maturation of cytosolic iron-sulfur (Fe/S) cluster-containing proteins. Previously, a missense mutation in the human ABC7 gene was shown to be the defect in members of a family affected with X-linked sideroblastic anemia with cerebellar ataxia (XLSA/A). Here, the promoter region and the intron/exon structure of the human ABC7 gene were characterized, and the function of wild-type and mutant ABC7 in cytosolic Fe/S protein maturation was analyzed. The gene contains 16 exons, all with intron/exon boundaries following the AG/GT rule. A single missense mutation was found in exon 10 of the ABC7gene in 2 affected brothers with XLSA/A. The mutation was a G-to-A transition at nucleotide 1305 of the full-length cDNA, resulting in a charge inversion caused by the substitution of lysine for glutamate at residue 433 C-terminal to the putative sixth transmembrane domain of ABC7. Expression of normal ABC7 almost fully complemented the defect in the maturation of cytosolic Fe/S proteins in a yeast strain in which the ATM1 gene had been deleted (Δatm1 cells). Thus, ABC7 is a functional orthologue of Atm1p. In contrast, the expression of mutated ABC7 (E433K) or Atm1p (D398K) proteins in Δatm1 cells led to a low efficiency of cytosolic Fe/S protein maturation. These data demonstrate that both the molecular defect in XLSA/A and the impaired maturation of a cytosolic Fe/S protein result from an ABC7 mutation in the reported family.
Introduction
The family of ABC transporters consists of a large group of adenosine triphosphate-dependent transmembrane proteins that specifically transport a wide variety of substrates across cell and organelle membranes.1-3 Importantly, numerous genetic disorders result from alterations in the ability of these proteins to transport substrates such as cholesterol in Tangier disease,4 retinal in Stargardt disease,5,6Cl− in cystic fibrosis,7 and conjugated bilirubin in Dubin-Johnson syndrome.8 Other ABC transporter defects include adrenoleukodystrophy,9multiple drug resistance,10 and Zellweger syndrome.11 There is evidence that ABC transporters are also able to regulate other ion channels.2
Recently, the human ABC7 half-transporter gene was proposed12,13 to be the defect in a rare type of X-linked sideroblastic anemia associated with cerebellar ataxia, (XLSA/A; OMIM 301310). This disease has been linked to the long arm of the X chromosome at Xq13.14 It is characterized by the presence of nonprogressive cerebellar ataxia, diminished deep tendon reflex, incoordination, elevated free erythrocyte protoporphyrin, normal parenchymal iron stores, and lack of correction by pyridoxine supplementation.15 These characteristics distinguish it from the more common X-linked sideroblastic anemia (XLSA; OMIM 301300), which does not have a neurologic component, is typically at least partially pyridoxine-responsive, and is caused by mutations in the erythroid-specific 5-aminolevulinate synthase (ALAS2) gene at Xp11.21.16 17
The hypothesis that ABC7 was a candidate gene for XLSA/A was supported by its mapping to the q12-q13 region of the human X chromosome,13,18 the demonstration that mutations in the homologous yeast ABC transporter gene, ATM1, resulted in 30-fold higher accumulation of iron19 analogous to the elevation in mitochondrial iron in XLSA/A, and the normal iron levels that could be achieved in these Atm1p-deficient cells by complementation with human ABC7.12 That ABC7 is the defective gene has now been supported by the publication of the first mutation of this gene in a family with XLSA/A.20
A role of the yeast ABC transporter protein, Atm1p, in the generation of cytosolic Fe/S proteins has recently been identified.21It was proposed that Atm1p is involved in the export of a component required for cytosolic Fe/S protein assembly. Consistent with this idea, Atm1p was required for maturation of the cytosolic Fe/S cluster-containing protein, isopropyl malate isomerase (Leu1p), but not for the assembly of mitochondrial Fe/S proteins.
To investigate the involvement of ABC7 in a family with the clinical phenotype of XLSA/A, we determined the entire intron/exon organization of the human ABC7 gene and identified a new mutation in the gene in this family. We showed that the normal ABC7 protein participated in cytosolic Fe/S protein assembly whereas the mutant protein was defective in this function. Our data suggest that XLSA/A is caused by an impairment of the maturation of cytosolic Fe/S proteins.
Materials and methods
Characterization of the genomic structure of the humanABC7 gene
A Genome Systems (St Louis, MO) BAC library was screened with 2 sets of primers (852/854 and 778/779; Table1), and a single clone (gABC7-B1) positive for both sets was isolated and purified according to the manufacturer's protocol (brochure TS-QPT 05/98). Primers from the first 521 bp of the cDNA sequence (Genbank accession no. [GB]:AB005289) were used to sequence the relevant regions of the BAC clone by automated fluorescent dideoxy sequencing.
PCR primers and conditions for amplification of ABC7 sequences
| Amplified region . | Size (bp) . | ABC7 sequence* . | Primer coord† . | Primer no. . | Temp (°C) . | Oligonucleotides‡ . |
|---|---|---|---|---|---|---|
| Exon 1 | 484 | gABC7-B1 | UP(−)154 | 908 | 48 | 5′ GCTCTGCCCTAAGGAAAAA 3′ |
| LP(+)134 | 909 | 5′ TTCACTTACTGCTGCTCC 3′ | ||||
| Exon 1 | 116 | cABC7-A | UP3 | 852 | 53 | 5′ TCAAGAGGCGCTGCTCG 3′ |
| partial | LP119 | 854 | 5′ CGAGCAGCGCCATCTTGA 3′ | |||
| Exon 1 | NA | cABC7-A | UP102 | 933 | 68 | 5′ CGGACCTGAGCCGCTAACAGAGAC 3′ |
| 5′ Race | LP144 | 934 | 5′ TCGAGCGGTTCCCAAGGCGCCGAG 3′ | |||
| Exon 2 | 455 | gABC7-B1 | UP(−)186 | 911 | 62 | 5′ CAGGATTATAGGCGTGAG 3′ |
| LP(+)189 | 913 | 5′ CCAGGGGTAGAAAACATC 3′ | ||||
| Exon 3 | 353 | gABC7-B1 | UP(−)154 | 914 | 48 | 5′ GTGGAGGTGTTTTGCTTT 3′ |
| LP(+)115 | 917 | 5′ GTCCGAGAAAATATCATC 3′ | ||||
| Exon 4 | 454 | gABC7-B1 | UP(−)236 | 918 | 52 | 5′ GTGGCTAAAATTGAAAGT 3′ |
| LP(+)99 | 920 | 5′ ATGCTACCCCAAAAGTGA 3′ | ||||
| Exon 5 | 486 | AC002417 | UP17186 | 778 | 42 | 5′ AAGCAGAAGCATTTGACT 3′ |
| LP17671 | 779 | 5′ ACGCTAAGTTGTGTTTGT 3′ | ||||
| Exon 6 | 651 | AC002417 | UP18166 | 780 | 50 | 5′ GCACATTTATCCACAGTA 3′ |
| LP18816 | 781 | 5′ CATGCAACAGTACAACAG 3′ | ||||
| Exons | 601 | AC002417 | UP19900 | 782 | 45 | 5′ CCTATTAGTATTTGATGG 3′ |
| 7 + 8 | LP20500 | 783 | 5′ AACCTGAAGCTCTAATTT 3′ | |||
| Exon 9 | 711 | AC002417 | UP22068 | 774 | 51 | 5′ GGCATAATGTTGTGAGGG 3′ |
| LP22778 | 775 | 5′ GGCAGTGCTAAACAAAAG 3′ | ||||
| Exon 10 | 501 | AC002417 | UP23319 | 786 | 47 | 5′ CCTCACTTAAACCATTCT 3′ |
| LP23819 | 787 | 5′ GTTTGCTAATGTGGTGAT 3′ | ||||
| Exons | 838 | AC002417 | UP24376 | 788 | 50 | 5′ CATTATCCCTTCTCCCTC 3′ |
| 11 + 12 | LP25213 | 789 | 5′ GAATTTCACTGGTAGGGT 3′ | |||
| Exon 13 | 569 | AC002417 | UP28581 | 790 | 45 | 5′ GACCATACTTGTGTTAGG 3′ |
| LP29149 | 791 | 5′ GTAACCCAATCAAATGTG 3′ | ||||
| Exon 14 | 494 | AC002417 | UP31408 | 792 | 40 | 5′ GCCTCATTCTCATTCTTC 3′ |
| LP31901 | 793 | 5′ TGGAAAAAGGGGGATAGG 3′ | ||||
| Exon 15 | 468 | AC002417 | UP33505 | 794 | 47 | 5′ GGAGAAATGTTGCCTAAG 3′ |
| LP33972 | 795 | 5′ CAGGATTCAGTCTCAACC 3′ | ||||
| Exon 16 | 644 | AC002417 | UP40261 | 796 | 45 | 5′ GGTGTTTATGGTATTAGA 3′ |
| LP40904 | 797 | 5′ TTGGGGATAATACAGATG 3′ | ||||
| Intron 2 | 2000 | cABC7-A | UP206 | 857 | 60 | 5′ GCAGAGATTGGGAAAAGG 3′ |
| LP321 | 858 | 5′ TTCTTTTGGGTCTGTGTG 3′ | ||||
| Intron 10 | 175 | AC002417 | UP23588 | 925 | 44 | 5′ GTAATGGTGAATGGACTG 3′ |
| partial | LP23762 | 926 | 5′ GTTCACATACTTTTGAAAAGGTACTATCTA 3′ | |||
| PRS424 | 249 | AC002417 | UP22443 | 844 | 48 | 5′ GCCGCCGGATCCGGATTTTTGAAGACGTATGAGA 3′ |
| construct | In vector | 851 | 5′ TTGTCTAGTCTCTCTATATAGAG 3′ | |||
| PRS424 | 231 | AC002417 | In vector | 850 | 48 | 5′ CTGTATATAGAAAGACTAGACAA 3′ |
| construct | UP24752 | 845 | 5′ GCCGCCGAGCTCCCACTTTCTTTCCTGCAGGG 3′ |
| Amplified region . | Size (bp) . | ABC7 sequence* . | Primer coord† . | Primer no. . | Temp (°C) . | Oligonucleotides‡ . |
|---|---|---|---|---|---|---|
| Exon 1 | 484 | gABC7-B1 | UP(−)154 | 908 | 48 | 5′ GCTCTGCCCTAAGGAAAAA 3′ |
| LP(+)134 | 909 | 5′ TTCACTTACTGCTGCTCC 3′ | ||||
| Exon 1 | 116 | cABC7-A | UP3 | 852 | 53 | 5′ TCAAGAGGCGCTGCTCG 3′ |
| partial | LP119 | 854 | 5′ CGAGCAGCGCCATCTTGA 3′ | |||
| Exon 1 | NA | cABC7-A | UP102 | 933 | 68 | 5′ CGGACCTGAGCCGCTAACAGAGAC 3′ |
| 5′ Race | LP144 | 934 | 5′ TCGAGCGGTTCCCAAGGCGCCGAG 3′ | |||
| Exon 2 | 455 | gABC7-B1 | UP(−)186 | 911 | 62 | 5′ CAGGATTATAGGCGTGAG 3′ |
| LP(+)189 | 913 | 5′ CCAGGGGTAGAAAACATC 3′ | ||||
| Exon 3 | 353 | gABC7-B1 | UP(−)154 | 914 | 48 | 5′ GTGGAGGTGTTTTGCTTT 3′ |
| LP(+)115 | 917 | 5′ GTCCGAGAAAATATCATC 3′ | ||||
| Exon 4 | 454 | gABC7-B1 | UP(−)236 | 918 | 52 | 5′ GTGGCTAAAATTGAAAGT 3′ |
| LP(+)99 | 920 | 5′ ATGCTACCCCAAAAGTGA 3′ | ||||
| Exon 5 | 486 | AC002417 | UP17186 | 778 | 42 | 5′ AAGCAGAAGCATTTGACT 3′ |
| LP17671 | 779 | 5′ ACGCTAAGTTGTGTTTGT 3′ | ||||
| Exon 6 | 651 | AC002417 | UP18166 | 780 | 50 | 5′ GCACATTTATCCACAGTA 3′ |
| LP18816 | 781 | 5′ CATGCAACAGTACAACAG 3′ | ||||
| Exons | 601 | AC002417 | UP19900 | 782 | 45 | 5′ CCTATTAGTATTTGATGG 3′ |
| 7 + 8 | LP20500 | 783 | 5′ AACCTGAAGCTCTAATTT 3′ | |||
| Exon 9 | 711 | AC002417 | UP22068 | 774 | 51 | 5′ GGCATAATGTTGTGAGGG 3′ |
| LP22778 | 775 | 5′ GGCAGTGCTAAACAAAAG 3′ | ||||
| Exon 10 | 501 | AC002417 | UP23319 | 786 | 47 | 5′ CCTCACTTAAACCATTCT 3′ |
| LP23819 | 787 | 5′ GTTTGCTAATGTGGTGAT 3′ | ||||
| Exons | 838 | AC002417 | UP24376 | 788 | 50 | 5′ CATTATCCCTTCTCCCTC 3′ |
| 11 + 12 | LP25213 | 789 | 5′ GAATTTCACTGGTAGGGT 3′ | |||
| Exon 13 | 569 | AC002417 | UP28581 | 790 | 45 | 5′ GACCATACTTGTGTTAGG 3′ |
| LP29149 | 791 | 5′ GTAACCCAATCAAATGTG 3′ | ||||
| Exon 14 | 494 | AC002417 | UP31408 | 792 | 40 | 5′ GCCTCATTCTCATTCTTC 3′ |
| LP31901 | 793 | 5′ TGGAAAAAGGGGGATAGG 3′ | ||||
| Exon 15 | 468 | AC002417 | UP33505 | 794 | 47 | 5′ GGAGAAATGTTGCCTAAG 3′ |
| LP33972 | 795 | 5′ CAGGATTCAGTCTCAACC 3′ | ||||
| Exon 16 | 644 | AC002417 | UP40261 | 796 | 45 | 5′ GGTGTTTATGGTATTAGA 3′ |
| LP40904 | 797 | 5′ TTGGGGATAATACAGATG 3′ | ||||
| Intron 2 | 2000 | cABC7-A | UP206 | 857 | 60 | 5′ GCAGAGATTGGGAAAAGG 3′ |
| LP321 | 858 | 5′ TTCTTTTGGGTCTGTGTG 3′ | ||||
| Intron 10 | 175 | AC002417 | UP23588 | 925 | 44 | 5′ GTAATGGTGAATGGACTG 3′ |
| partial | LP23762 | 926 | 5′ GTTCACATACTTTTGAAAAGGTACTATCTA 3′ | |||
| PRS424 | 249 | AC002417 | UP22443 | 844 | 48 | 5′ GCCGCCGGATCCGGATTTTTGAAGACGTATGAGA 3′ |
| construct | In vector | 851 | 5′ TTGTCTAGTCTCTCTATATAGAG 3′ | |||
| PRS424 | 231 | AC002417 | In vector | 850 | 48 | 5′ CTGTATATAGAAAGACTAGACAA 3′ |
| construct | UP24752 | 845 | 5′ GCCGCCGAGCTCCCACTTTCTTTCCTGCAGGG 3′ |
UP, upper primer; LP, lower primer. Numbers correspond to the 5′ ends of the primers; coordinates identify the nucleotide in the cDNA or genomic sequences except for gABC7-B1 where coordinates indicate the distance from the ends of the exons.
Mismatch nucleotide is underlined.
Mismatch polymerase chain reaction (PCR) was used for analysis of the presence of a base change found in intron 10 just downstream from the intron/exon boundary. After amplification using the Expand High Fidelity System (Roche, Indianapolis, IN) with primers 925 and 926 (Table 1), a mismatch in the antisense primer (926) plus the presence of the base change created an XbaI site. The restriction digests were electrophoresed in 2.5% Ultrapure agarose (Gibco BRL, Rockville, MD).
Characterization of the 5′ end of theABC7 cDNA
Rapid amplification of cDNA ends (RACE) was used to characterize the 5′ sequence of the ABC7 gene. Marathon-ready cDNAs of human skeletal muscle (Clontech, Carlsbad, CA) were used as templates in RACE PCR to obtain 5′-end cDNA fragments. The first PCR was performed using Marathon adaptor primer AP1 (sense primer) and a gene-specific oligonucleotide (primer 934; Table 1). A nested PCR reaction used Marathon adaptor primer AP2 (sense primer) and a gene-specific oligonucleotide (primer 933; Table 1). A major band of approximately 200 bp and a smear between 200 bp and 1 kb were observed after agarose gel electrophoresis. The PCR product was cloned into the PCR II-TOPO vector (Invitrogen) according to the manufacturer's protocol. Nineteen plasmids carrying an insert were sequenced using the T7 and SP6 primers. The major band was purified in 2% low-melting agarose (Gibco BRL) and ligated into the pGEM-T Easy vector (Promega, Madison, WI). Ten plasmids carrying an insert were sequenced using the T7 and SP6 primers.
Molecular analysis of the ABC7 gene in a family with XLSA/A
Genomic DNA was isolated by standard techniques from peripheral blood of the proband and other family members. PCR amplification primer sequences (Table 1) were selected based on the sequence of the BAC clone gABC7-B1 and the sequence of the htgs contig GB:AC002417. Sense primers were designed to amplify from 137 to 319 nt of 5′ flanking intronic sequence. Because intron 7 measured only 91 nt, exons 7 and 8 were amplified together; similarly, exons 11 and 12 were amplified together because intron 11 measured 154 nt. Antisense primers were designed to amplify from 86 to 200 nt of 3′ flanking intronic sequence. The amplification of ABC7 gene exons 1 to 16 was performed with the Expand High Fidelity System (Roche). The PCR products were purified using the Qiagen PCR Purification kit and sequenced in both orientations using the same primers used for amplification. For confirmation of the point mutation in exon 10 of the ABC7gene by restriction analysis, the PCR product was cut byBsmAI (New England Biolabs, Beverly, MA). Restriction digests were electrophoresed in 2.5% Ultrapure agarose (Gibco BRL).
Generation of yeast expression constructs
The full length ATM1 gene, including 5 nucleotides of the 5′-untranslated region, was amplified by PCR and cloned to theEcoRI and SalI sites of the centromeric yeast shuttle vector pRS416,22 resulting in plasmid pRS416ATM1 and sequence confirmed. Site-directed mutagenesis of amino acid residue 398 of the ATM1 gene from aspartate to lysine was performed using the QuikChange kit (Stratagene, La Jolla, CA), using the plasmid pRS416ATM1 as a template. For introduction of the mutations, the oligonucleotides 5′-AGT GTT TAC AGA AAG TTA AAG CAG TCT CTG-3′ and 5′-CAG AGA CTG CTT TAA CTT TCT GTA AAC ACT-3′ were used as PCR primers. Synthesis of the wild-type or mutant ABC7 proteins in Δatm1 yeast cells was achieved by inserting the full-length coding sequences of the wild-type or mutant ABC7gene plus 5 upstream noncoding nucleotides into the yeast expression vector pRS424-GPD.23 Full-length wild-typeABC7 cDNA was cloned into pRS424-GPD in 2 steps. The EST clone GB:AA305099 was cut with XhoI, and the piece containing the 3′ ABC7 fragment was cloned to theXhoI site of pRS424-GPD to yield the plasmid pRS424-GPD/ABC7partial. The 5′ piece of ABC7 was amplified using 5′-AAA GAA TTC TCA AGA TGG CGC TGC TCG CGA TGC ATT CTT GGC GC-3′ and 5′-CAC TAC CTC CTA CAA TGG-3′ as primers. The PCR product was trimmed with EcoRI and cloned to theEcoRI-digested pRS424-GPD/ABC7partial plasmid. Orientation of the integrated 5′ EcoRI piece of ABC7 was determined by restriction mapping and by DNA sequencing.
The point mutation found in the XLSA/A proband was introduced into the pRS424-GPD/ABC7 expression construct by site-directed mutagenesis using unique flanking BbsI and PstI sites. PCR products from 2 initial reactions using mutagenic primer 851 and theBbsI site-containing primer 844 with an addedEcoRI site and mutagenic primer 850 and the PstI site-containing primer 845 flanked by an added EcoRI site were purified, treated with T4 DNA polymerase, and used as megaprimers and template in a PCR reaction with primers 844 and 845. The 450-bp product was digested with EcoRI and subcloned to pBS-II SK for sequence confirmation of the mutated site. In addition, the 2.2-kbXhoI fragment containing ABC7 from pRS424-GPD/ABC7 was subcloned to XhoI-cut pGEM7Z. Subsequently, the pGEM7Z and the pBS-II SK constructs were cut byBbsI and PstI, and the inserts were swapped. The mutated XhoI fragment in the new pGEM7Z construct was then ligated into the purified vector fragment from XhoI-cut pRS424-GPD/ABC7. A clone with the correct orientation was sequenced to confirm the introduction of the desired mutation.
Complementation studies in yeast
Results
Patient data
The proband in family 1 (II-1; Figure1D) was a white male (DOB, 12/22/66) born at a low birthweight (2495 g) to nonconsanguineous parents. From early infancy, the proband had mild postnatal growth retardation and substantially impaired gross motor and cognitive development. He did not sit unsupported until age 4, when congenital ataxia was diagnosed. By age 6, he managed to walk with support and subsequently could walk short distances unsupported. He has used simple language from age 5, and he attended a special school where he exhibited severe, static intellectual impairment. His karyotype and plasma amino acid levels were normal. In 4 decades there has been no evidence of progression of ataxia or intellectual impairment, and the proband has been in good physical and mental health. At age 13, his hemoglobin level measured 10.1 g/dL, and his erythrocytes were microcytic. Computed tomography of the brain at age 18 showed striking, selective cerebellar hypoplasia, but no further action was taken until the proband was brought for examination at age 22 year with pallor and weight loss; sideroblastic anemia and nonprogressive cerebellar ataxia were diagnosed. At this time he began pyridoxine supplementation at 200 mg/d, which was slowly reduced to 25 mg/d. Although the hemoglobin deficiency was refractory to this treatment, supplementation has been maintained at 25 mg/d. A younger brother (II-2, DOB 7/26/69), who had a similar phenotype, underwent diagnosis at this time and also was found to be affected but has not received pyridoxine supplementation. At age 25, the proband's blood studies (Table2) revealed anemia (Hb, 10.8 g/dL) with hypochromic (MCH, 20.0 pg) microcytic (MCV, 62.2 fL) erythrocytes, and normal serum ferritin (156 μg/L). His younger brother was slightly more anemic (Hb, 9.9 g/dL; MCH, 19.6 pg; MCV, 60.2 fL). His serum ferritin level was slightly above normal at 331 μg/L. Their mother's (I-1, DOB 9/20/42) blood film showed dimorphism, consistent with an X-linked defect. In the family history, the proband's mother and her deceased father have been affected by adult-onset retinitis pigmentosa. The proband's maternal aunt had one son who died at 19 of Duchenne muscular dystrophy.
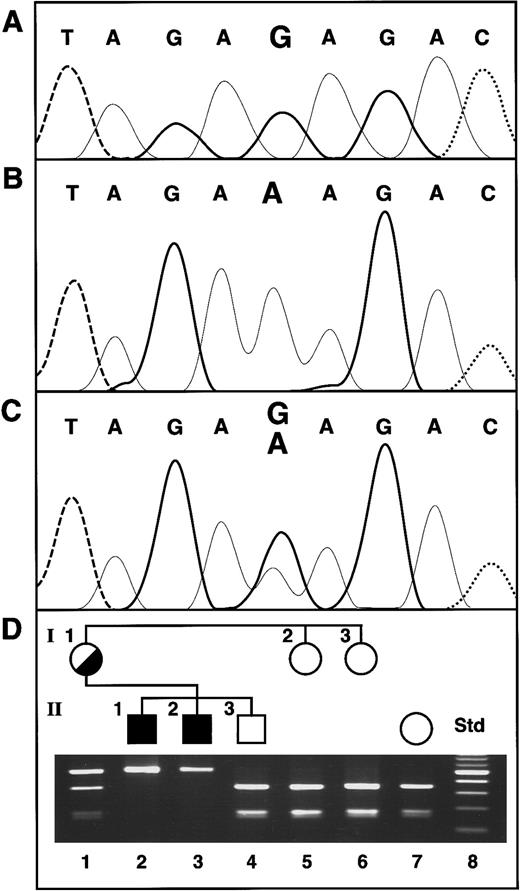
Sequence of the
ABC7 gene mutation in XLSA/A and confirmation of the presence of the mutation in family members. (A) Normal female control. The bases are distinguished by heavy solid lines, G; light solid lines, A; dashed lines, T; and dotted lines, C. (B) Proband II-1. (C) Heterozygous mother of the proband. (D) Restriction analysis for confirmation of the exon 10 mutation. The Std lane contains a 100-bp increment ladder, with the brighter band 500 bp. The presence of the G1305A mutation was assayed by restriction analysis withBsmAI, which digests only the normal allele. Thus, in the absence of the mutation, the 501-bp PCR product is cleaved into fragments of 330 and 171 bp.
Sequence of the
ABC7 gene mutation in XLSA/A and confirmation of the presence of the mutation in family members. (A) Normal female control. The bases are distinguished by heavy solid lines, G; light solid lines, A; dashed lines, T; and dotted lines, C. (B) Proband II-1. (C) Heterozygous mother of the proband. (D) Restriction analysis for confirmation of the exon 10 mutation. The Std lane contains a 100-bp increment ladder, with the brighter band 500 bp. The presence of the G1305A mutation was assayed by restriction analysis withBsmAI, which digests only the normal allele. Thus, in the absence of the mutation, the 501-bp PCR product is cleaved into fragments of 330 and 171 bp.
Hematologic values for the XLSA/A family
| Patient . | Age (y) . | Hb . | MCV . | MCH . | Ferritin . | Serum iron . | TIBC . | Transferrin saturation . | sTfR . | EP* . |
|---|---|---|---|---|---|---|---|---|---|---|
| Mother | 49 | 13.4 | 80.9 | 27.0 | 76 | |||||
| 57 | 11.2 | 81.3 | 26.9 | 105 | 9 | 75 | 12 | 25.1 | ||
| Proband | 25 | 10.8 | 62.2 | 20.0 | 156 | |||||
| 33 | 10.0 | 61.8 | 18.7 | 277 | 7 | 45 | 16 | 34.7 | 25.7 | |
| Brother | 22 | 9.9 | 60.2 | 19.6 | 331 | |||||
| 30 | 8.4 | 58.7 | 16.8 | 622 | 7 | 41 | 17 | 52.4 | 19.5 | |
| Normal range | 12.5-16.5 | |||||||||
| 80-100 | 27-32 | 15-300 | 10-40 | 45-77 | 15-30 | 8.7-28.1 | 0.4-1.7 | |||
| 13.5-17.5 |
| Patient . | Age (y) . | Hb . | MCV . | MCH . | Ferritin . | Serum iron . | TIBC . | Transferrin saturation . | sTfR . | EP* . |
|---|---|---|---|---|---|---|---|---|---|---|
| Mother | 49 | 13.4 | 80.9 | 27.0 | 76 | |||||
| 57 | 11.2 | 81.3 | 26.9 | 105 | 9 | 75 | 12 | 25.1 | ||
| Proband | 25 | 10.8 | 62.2 | 20.0 | 156 | |||||
| 33 | 10.0 | 61.8 | 18.7 | 277 | 7 | 45 | 16 | 34.7 | 25.7 | |
| Brother | 22 | 9.9 | 60.2 | 19.6 | 331 | |||||
| 30 | 8.4 | 58.7 | 16.8 | 622 | 7 | 41 | 17 | 52.4 | 19.5 | |
| Normal range | 12.5-16.5 | |||||||||
| 80-100 | 27-32 | 15-300 | 10-40 | 45-77 | 15-30 | 8.7-28.1 | 0.4-1.7 | |||
| 13.5-17.5 |
Total erythrocyte protoporphyrin. The proband's total was 65% zinc EP and 35% FEP, whereas in his brother only the total was measured.
On recent examination, both affected brothers are nondysmorphic and have reduced heights (157.5 and 160 cm, respectively). Head circumferences, 57.5 and 59 cm, respectively, are within +2 SD. The brothers have cerebellar signs including nystagmus, dysarthria, past pointing, and dysdiadochokinesis. Tendon reflexes could not be elicited at the elbows or ankles and were diminished at the knees. Plantar responses were flexor. For comparison, their unaffected older brother (II-3) was of normal birthweight (3204 g), has no motor or mental impairment, and is 165.24 cm tall.
When the proband's blood was recently evaluated (Western Infirmary, Glasgow, UK) at age 33, he continued to display hypochromic, microcytic anemia, and his affected younger brother continued to exhibit slightly more anemia (Table 2). The proband's distribution of erythrocyte sizes was monomodal but broad (RDW, 29.3%) (Figure2B). Evaluation of the proband for iron revealed a serum ferritin level of 277 μg/L, serum iron level of 7 μmol/L, TIBC of 47 μmol/L, and a transferrin saturation of 16%. The proband's affected younger brother showed evidence of increased iron stores (serum ferritin level, 622 μg/L). However, transferrin saturation values were low in both affected brothers, whereas the serum soluble transferrin receptor (sTfR) concentrations were elevated (Table2). Total erythrocyte protoporphyrin (EP) was markedly elevated in the proband to 25.7 μmol/L, with zinc EP comprising 65% of the total and free erythrocyte protoporphyrin (FEP) accounting for 35% (Table 2). Cutaneous photosensitivity was not observed or reported, consistent with most of the accumulated porphyrin being zinc EP, which is not a photosensitizer. At this time the proband's mother also had hypochromic and microcytic erythrocytes (Table 2). She exhibited a dimorphic erythrocyte pattern with a clearly bimodal histogram yielding an RDW of 28.7% (Figure 2A).
Erythrocyte size distribution in normal and
ABC7 mutant cells. Shaded bands are the ranges of values for the volumes of normal erythrocytes. Solid lines are the size distributions for erythrocytes from the heterozygous mother (A) and her affected son (B).
Erythrocyte size distribution in normal and
ABC7 mutant cells. Shaded bands are the ranges of values for the volumes of normal erythrocytes. Solid lines are the size distributions for erythrocytes from the heterozygous mother (A) and her affected son (B).
Characterization of the genomic structure of the humanABC7 gene
The structure of the ABC7 gene was determined to characterize the molecular defect in a family with XLSA/A. The High Throughput Genomic Sequences (htgs) section of GenBank contained an unpublished sequence of 100639 nt (GB:AC002417) consisting of 3 unordered contigs that matched part of the previously publishedABC7 cDNA (GB:AB005289).13 The match began with nucleotide 522 of the cDNA and nucleotide 17437 of the second contig (nt 12201 to 50410) and extended to the end of the 3′ untranslated sequence in the cDNA corresponding to nt 40804 of the genomic clone. Comparison of these cDNA and genomic sequences identified 12 exons in this portion of the ABC7 gene (Table3). To obtain the genomic sequence corresponding to the 5′ end of the cDNA (GB:AB005289), a BAC library was screened for clones positive with 2 sets of primer pairs. The first primer pair amplified a region at the most 5′ sequence of this cDNA for which the same-sized fragment was obtained in both the cDNA and the genomic DNA (ie, no intronic sequence). The second primer pair amplified the first ABC7 exonic region of the genomic sequence described above (GB:AC002417), including approximately 250 bp of 5′- and 100 bp of 3′-flanking sequences. A single BAC clone (gABC7-B1) was isolated that was positive for primer pairs. Four additional exons were identified by sequencing this clone with primers (Table 1) from nt 61 to 522 of the cDNA, for a total of 16 exons (Table3). Attempts to determine the sizes of the first 4 introns using long-range PCR revealed that intron 2 measured 2.0 kb (Table 3), suggesting that the remaining introns are large. Each of the 16 exons was amplified using primers designed to include 5′- and 3′-flanking intron sequences. Each of these 16 PCR products was sequenced in both orientations with the same primers used for amplification. The first 4 exon sequences were identical to nt 61 to 522 of the cDNA. The remaining 12 exon sequences were identical to the unpublished genomic sequence (clone GB:AC002417, htgs section). A single sequence difference from this genomic sequence was found in the third nucleotide of intron 10. Mismatch PCR analysis with XbaI digestion was used to test for the presence of this G-to-C base change in genomic DNA. The predicted restriction fragments in the presence of a C were 145 and 30 bp, whereas the PCR product would remain uncut in the presence of a G (175 nt). Analysis of 36 alleles revealed that the normal sequence had a C in this position. The difference seen in the database sequence was probably due to a rare mutation or a sequence problem in the genomic clone GB:AC002417. The assembly of all the exonic sequences determined from amplified genomic DNA resulted in a 2356-nt ABC7 conceptual cDNA sequence (GenBank accession numbers AF241872 to AF241887). This sequence contained 8 5′ untranslated (UT) nucleotides, 2256 nt of coding sequence and 92 nt of 3′ UT sequence. The recently published cDNA (GB:AF13365920) confirmed nt 9 through nt 2353 of this sequence. The GB:AB005289 sequence used above to identify intron/exon boundaries by comparison with genomic sequences had 3 sequence differences and contained a 60 bp 5′ extension that was not in the genomic clone gABC7-B1 or in any of the 4 ABC7 cDNAs present in GenBank.
Intron-exon boundaries of the human ABC7 gene
| Exon . | 5′ splice donor . | Intron . | 3′ splice acceptor . | Exon . | ||
|---|---|---|---|---|---|---|
| (bp) | (kb) | Promoter ‖CGCTCAAG | 1 | |||
| 1 | (176) | CCTACCAG‖gtgaga | 1 | (?) | attaagtgtccactctgaaacttccccctactttttttccttgtcagcag‖ATTCCAGG | 2 |
| 2 | (78) | CTGCAAAG‖gtaaga | 2 | (2000) | tctgaaattctgtaatagctaacggactgcttggtatcatcttcaattag‖GCTCTCCA | 3 |
| 3 | (87) | AAGAAGGG‖gtaaga | 3 | (?) | atgtttgaaatgacatgttattcactgctttgttttctctctttcaatag‖TTAAAAGA | 4 |
| 4 | (120) | GTGCAAAG‖gtaaga | 4 | (?) | taaataagtattttgtgtataatgggtaattgatgtttttatgtttttag‖GCCATGAA | 5 |
| 5 | (133) | GATTGGCT‖gtatgt | 5 | (891) | ttgattctaatattgttacatcctattttttcttttgaatgtattgttag‖ATGGTGTA | 6 |
| 6 | (269) | GTGTTTTG‖gtaagt | 6 | (1405) | gggaagaacaataactgtttcacgtttcttctttggacttccttttctag‖TATTACAA | 7 |
| 7 | (89) | CGGTGGAG‖gtaatt | 7 | (91) | tttgcttttataacatgtatttcatgcctatgcaaatctttgtcttacag‖AACTAGAT | 8 |
| 8 | (88) | CTGTGAAG‖gtaata | 8 | (2001) | gaataaattctaattatacttgcctatagttatttttatatattttgtag‖TATTTTAA | 9 |
| 9 | (175) | TGTGGCAG‖gtaatg | 9 | (986) | aagtatgtaacatttatctaaaataccctttttttcttctgccttttaag‖GTACCCTT | 10 |
| 10 | (158) | AAATTAAA‖gtaagt | 10 | (910) | tgtactctaggcagtgcgtaaagtggctttttttttctcttccctgctag‖GACAAAGT | 11 |
| 11 | (164) | GGGTCAGG‖gtgggt | 11 | (154) | ttcctaacattaataggagctaaataacctttcttcatctccaatttcag‖GAAAAGCA | 12 |
| 12 | (130) | TACCTCAG‖gtattt | 12 | (3764) | agagagatatatagatatagatatatattcattcagtatatctgcttcag‖GATGCTGT | 13 |
| 13 | (172) | GCTTTCAG‖gtgatc | 13 | (2638) | taaaatatgcatgcactatgcttccttaaagtaaaattttcctttaccag‖GAGGAGAA | 14 |
| 14 | (104) | CTGAAGAG‖gtaagt | 14 | (1997) | tgtgtacttctttttattttctcctgttcttcatattaccaaacttgcag‖ACTATTCT | 15 |
| 15 | (108) | TGGATCAG‖gtaaga | 15 | (6636) | atgctagttaaatgacctttctccgtgtttctcatgtgtttactttttag‖GGTAAGGT | 16 |
| 16 | (305) | ACATTCCC‖attttctataatccttcttttagataagatttattt 3′ Flanking Region | ||||
| Consensus: | a | |||||
| Exon . | 5′ splice donor . | Intron . | 3′ splice acceptor . | Exon . | ||
|---|---|---|---|---|---|---|
| (bp) | (kb) | Promoter ‖CGCTCAAG | 1 | |||
| 1 | (176) | CCTACCAG‖gtgaga | 1 | (?) | attaagtgtccactctgaaacttccccctactttttttccttgtcagcag‖ATTCCAGG | 2 |
| 2 | (78) | CTGCAAAG‖gtaaga | 2 | (2000) | tctgaaattctgtaatagctaacggactgcttggtatcatcttcaattag‖GCTCTCCA | 3 |
| 3 | (87) | AAGAAGGG‖gtaaga | 3 | (?) | atgtttgaaatgacatgttattcactgctttgttttctctctttcaatag‖TTAAAAGA | 4 |
| 4 | (120) | GTGCAAAG‖gtaaga | 4 | (?) | taaataagtattttgtgtataatgggtaattgatgtttttatgtttttag‖GCCATGAA | 5 |
| 5 | (133) | GATTGGCT‖gtatgt | 5 | (891) | ttgattctaatattgttacatcctattttttcttttgaatgtattgttag‖ATGGTGTA | 6 |
| 6 | (269) | GTGTTTTG‖gtaagt | 6 | (1405) | gggaagaacaataactgtttcacgtttcttctttggacttccttttctag‖TATTACAA | 7 |
| 7 | (89) | CGGTGGAG‖gtaatt | 7 | (91) | tttgcttttataacatgtatttcatgcctatgcaaatctttgtcttacag‖AACTAGAT | 8 |
| 8 | (88) | CTGTGAAG‖gtaata | 8 | (2001) | gaataaattctaattatacttgcctatagttatttttatatattttgtag‖TATTTTAA | 9 |
| 9 | (175) | TGTGGCAG‖gtaatg | 9 | (986) | aagtatgtaacatttatctaaaataccctttttttcttctgccttttaag‖GTACCCTT | 10 |
| 10 | (158) | AAATTAAA‖gtaagt | 10 | (910) | tgtactctaggcagtgcgtaaagtggctttttttttctcttccctgctag‖GACAAAGT | 11 |
| 11 | (164) | GGGTCAGG‖gtgggt | 11 | (154) | ttcctaacattaataggagctaaataacctttcttcatctccaatttcag‖GAAAAGCA | 12 |
| 12 | (130) | TACCTCAG‖gtattt | 12 | (3764) | agagagatatatagatatagatatatattcattcagtatatctgcttcag‖GATGCTGT | 13 |
| 13 | (172) | GCTTTCAG‖gtgatc | 13 | (2638) | taaaatatgcatgcactatgcttccttaaagtaaaattttcctttaccag‖GAGGAGAA | 14 |
| 14 | (104) | CTGAAGAG‖gtaagt | 14 | (1997) | tgtgtacttctttttattttctcctgttcttcatattaccaaacttgcag‖ACTATTCT | 15 |
| 15 | (108) | TGGATCAG‖gtaaga | 15 | (6636) | atgctagttaaatgacctttctccgtgtttctcatgtgtttactttttag‖GGTAAGGT | 16 |
| 16 | (305) | ACATTCCC‖attttctataatccttcttttagataagatttattt 3′ Flanking Region | ||||
| Consensus: | a | |||||
Characterization of the 5′ end of theABC7 gene
The sequence of the genomic BAC clone, gABC7-B1, did not contain either the sequence from nt 1 to 60 of cDNA sequence GB:AB005289 or a consensus sequence for a 5′ intron/exon boundary, consistent with nucleotide 61 of sequence GB:AB005289 being the start of transcription. Examination of EST databases revealed 14 sequences, all initiating between nt 15 and 119 of the ABC7 gene as shown in Figure3. Although these and 4 additional published cDNAs began at similar sites, 5′ RACE was used to help establish the transcription start site to see whether any sequences contained the 5′ extension of cDNA GB:AB005289 or any other 5′ UT nucleotides. Eleven 5′ RACE products with ABC7 sequences of the 5′ ends began between nt 17 and 96 of the ABC7 gene (Figure 3). All attempts to amplify the 60 nt extension sequence of cDNA GB:AB005289 from genomic DNA, our BAC clone, and from Marathon muscle cDNA using various primers were unsuccessful. Because most ESTs and the 5′ race ends began in this region (Figure 3) and because the most 5′ base (nt 60) of ABC7 cDNA AB005289 matched our genomic ABC7 sequence, this was designated the transcription start site.
Sequence of the
ABC7 promoter and first exon. Putative transcription factor-binding sites are boxed with the factor identified above and a caret indicating the orientation. The 5′ ends of cDNA and EST sequences from GenBank are indicated by letters a to j, where a = AF038950; b = AF133659; c = AA226777; d = AF078777 andAA305099; e = AI34738; f = AI241249, AA971 952, AA282065,AA810484, AA810091, AA923765, and AI873369; g = AA815080; h = AI016955; i = AA766839; and j = AA732107. 5′-RACE products are indicated by 1 or 2, indicating the number of RACE ends found at each position. The first exon/intron boundary is indicated by a vertical line flanked with the exon numbers. This sequence has been deposited in GenBank with the accession number GB:AF241872.
Sequence of the
ABC7 promoter and first exon. Putative transcription factor-binding sites are boxed with the factor identified above and a caret indicating the orientation. The 5′ ends of cDNA and EST sequences from GenBank are indicated by letters a to j, where a = AF038950; b = AF133659; c = AA226777; d = AF078777 andAA305099; e = AI34738; f = AI241249, AA971 952, AA282065,AA810484, AA810091, AA923765, and AI873369; g = AA815080; h = AI016955; i = AA766839; and j = AA732107. 5′-RACE products are indicated by 1 or 2, indicating the number of RACE ends found at each position. The first exon/intron boundary is indicated by a vertical line flanked with the exon numbers. This sequence has been deposited in GenBank with the accession number GB:AF241872.
Sequence analysis of the ABC7 proximal promoter region
The sequence of the promoter region containing 499 bp of 5′-flanking sequence and the first exon is shown in Figure 3. This sequence has been deposited in GenBank (AF241872). The promoter region was analyzed for putative transcription factor-binding motifs using the MatInspector program, version 2.2, and the Transfac, version 3.5, databases based on perfect matches to the core sequences with 85% overall homology.25 Basal promoter elements identified included a TATA-like box found at −26 bp, Inr initiator consensus sequences at −34 and −42 bp, PEA3 initiator sequences at −1 and −8 bp, and an NFY (CAAT box) motif at −205 (Figure 3). Putative enhancer or repressor motifs included NFAT (−80, −155, −181, −227, −414, −458, and −481 bp), NKX2.5 (−59 bp), ETS (−100 bp), GFI1 (−172 bp), E-Box (−243 bp), deltaEF1 (−264 and −377 bp), HNF2 (−289 bp) GKLF (−405 bp), OCT-1 (−424 bp), and GATA1 (−435 bp).
Identification of a missense mutation in theABC7 gene in a patient with XLSA/A
Genomic DNA was isolated from the XLSA/A proband, his affected brother, and other family members. The promoter region (137 nt) and each of the 16 exons of the ABC7 gene, including typically 250 nt of 5′ flanking and 100 nt of 3′ flanking sequence, were PCR amplified and sequenced in both orientations. A point mutation found in exon 10 of the gene was the only base change from the normalABC7 genomic sequence (Figure 1A). This G-to-A transition at nt 1305 of the cDNA was also present in the proband's affected brother (Figure 1B) and on one allele in their mother (Figure 1C). The mutation resulted in a substitution of lysine for the wild-type glutamate at amino acid 433 (E433K) of the ABC7 protein and abolished aBsmAI restriction site (Figure 1D). Restriction of the PCR product from normal genomic DNA resulted in fragments of 330 and 171 bp, whereas analysis of the 2 affected brothers' DNA yielded a single undigested PCR product of 501 bp. Their mother was heterozygous, showing all 3 fragment sizes, but the proband's older brother and 2 maternal aunts were normal. The absence of this mutation in any of 120 alleles in unrelated white females indicated that it was not a polymorphism (data not shown).
Complementation analysis in yeast
To analyze the function of the wild-type and mutant ABC7 proteins, we expressed them in yeast cells from which the ATM1 gene had been deleted (Δatm1 cells19). In addition, we performed similar studies with Atm1p into which the mutation corresponding to the human ABC7 mutant (E433K) was introduced (Atm1p D398K). Wild-type Atm1p and, to a slightly lesser extent, ABC7 complemented the growth defect of Δatm1 cells and the defect leading to mitochondrial iron accumulation.12 Similarly, both the Atm1p D398K and the ABC7 E433K mutant proteins restored growth of Δatm1 cells to a similar extent as the corresponding wild-type proteins (data not shown). This indicates that the mutations have only a mild effect on cell growth.
To analyze the effects of the mutations more specifically, the de novo formation of the cytosolic Fe/S protein, isopropyl malate isomerase (Leu1p), was measured. Atm1p function has been shown to be crucial for the assembly of this protein.21 To determine whether wild-type ABC7 could functionally replace Atm1p in cytosolic Fe/S protein synthesis, Δatm1 yeast cells expressing either no Atm1 homologue, yeast Atm1p, or human ABC7 were incubated with radioactive55Fe for 1 hour. Leu1p was immunoprecipitated from cell extracts using specific antibodies. The amount of radioactive55Fe associated with Leu1p served as a direct measure for the incorporation of the Fe/S cluster into Leu1p.21Expression of ABC7 in Δatm1 cells restored the formation of the Fe/S cluster of Leu1p to approximately 75% of the levels observed on expression of wild-type Atm1p in Δatm1 cells (Figure4). This demonstrated that ABC7 is the human orthologue of Atm1p with respect to its function in the biosynthesis of cytosolic Fe/S proteins.
ABC7 can replace yeast Atm1p in its function in the biogenesis of cytosolic Fe/S proteins.
Yeast cells lacking the ATM1 gene (Δatm1 cells19) were transformed with the yeast expression plasmid pRS424-GPD carrying either no DNA insert or coding sequences for wild-type and mutant ATM1 or ABC7 genes as indicated. Cells were grown in minimal media without added iron in the presence of dextrose. To measure the de novo formation of the Fe/S cluster in cytosolic Leu1p, cells were radiolabeled with (55Fe) iron for 1 hour.21 After isolation of the cells, an extract was prepared by breaking cells with glass beads. Immunoprecipitation was performed using antiserum raised against Leu1p, and the co-precipitated 55Fe was measured by scintillation counting. The data represent the average of 3 independent experiments, and the bars represent the standard error. The data are given relative to the signal obtained after expression of wild-type ATM1. A background signal for immunoprecipitation with preimmune serum was subtracted (approximately 2%).
ABC7 can replace yeast Atm1p in its function in the biogenesis of cytosolic Fe/S proteins.
Yeast cells lacking the ATM1 gene (Δatm1 cells19) were transformed with the yeast expression plasmid pRS424-GPD carrying either no DNA insert or coding sequences for wild-type and mutant ATM1 or ABC7 genes as indicated. Cells were grown in minimal media without added iron in the presence of dextrose. To measure the de novo formation of the Fe/S cluster in cytosolic Leu1p, cells were radiolabeled with (55Fe) iron for 1 hour.21 After isolation of the cells, an extract was prepared by breaking cells with glass beads. Immunoprecipitation was performed using antiserum raised against Leu1p, and the co-precipitated 55Fe was measured by scintillation counting. The data represent the average of 3 independent experiments, and the bars represent the standard error. The data are given relative to the signal obtained after expression of wild-type ATM1. A background signal for immunoprecipitation with preimmune serum was subtracted (approximately 2%).
Finally, we investigated the consequences of the XLSA/A mutation E433K in ABC7 and the corresponding mutation in Atm1p (D398K) for the function of these proteins in cytosolic Fe/S protein maturation. Both proteins were active in supporting the de novo incorporation of the Fe/S cluster into Leu1p (Figure 4). However, compared with the respective wild-type proteins, their activities were reduced by approximately 50%. The findings demonstrate that the mutations significantly disturb the function of both proteins in cytosolic Fe/S protein maturation, yet the alterations do not completely destroy their activity. Thus, these results indicate the participation of human ABC7 in the maturation of cytosolic Fe/S proteins and document the functional impairment of cytosolic Fe/S protein assembly resulting from ABC7/Atm1p mutations.
Discussion
Recent evidence that the ABC7 transporter was a candidate gene for XLSA/A12 13 and our identification of a family with most of the clinical and molecular characteristics of this rare disorder prompted us to search for molecular lesions in theABC7 gene in an affected family member. Therefore, we determined the genomic structure of ABC7. Exons 5 to 16 were represented in an unannotated 101-kb contig deposited in the htgs database section of GenBank (GB:AC002417). The remaining 5′ end of the gene was obtained in this study from a clone identified by screening a human genomic BAC library with PCR primer pairs that amplified a region at the most 5′ end of the published candidate cDNA (GB:AB005289) and a region including exon 5. Sequence analysis of this clone identified a putative promoter region and the first 4 exons, completing the characterization of the genomic structure of the humanABC7 gene (Figure 3, Table 3)
The genomic sequence upstream from exon 1 does not contain the first 60 nt of the previously published ABC7 cDNA.13This 60-nt sequence seems to be novel and not represented by any known human EST or cDNA clone. Repeated PCR amplifications of human muscle-derived Marathon-Ready cDNA, genomic DNA, and the BAC clone DNA using primers derived from this 60-nt extension did not result in any PCR products. All the 5′ ends of EST and all the ends obtained after 5′ RACE were within exon 1 (Figure 3). This suggests that the 60-nt extension is possibly an artifactual sequence not present in normal ABC7 transcripts.
Interestingly, the promoter region lacked canonical TATA and Sp1 sequences, though a TATA-like sequence was noted 26 nt upstream of the designated transcription start site. Importantly, there were multiple initiator sites, including 2 Inr sites, 34 and 42 nt upstream of the transcription start site. The Inr sequence is a strong promoter core initiation element that functions like a TATA box.26,27 In addition, 2 PEA3 sites occurred at −1 and −8 that can bind GABP, an initiator for TATA-less promoters.28 In this promoter, the lack of elements usually associated with high-level expression is consistent with the observed low level of expression of the normalABC7 transcript in many tissues.13 The high level of ABC7 expression seen in muscle tissue is consistent with the identification of a MyoD consensus E-Box at −243 bp. Related to this is the notable presence of 7 NFAT sites, a GATA1 site, and an NKx2.5 site in the promoter region (Figure 3), all of which have been shown to be important for heart muscle development.29-32On the other hand, the GATA1 binding site may participate in erythroid expression of ABC7, as may the binding of the erythroid-specific Tal1-E47 factor at the consensus E-Box in theABC7 promoter.33 Interestingly, HNF3 (−389 bp) and GKLF (−405 bp) factors are both active in the intestine, and HNF3 is found in the transferrin and hemochromatosis (HFE) gene promoters.34,35 GFI1 and PEA3 sites are also present in the HFE promoter.36 Future dissection of the putative ABC7 promoter elements will require functional studies such as electrophoretic mobility shift and footprinting analyses in a variety of cell types.
PCR primer pairs designed to encompass the promoter region and all 16 exons and flanking intron sequences permitted detection of the single base change in the ABC7 gene in this family. This G-to-A point mutation at nt 1305 of the cDNA (Figure 1) occurred in the middle of exon 10 and resulted in the substitution of the basic residue lysine for the acidic wild-type residue, glutamic acid, at codon 433. This dramatic change from a negative to a positive charge after the carboxy terminus of the putative sixth transmembrane domain20 is consistent with the presence of biochemical and clinical phenotypes. The mutation was present in both affected brothers and on one allele in their mother, and it was absent in their unaffected brother and 2 unaffected maternal aunts. Thus, inheritance of the mutation in the hemizygous state was always coincident with the disease phenotype, supporting its causative effect. These results represent the second mutation in the human ABC7 gene and therefore confirm and extend the previous identification of an ABC7 mutation (1208T→G) that substituted a methionine for an isoleucine at codon 400 in exon 9 as the cause of XLSA/A.
In an earlier investigation, human ABC7 was demonstrated to be the functional orthologue of yeast Atm1p.12 Expression of ABC7 almost fully restored the phenotypical defects of Atm1p-deficient cells, such as the iron accumulation within mitochondria, the high cellular concentration of glutathione, and the lack of cytochromes. Subsequent work has shown that these phenotypical characteristics of Δatm1 cells are late consequences of the failure of Atm1p in its function in the biogenesis of cytosolic Fe/S proteins.21ABC7 expression complemented to a large extent the failure of Δatm1 cells to yield cytosolic Fe/S proteins, suggesting that like Atm1p in yeast, ABC7 in the human cell functions in the maturation of cytosolic Fe/S proteins (Figure 4). Our working hypothesis for ABC7 function is that it participates in the export of Fe/S clusters generated in the mitochondria for assembly of cytoplasmic Fe/S proteins.37 38 ABC7 mutations block export and result in mitochondrial iron overload and disease.
The E433K mutation in ABC7 identified in this XLSA/A family impaired the function of the protein in cytosolic Fe/S protein assembly. The same result was observed for the corresponding mutation in yeast Atm1p. The effect of the mutation on cytosolic Fe/S protein biosynthesis was mild (a 2-fold decrease), indicating that a human cell can tolerate only slight disturbances of this process. Fe/S protein biogenesis in yeast is the only known process that renders mitochondria essential (for reviews, see Lill and Kispal38 and Lill et al39). Therefore, it may not be surprising that even relatively mild mutations in ABC7 (see Allikmets et al20) severely affect the normal well-being of a human cell.
Reduced ABC7 activity and thus impaired cytosolic Fe/S cluster maturation should affect the complex regulation of cellular iron homeostasis mediated by iron-regulatory protein 1 (IRP-1).40 41 When IRP-1 is bound in low-iron states to the 5 IREs in the TfR 3′ untranslated region, the mRNA is protected from degradation, resulting in increased TfR synthesis and iron uptake. Conversely, ferritin and ALAS2 synthesis is decreased because of IRP-1 binding to single IREs in the 5′ untranslated regions of the mRNAs. In iron-replete states, Fe/S cluster assembly on IRP-1 converts it to an aconitase-like protein that does not bind to IRE. This predicts that failure of cytosolic Fe/S cluster assembly from ABC7 mutation should increase TfR synthesis and decrease ferritin and ALAS2 synthesis. Indeed, in both affected brothers in this family, serum sTfR is increased.
At present, the connections between the ABC7 defect, the mitochondrial iron overloading evidenced by the presence of ringed sideroblasts, and the XLSA/A disease phenotype cannot be derived in molecular detail. This and previous studies19,37 suggest that the severe changes in mitochondrial iron homeostasis and likely intracellular iron distribution are caused by defects in the maturation of cytosolic Fe/S proteins. Because yeast does not have an IRP-1 homologue but still displays mitochondrial iron overloading,38 a defect in the maturation of cytosolic IRP-1 is unlikely. The relationship between ABC7-dependent Fe/S protein maturation in the cytosol and heme synthesis is also far from being understood. Heme synthesis is clearly impaired in the proband, but ALAS2 activity may only be partially deficient because protoporphyrin is considerably elevated. Ferrochelatase cannot be markedly deficient either, because it is required for the synthesis of zinc protoporphyrin, which was 65% of the total accumulated protoporphyrin in the proband. The combined hematologic evidence for functional iron deficiency in the proband (microcytic, hypochromic anemia with low serum iron, low transferrin saturation, and elevated sTfR) suggests that the iron imported into the mitochondria is unavailable for ferrochelatase-catalyzed insertion into protoporphyrin IX. It is possible that some cytosolic Fe/S protein is critical to this process or that oxidative stress induced by iron overload might interfere with heme biosynthesis or stability.19,42 43 The anemia caused by this decreased heme concentration could drive ineffective erythropoiesis, leading to increased gastrointestinal iron absorption. Reticuloendothelial iron loading with increased serum ferritin, as seen in the probands, would result from the scavenging of ineffective red cell precursors. The presence of cerebellar ataxia in XLSA/A indicates that, as evidenced in Friedreich's ataxia, iron plays an important role in cerebellar cells. Delineation of the underlying mechanisms requires further investigations of the molecular pathology of the XLSA/A erythroblast and neural cells.
In summary, these studies provide the complete gene structure of humanABC7, confirm that mutations in this gene cause XLSA/A, and demonstrate that this mutation impairs cytosolic Fe/S protein assembly. Thus, ABC7 joins the growing list of proteins to be considered (eg, frataxin, HFE, DMT1, IRP-1, IRP-2, transferrin, transferrin receptor-1, receptor-2, and ferritin) in arriving at a full understanding of iron metabolism.
Acknowledgments
We thank Dr Alison May (Cardiff, UK) for alerting us to the existence of the partial genomic sequence for ABC7 in GenBank. We thank Dr D. Ramsay (Stirling, UK) for initial diagnosis and clinical evaluations, Martin Kampmann (Marburg, Germany) for help during the generation of the Atm1 mutant, and Livia Lam and Shanthini Kasturi (New York, NY) for technical assistance.
Supported in part by a research grant from the National Institutes of Health (R01 DK40895) to D.F.B., a grant from the Association Francaise Contre les Myopathies (5897) to S.B., and a grant from the Scottish Home and Health Department (K/MRS/50/C2012) to E.F. G.K., H.L., and R.L. gratefully acknowledge grant support of the Sonderforschungsbereich 286 from the Deutsche Forschungsgemeinschaft, the Volkswagen-Stiftung, the Fonds der Chemischen Industrie, and the Hungarian Funds OKTA.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David F. Bishop, Department of Human Genetics, Box 1498, Mount Sinai School of Medicine, New York, NY 10029; e-mail:bishop@msvax.mssm.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal