Abstract
The c-fes proto-oncogene encodes a 92-kd protein tyrosine kinase whose expression is restricted largely to myeloid and endothelial cells in adult mammals. A 13.2-kilobase (kb) humanc-fes genomic fragment was previously shown to containcis-acting element(s) sufficient for a locus control function in bone marrow macrophages. Locus control regions (LCRs) confer transgene expression in mice that is integration site independent, copy number dependent, and similar to endogenous murine messenger RNA levels. To identify sequences required for this LCR,c-fes transgenes were analyzed in mice. Myeloid-cell–specific, deoxyribonuclease-I–hypersensitive sites localized to the 3′ boundary of exon 1 and intron 3 are required to confer high-level transgene expression comparable to endogenous c-fes, independent of integration site. We define a minimal LCR element as DNA sequences (nucleotides +28 to +2523 relative to the transcription start site) located within intron 1 to intron 3 of the human locus. When this 2.5-kb DNA fragment was linked to a c-fes complementary DNA regulated by its own 446–base-pair promoter, integration-site–independent, copy-number–dependent transcription was observed in myeloid cells in transgenic mice. Furthermore, this 2.5-kb cassette directed expression of a heterologous gene (enhanced green fluorescent protein) exclusively in myeloid cells. The c-fes regulatory unit represents a novel reagent for targeting gene expression to macrophages and neutrophils in transgenic mice.
Introduction
Hematopoietic cells of the myeloid lineages (monocytes/macrophages and neutrophils) are likely to be derived from a common multipotent progenitor cell. c-fes, the cellular homologue of an oncogene transduced in numerous feline and avian retroviruses, is preferentially expressed in hematopoietic progenitor cells and mature cells of the myeloid lineages.1-7The mammalian c-fes proto-oncogene encodes a 92-kd cytoplasmic protein tyrosine kinase (p92c-fes) thought to regulate proliferation and differentiation during myelopoiesis. In adult animals, peritoneal macrophages and bone-marrow–derived monocytes, macrophages, and granulocytes demonstrate high levels of c-fes messenger RNA (mRNA) and p92c-fes protein.1,4,5,8 There has also been detection of c-fes mRNA in highly purified CD34+ hematopoietic stem cells.8Interestingly, c-fes expression remains constant during myelomonocytic differentiation but decreases and is extinguished upon erythroid maturation. Greer et al7 have also demonstratedc-fes expression in adult human and murine vascular endothelial cells. During early embryonic development, c-fesmRNA has been detected in multiple fetal tissues derived from all 3 germ layers.8 However, prominent p92c-fes expression becomes more limited at later stages of development and is largely restricted to myeloid and vascular endothelial cells in the adult.9
A critical role for p92c-fes in myeloid development has been suggested by a variety of experiments. For example, K562 leukemic cells (expressing undetectable levels of p92c-fes) spontaneously undergo myeloid differentiation upon stable transfection with a 13.2-kilobase (kb) human c-fes genomic construct.10 Inhibition ofc-fes expression in HL60 cells with antisense oligonucleotides results in apoptosis during granulocytic differentiation or a block in macrophage production during treatment with vitamin D3.11-13 The protein p92c-fes is tyrosine phosphorylated and catalytically activated in response to granulocyte-macrophage colony-stimulating factor (GM-CSF),13,14 which (along with interleukin [IL]–3) is a potent enhancer of neutrophilic and monocytic development from hematopoietic progenitors. A direct association between phosphorylated p92c-fes and the common β chain shared by the IL-3 and GM-CSF receptors has previously been reported. GM-CSF treatment induces the formation of a multiprotein complex (consisting of the β subunit, c-fes, JAK2, STAT1, and STAT3) that results in tyrosine phosphorylation and activation of STAT3 by c-fes.15 These results identify a signal transduction pathway initiated by GM-CSF (or IL-3) that stimulates p92c-fes kinase activity in myeloid cells. A role for c-fes in myeloid signal transduction has also been confirmed with the use of targeted mutations of the c-fes locus in mice (Senis et al16 and Hackenmiller et al61).
The transcription of most genes introduced into transgenic mice is influenced by the surrounding chromatin at the site of integration. Remarkably, a 13.2-kb human c-fes transgene is expressed in mice in a tissue-specific manner irrespective of integration site and proportional to transgene copy number.1 Therefore, the human c-fes transcription unit includescis-regulatory DNA elements sufficient for a locus control region (LCR). LCRs were first described for the human β-globin gene cluster17 and have now been detected in a variety of genes, including human α-globin,18,19 chicken lysozyme,20 human CD2,21,22 human keratin 18,23,24 mouse metallothionein,25 human adenosine deaminase,26 and the mouse T-cell receptor α/δ locus.27 We demonstrated in transient-transfection experiments that luciferase reporter plasmids containing 446 base pairs (bp) of c-fes 5′ flanking sequences are active exclusively in myeloid cells.28 This myeloid-specific promoter is regulated by Sp1, PU.1, and a novel 70-kd transcription factor, termedc-fes expression factor (FEF).29 However,cis-acting elements required for locus control function have not been functionally delineated.
Active genes are typically located within regions of general deoxyribonuclease I (DNase I) sensitivity. Interestingly, thec-fes locus contains 3 myeloid-cell–specific Dnase-I–hypersensitive sites (HSs).30 Here, we show that all 3 sites are essential for full locus control activity. Thec-fes LCR is located within DNA sequences +28 to +2523. Thus, like many other genes containing LCRs (β-globin17,22 and rat liver–enriched activator protein31), the c-fes LCR colocalizes with tissue-specific HSs. To test the usefulness of the LCR to direct expression of a heterologous gene in the myeloid compartment, we developed transgenic mice that express the gene encoding the enhanced green fluorescent protein (EGFP) from the c-fes minimal cassette. Flow cytometry analysis of cells from bone marrow, spleen, and thymus show that the transgene is expressed in a myeloid-restricted manner.
Materials and methods
Generation of transgenic constructs and animals
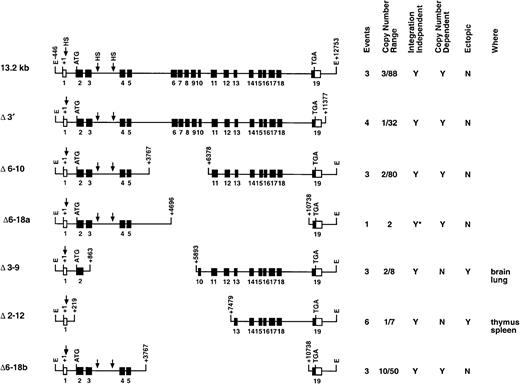
The 13.2-kb human c-fes locus cloned into theEcoRI site of plasmid pSVBR91 was kindly provided by Dr Anton Roebroek.32 Plasmid p13.2 (c-fessubcloned into the EcoRI site of pBluescript II KS) was used to generate all constructs depicted in Figure 2. We derived Δ3′ from p13.2 by double digestion with EcoRI andNsiI, producing an 11.8-kb transgene missing 1.4 kb of 3′ flanking sequences. Additional transgenic constructs were generated by using naturally occurring restriction enzyme sites. For example, Δ6-10 was created by digestion-eliminated genomic sequences between +3767 and +6370 (nucleotide assignments according to Roebroek et al32), deleting exons 6 to 10, introns 6 to 9, and parts of introns 5 and 10. The resulting 10.6-kb construct was isolated withEcoRI digestion. The 7.2-kb Δ6-18, 8.2-kb Δ3-9, and 5.9-kb Δ2-10 constructs were generated by means of the same strategy. The 6.2-kb Δ6-18a transgene was generated by digestion withNheI followed by Klenow-mediated blunting of 5′ ends, ligation to SphI linkers, and SphI digestion. A final ligation step joined the natural SphI site at position +3767 to a novel site at +10738. All constructs were subjected to thorough restriction analysis and DNA sequencing across ligated joints. Finally, each transgene was isolated from pBluescript byEcoRI digestion.
An alternative approach was used to develop a second series of transgenic constructs (Figure 5). All of these plasmids contain thec-fes promoter (nucleotides −446 to +71), the humanc-fes complementary DNA (cDNA) (kindly provided by Ricardo Feldman, University of Maryland, College Park), and human growth hormone (hGH) exons and polyadenylation signals (Nichols Institute, San Juan Capistrano, CA), totaling 5.1 kb. This basic construct is designated 0.5 kb as it contains approximately 0.5 kb of 5′ regulatory sequences. We also analyzed 3 other constructs containing genomic c-fes fragments placed behind thehGH gene. This strategy has worked for other LCR elements that function regardless of their orientation or proximity to the promoter.22 The largest construct (0.5 kb, HSabc) contains c-fes DNA sequences between +28 and +2523, inserted into the 0.5-kb construct via polymerase chain reaction with the primers 5′-TAAGCATGCGTCGGTCCGAGGCCGTCCCAG-3′ (forward) and 5′-TAAGAATTCGCCAGAGCTCGGTACTGGCTC-3′ (reverse): these primers introduce SphI and EcoRI sites and allow subcloning into the 0.5-kb plasmid. We sequenced 0.5-kb HSabc to determine that no mutations were introduced. The 0.5-kb Hsa includes sequences between nucleotides +447 and +863 introduced into 0.5 kb by digestion with RsrII and BglII and ligation to SphI and EcoRI linkers. Construct 0.5-kb HSbc contains c-fes nucleotides +1094 and +2519, generated with SacI digestion and subsequent linking.
Ribonuclease protection assays
We analyzed c-fes expression by ribonuclease protection assays (RPAs), which allowed a sensitive and quantitative comparison of endogenous murine and exogenous human transcripts.35 36 Hybridization analysis of mousec-fes mRNA levels controls for myeloid cell content in the various tissues sampled (brain, lung, thymus, spleen, and bone marrow). Levels of human and murine c-fes transcripts were quantitated by densitometry with a Phosphorimager (Molecular Dynamics, Sunnyvale, CA). All results are reported as humanc-fes/murine c-fes normalized to the expression level of 88 copies of the 13.2-kb human c-festransgene.
RNA was isolated from 6-week-old transgenic founder animals with the exception of the 13.2 and Δ3′ constructs, where 6-week-old progeny obtained by mating 13.2-kb or Δ3′ transgenics were used. RNA was extracted from bone marrow, spleen, brain, lung, and thymus by Trizol (Gibco-BRL, Rockville, MD) according to the manufacturer's instructions, and 10 μg of total RNA were used for detection of murine or human c-fes transcripts. RPAs for murine β-actin were performed with 5 μg of RNA. The human c-fes probe extends from genomic nucleotides +11273 to +11645 and protects a fragment of 273 or 180 nucleotides from exon 19.37 In the process of cloning thehGH gene behind a c-fes cDNA, a portion of exon 19 was deleted, leading to protection of a shorter, 180-nucleotide RNA fragment in the c-fes cDNA constructs. The murinec-fes probe extends from cDNA nucleotide 2223 to 2537 according to the number of Wilks and protects an exon-19 fragment of 288 bp.38 The 3′ ends of the c-fes genes were used because they are the most divergent between the human and mouse species. The murine β-actin probe protects a fragment of 250 bp. RNAs were analyzed by means of the Ambion (Austin, TX) RPA II kit with 500 000 disintegrations per minute of riboprobe, 5 μg/mL ribonuclease A (RNase A), and 100 U/mL RNase T1. Importantly, all assays were performed in probe excess and in the linear range of the assay, as determined by adding increasing amounts of RNA over a range of 2 to 20 μg.
Generation of the c-fes EGFP construct and transgenic animals
The 0.5-kb HSabc construct was digested completely withSpeI and partially with XbaI to remove thec-fes cDNA sequence and to insert a new polylinker. This polylinker re-established the SpeI site and disrupted theXbaI site and consists of recognition sites for the following unique restriction enzymes:SpeI-SalI-MluI-ClaI-NotI-XhoI. For simplicity, this construct is now called the c-fescassette. An NheI-XhoI fragment from pEGFP-C1 (Clontech) containing the EGFP gene was cloned into the SpeI/XhoI sites of the c-fescassette. The function of the c-fes EGFP construct was verified by transient transfection of the murine myeloid FDC-P1 cell line (from ATCC; Rockville, MD) by means of electroporation, essentially as described,39 followed by fluorescence microscopy after 24 hours. We identified 8 EGFP transgenic founders by dot blot analysis on purified tail DNA (Dneasy tissue kit, Qiagen, Valencia, CA) using an EGFP-specific probe. The transgenic status of the founders was subsequently verified by Southern analysis.
Flow cytometry analysis of c-fes EGFP mice
Cells from bone marrow, spleen, or thymus were hemolyzed (with the use of NH4Cl) to remove erythrocytes and washed twice in flow buffer (phosphate-buffered saline with 2% fetal calf serum and 2 mmol/L NaN3), and 1 million leukocytes were preincubated for 5 minutes on ice with Fc-Block (Becton Dickinson, San Diego, CA) to prevent nonspecific binding of antibody and were subsequently incubated for 30 to 45 minutes on ice with either the monoclonal antibodies Gr-1 (phycoerythrin [PE] conjugated), Mac-1 (PE or PE-Cy5), and B220 (PE-Cy5), or the matching isotype controls (Cedarlane Laboratories, Hornby, ON, Canada). The cells were washed twice in flow buffer and dissolved in flow buffer containing 1% formaldehyde to fix the cells prior to flow cytometry analysis. Initial screening for transgene-expressing lines was done by analyzing bone marrow cells for EGFP expression. The samples were analyzed on a Coulter XL flow cytometer (Beckman Coulter, Fullerton, CA) by means of the fluorescein isothiocyanate channel green fluorescence. We analyzed 50 000 counts from each sample. List-mode analysis was done with the use of Coulter software version 2.
Results
Generation of transgenic mice carrying the humanc-fes locus
A human 13.2-kb EcoRI restriction fragment contains all 18 c-fes coding exons, the first noncoding exon, 446-bp 5′ flanking sequences, and 1.5-kb 3′ flanking nucleotides (Figure1A). Greer et al1 determined that this relatively short 13.2-kb construct is likely to contain an LCR when introduced into transgenic animals. Their report analyzed transgene expression in a large number of murine tissues, including bone marrow, spleen, thymus, heart, lung, kidney, liver, brain, testes, and muscle. We generated 3 13.2-kb transgenic founder lines with 3, 23, and 88 copies each of the human c-fes genomic fragment and tested bone marrow RNA for human c-fes transcripts by RPAs. Transgene copy number for each founder animal was determined by DNA hybridization analysis of tail DNA samples and comparison with the endogenous murine gene. The human c-fes–specific riboprobe was prepared with the use of a 372-bp AflII-NaeI genomic fragment that includes most of exon 19; human transcripts protect a 273-nucleotide fragment derived from this probe (see “Materials and methods”). A 314-bp murine c-fes cDNA fragment yielding a 288-nucleotide protected band was used to detect endogenous mouse mRNA. As shown in Figure 1B, all 3 founders expressed human c-fes mRNA in the bone marrow. Consistent with previous observations,1 transgene expression correlated well with copy number. For example, compared with 2 copies of the murine c-fes locus, founder line 3 carrying 88 copies of the human transgene expressed 43 to 44 times as much human c-fesRNA. No human c-fes transcripts were detected in nontransgenic CD1 mice (Figure 3A).
Expression of the 13.2-kb transgene in murine tissues.
(A) Schematic representation of the human c-fes locus. All 19 exons are indicated along with coding regions (▪), noncoding regions (■), and myeloid-cell–specific DNase I HS sites (HSa, HSb, and HSc). The positions of translational initiation and termination codons are also shown. The 5′ and 3′ EcoRI restriction sites (E) are located 0.446 kb upstream of exon 1 and 1.5 kb downstream of exon 19, respectively. Multiple transcription initiation sites occur within the first exon; +1 corresponds to the first and most prominent mRNA cap site. (B) Production ofc-fes mRNA in bone marrow obtained from multiple transgenic mice generated with the 13.2-kb EcoRI human genomic fragment depicted in panel A. Two progeny animals obtained from founder no. 3 (88 copies) in addition to founders harboring 23 and 3 copies each of the 13.2-kb construct were analyzed. Ten micrograms of total RNA were hybridized to the 372-bp human c-fes and the 314-bp murinec-fes riboprobes. The 273-bp and 288-bp protected fragments are indicated. Included as negative and positive controls, respectively, were 25 μg yeast transfer-RNA and total RNA harvested from a mouse macrophage cell line (J774.1) and a human monoblastic cell line (THP-1). (C) Production of c-fes mRNA in various tissues harvested from the same transgenic mouse analyzed in lane 1 of panel B. To control for RNA loading, 5 μg RNA were hybridized to a murine β-actin probe.
Expression of the 13.2-kb transgene in murine tissues.
(A) Schematic representation of the human c-fes locus. All 19 exons are indicated along with coding regions (▪), noncoding regions (■), and myeloid-cell–specific DNase I HS sites (HSa, HSb, and HSc). The positions of translational initiation and termination codons are also shown. The 5′ and 3′ EcoRI restriction sites (E) are located 0.446 kb upstream of exon 1 and 1.5 kb downstream of exon 19, respectively. Multiple transcription initiation sites occur within the first exon; +1 corresponds to the first and most prominent mRNA cap site. (B) Production ofc-fes mRNA in bone marrow obtained from multiple transgenic mice generated with the 13.2-kb EcoRI human genomic fragment depicted in panel A. Two progeny animals obtained from founder no. 3 (88 copies) in addition to founders harboring 23 and 3 copies each of the 13.2-kb construct were analyzed. Ten micrograms of total RNA were hybridized to the 372-bp human c-fes and the 314-bp murinec-fes riboprobes. The 273-bp and 288-bp protected fragments are indicated. Included as negative and positive controls, respectively, were 25 μg yeast transfer-RNA and total RNA harvested from a mouse macrophage cell line (J774.1) and a human monoblastic cell line (THP-1). (C) Production of c-fes mRNA in various tissues harvested from the same transgenic mouse analyzed in lane 1 of panel B. To control for RNA loading, 5 μg RNA were hybridized to a murine β-actin probe.
Further analysis revealed that human c-fes mRNA was present in bone marrow, spleen, brain, lung, and thymus (Figure 1C). On the basis of numerous in situ hybridization and immunohistochemical analyses, these transcripts arise from infiltrating myeloid cells, such as alveolar macrophages in the lung and microglial cells in the central nervous system.1,7,9 RNase assays for murinec-fes transcripts allow careful quantitation of circulating myeloid cells within these tissues (Figure 1C). Densitometric scanning with a Phosphorimager revealed that human transcripts were proportional to murine c-fes mRNA (within a factor of 3) for all 3 founder lines. Therefore, the 13.2-kb construct appears to be expressed independently of integration site and dependent on copy number. These results are consistent with those of Greer et al1 and support their observation that an LCR active in myeloid cells resides within the human c-fes locus. Because c-fes is a proto-oncogene, careful histopathological assessment of all transgenic mice was performed; however, no tissue hyperplasia, neoplasia, or abnormal morphology was detected.
Genomic sequences downstream of exon 19 and between nucleotides +3767 and +10738 are not required for c-fesLCR activity
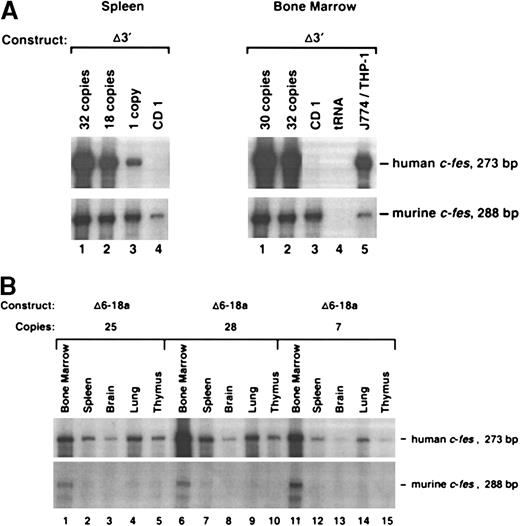
To determine if sequences 3′ to exon 19 includecis-acting elements that contribute to locus control function, we generated transgenic mice with the 3′ region deleted (Δ3′) (Figure 2). As shown in Figure3A, this construct still contains an intact LCR. We found that Δ3′ transgenic animals, harboring 1, 18, 30, and 32 copies of transgenic DNA, expressed human c-fesat levels comparable to the full-length 13.2-kb construct (Figures 2and 3A). Furthermore, based on RPAs of additional tissues (brain, lung, and thymus), the Δ3′ construct was expressed only in cells where the endogenous murine c-fes message was also detected (data not shown). We concluded that DNA 3′ to c-fes exon 19 is dispensable for LCR activity.
Deletion series of transgenic constructs.
This series analyzed various genomic segments of the c-fesgene for LCR activity. The columns at right summarize the data obtained for each construct. Events represents the number of founders generated and analyzed. Copy number range shows the lowest and highest number of transgene copies for each construct. A Y in the integration-independent column indicates that all of the founders expressed the transgene. A Y in the copy-number–dependent column indicates that all of the founders expressed the human gene at the expected levels in all tissues assayed. A Y in the ectopic column indicates that human c-fes mRNA detected was above the level expected (on the basis of copy number) in at least one of the tissues assayed. The last column lists the tissues where ectopic c-fes expression was observed. *Shows that a single founder was analyzed.
Deletion series of transgenic constructs.
This series analyzed various genomic segments of the c-fesgene for LCR activity. The columns at right summarize the data obtained for each construct. Events represents the number of founders generated and analyzed. Copy number range shows the lowest and highest number of transgene copies for each construct. A Y in the integration-independent column indicates that all of the founders expressed the transgene. A Y in the copy-number–dependent column indicates that all of the founders expressed the human gene at the expected levels in all tissues assayed. A Y in the ectopic column indicates that human c-fes mRNA detected was above the level expected (on the basis of copy number) in at least one of the tissues assayed. The last column lists the tissues where ectopic c-fes expression was observed. *Shows that a single founder was analyzed.
Expression of c-fes transgenes in murine tissues.
(A) Expression of c-fes in the spleen and bone marrow of a nontransgenic CD1 animal and 4 founders generated with the Δ3′ construct. Transgenics containing 1, 30, 32, and 18 copies of Δ3′ DNA were assayed in this experiment. (B) RNase protection analysis of human c-fes mRNA in transgenic tissues harvested from 3 founder mice generated with the Δ6-18a genomic construct (see Figure 2). As described in Figure 1, RNA samples were assayed with both the human- and murine-specific c-fes riboprobes. Transgene copy numbers were determined by Phosphorimager densitometric analysis of Southern blots prepared with transgenic tail DNAs.
Expression of c-fes transgenes in murine tissues.
(A) Expression of c-fes in the spleen and bone marrow of a nontransgenic CD1 animal and 4 founders generated with the Δ3′ construct. Transgenics containing 1, 30, 32, and 18 copies of Δ3′ DNA were assayed in this experiment. (B) RNase protection analysis of human c-fes mRNA in transgenic tissues harvested from 3 founder mice generated with the Δ6-18a genomic construct (see Figure 2). As described in Figure 1, RNA samples were assayed with both the human- and murine-specific c-fes riboprobes. Transgene copy numbers were determined by Phosphorimager densitometric analysis of Southern blots prepared with transgenic tail DNAs.
To rapidly localize domains within the 13.2-kb DNA element encompassing the c-fes LCR, we generated constructs with internal DNA sequences deleted by restriction-enzyme digestion of the full-length plasmid. Importantly, all constructs in this series maintain the integrity of exon 19 and could be assayed for expression by means of the probe descibed in Figure 1. Analyses of mice harboring 3 integration events of the Δ6-10 construct and one mouse containing the Δ6-18a construct (deleting exons 6 through 10 and 6 through 18, respectively) indicated that both transgenes still include locus control function (Figures 2 and 3B). Both transgenes produced high levels of human c-fes transcripts in the bone marrow, lung, and spleen. Importantly, each construct was also expressed in a copy-number–dependent manner proportional to murinec-fes mRNAs for all tissues analyzed (Figure 3B). In direct contrast, further deletions within 5′ sequences of the 13.2-kb construct resulted in a loss of locus control capability. As clearly shown in Figure 4A, the Δ3-9 construct (which deletes nucleotides +863 to +5893) and the Δ2-12 construct (deleting nucleotides +219 to +7479) exhibited humanc-fes mRNA production, suggesting that these sequences may be sufficient for integration-site–independent expression. However, human mRNA expression was not proportional to either transgene copy number or endogenous murine transcripts (Figures 2 and 4A). Importantly, both the Δ3-9 and Δ2-12 plasmids lack the myeloid-cell–specific DNase I HSs in intron 3; the Δ6-10 and Δ6-18 constructs include these sites. To test the importance of such sequences in intron 3 for locus control function, we generated the Δ6-18b plasmid shown in Figure 2. In 3 independent integration events, Δ6-18b was appropriately expressed in transgenic mice in a manner consistent with the presence of an LCR (shown in Figure 4B with the Δ3′ construct for comparison). Analysis of the Δ6-10, Δ6-18a, and Δ6-18b constructs demonstrated that genomic sequences between +3767 and +10738 of human c-fes are also not required for locus control activity. However, nucleotides between positions +219 and +3767 appear to be necessary for the copy-number–dependent characteristic of locus control activity.
RNase assays on tissues harvested from c-festransgenic mice.
(A) The Δ3-9 and Δ2-12 transgenic constructs are not expressed exclusively in myeloid cells. Ten micrograms of RNA from the indicated tissues were analyzed by RPAs. Southern blot analysis revealed that the Δ3-9 founder exhibited 2 copies of the transgene and the Δ2-12 transgenic mouse had 5 copies (data not shown). Various tissues obtained from both animals displayed ectopic expression: note the high levels of human c-fes in the spleen, brain, and lung of the Δ3-9 mouse and in the spleen, lung, and thymus in the Δ2-12 transgenic mouse. (B) The Δ6-18b construct exhibits LCR activity. Bone marrow RNAs from 3 founder animals with the indicated copy numbers of the Δ6-18b transgene are shown with Δ3′ founders for comparison. (C) The c-fes promoter linked to the human c-fescDNA (0.5-kb plasmid) is not sufficient for tissue-specific, copy-number–dependent transgene expression. The human c-fesriboprobe detects a 180-bp protected fragment in animals prepared with constructs based on the human c-fes cDNA because 93 bp fewer exon-19–encoded sequences are included in these plasmids (see “Materials and methods”). Note the inappropriately high levels of human c-fes transcripts apparent in the lung specimen.
RNase assays on tissues harvested from c-festransgenic mice.
(A) The Δ3-9 and Δ2-12 transgenic constructs are not expressed exclusively in myeloid cells. Ten micrograms of RNA from the indicated tissues were analyzed by RPAs. Southern blot analysis revealed that the Δ3-9 founder exhibited 2 copies of the transgene and the Δ2-12 transgenic mouse had 5 copies (data not shown). Various tissues obtained from both animals displayed ectopic expression: note the high levels of human c-fes in the spleen, brain, and lung of the Δ3-9 mouse and in the spleen, lung, and thymus in the Δ2-12 transgenic mouse. (B) The Δ6-18b construct exhibits LCR activity. Bone marrow RNAs from 3 founder animals with the indicated copy numbers of the Δ6-18b transgene are shown with Δ3′ founders for comparison. (C) The c-fes promoter linked to the human c-fescDNA (0.5-kb plasmid) is not sufficient for tissue-specific, copy-number–dependent transgene expression. The human c-fesriboprobe detects a 180-bp protected fragment in animals prepared with constructs based on the human c-fes cDNA because 93 bp fewer exon-19–encoded sequences are included in these plasmids (see “Materials and methods”). Note the inappropriately high levels of human c-fes transcripts apparent in the lung specimen.
The promoter is required but not sufficient for LCR function
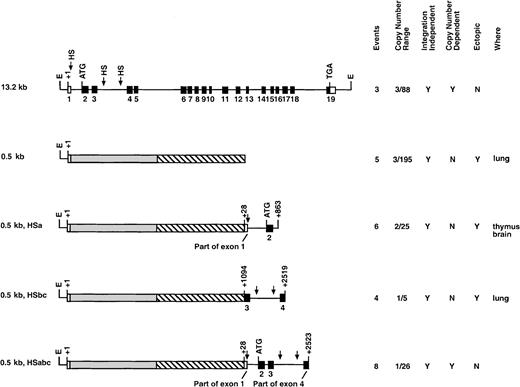
We have previously shown that the 446-bp c-fes 5′ flanking region contains a myeloid-cell–specific promoter element.28 To determine if the 446-bp promoter provided LCR activity, a construct was generated that includes c-fes5′ flanking sequences, a human c-fes cDNA, and 5 hGH exons to provide splicing and polyadenylation signals (0.5-kb, Figure 4C). 5 integration events were analyzed, and all transgenic animals demonstrated low levels of human c-fes mRNA and expression in ectopic locations (Figure 5). For example, the founder animal depicted in Figure 4C exhibited inappropriately high levels of human c-fes mRNA in the lung. Furthermore, regardless of copy numbers ranging from 3 to 195, each transgenic founder expressed equivalent amounts of humanc-fes (data not shown). Therefore, although the 446-bp c-fes promoter region is active when introduced into chromatin, it does not provide copy-number–dependent transcription in the appropriate cell types. These data are consistent with all 3 myeloid-cell–specific DNase I HSs being essential for locus control function.
Transgenic constructs analyzing genomic fragments in the human c-fes cDNA minilocus construct.
This series of transgenic mice analyzed various segments of thec-fes gene in conjunction with the c-fes cDNA regulated by the 446-bp c-fes promoter. ░ represents thec-fes cDNA, and ▧ represents hGH exons. The data columns are explained in Figure 2.
Transgenic constructs analyzing genomic fragments in the human c-fes cDNA minilocus construct.
This series of transgenic mice analyzed various segments of thec-fes gene in conjunction with the c-fes cDNA regulated by the 446-bp c-fes promoter. ░ represents thec-fes cDNA, and ▧ represents hGH exons. The data columns are explained in Figure 2.
Production of a c-fes minilocus that retains LCR activity
The production of a c-fes minilocus was accomplished by a combination of the 2 preceding transgenic strategies. To restore LCR activity to the c-fes 446-bp promoter/cDNA construct, we introduced sequences containing different HSs into plasmid 0.5 kb. Six transgenic founder animals containing the −446-bp 5′ region, the cDNA, and +28 bp to +863 bp of human c-fesstarting from exon 1 (0.5 kb HSa) were tested. However, as summarized in Figure 5, all 6 transgenics ranging in copy number from 2 to 25 exhibited ectopic expression in tissues such as the thymus and brain. Furthermore, transgene expression was not proportional to copy number. These results clearly indicate that the HSa site is not sufficient for LCR characteristics in transgenic mice. Therefore, the HS sites located within intron 3 appeared most likely to contribute to LCR function and were further analyzed. The 0.5-kb HSbc plasmid contains nucleotides +1094 to +2519 of the human locus, including small portions of exons 3 and 4 and all of intron 3. All 4 of the integration events expressed human c-fes in transgenic animals (Figure 5). However, transgene expression was neither copy number dependent nor tissue specific (data not shown). We concluded from these results that the HS sites in either intron 1 or intron 3 were not capable of conferring locus control function on their own.
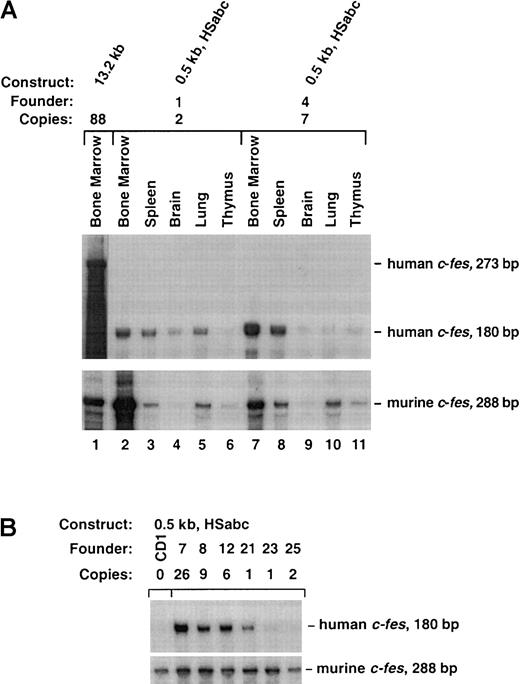
Finally, we tested the hypothesis that all 3 tissue-specific HS sites are necessary for full c-fes LCR activity. The 0.5-kb HSabc construct shown in Figure 5 harbors all 3 myeloid-specific HS sites. Eight transgenic founders ranging in copy number from 1 to 26 exhibited human c-fes expression that was copy number dependent, restricted to myeloid cells, and at levels similar to the murinec-fes locus (Figure6A). As shown in Figure 6B, splenic RNA samples from all 8 0.5-kb HSabc founders transcribed humanc-fes at levels consistent with their copy number within a factor of 4. Analysis of additional tissues (bone marrow, brain, lung, and thymus) confirmed that transgene expression was mostly in the appropriate cell types, on the basis of murine c-fesmRNA levels (Table 1). Three transgenic mice (numbers 1, 23, and 25) expressed only 10% of the expected levels of human c-fes RNA in the bone marrow (Table 1). However, all other tissues analyzed showed copy-number–dependent mRNA production. Comparison with murine c-fes mRNA levels demonstrated that the levels of human c-fes RNA are well within a factor of 3 of transgene copy numbers for all 8 of these founders (Table 1). In summary, the data from the transgenic strategy described above clearly support the notion that DNA localized around HS sites within introns 1 and 3, in conjunction with the promoter, contain the human c-fes LCR.
The 0.5-kb HSabc construct contains the locus control region.
(A) Ten micrograms of the indicated RNAs were separately analyzed by RPA. The tissues were collected from the bone marrow of an 88-copy-number 13.2-kb transgenic mouse (lane 1) and 2 founders containing the 0.5-kb HSabc construct (lanes 2-11). RNA from the 13.2-kb animal protected a human c-fes fragment of 273 bp, and the other RNAs protected a 180-bp fragment. Autoradiography was performed for various times to ease comparison between RPAs of mouse and human transcripts. (B) RPAs performed on splenic RNA samples derived from multiple transgenic mice. Total RNA harvested from the indicated founder strains was analyzed by means of the previously described riboprobes for human and murine c-fes transcripts. The designated number for each transgenic line is indicated so that these assays can be compared with the data presented in Table1.
The 0.5-kb HSabc construct contains the locus control region.
(A) Ten micrograms of the indicated RNAs were separately analyzed by RPA. The tissues were collected from the bone marrow of an 88-copy-number 13.2-kb transgenic mouse (lane 1) and 2 founders containing the 0.5-kb HSabc construct (lanes 2-11). RNA from the 13.2-kb animal protected a human c-fes fragment of 273 bp, and the other RNAs protected a 180-bp fragment. Autoradiography was performed for various times to ease comparison between RPAs of mouse and human transcripts. (B) RPAs performed on splenic RNA samples derived from multiple transgenic mice. Total RNA harvested from the indicated founder strains was analyzed by means of the previously described riboprobes for human and murine c-fes transcripts. The designated number for each transgenic line is indicated so that these assays can be compared with the data presented in Table1.
Percentage of expected human c-fes expression in various transgenic mouse tissues
| Construct . | Animal . | Copy no. . | Bone marrow . | Spleen . | Brain . | Lung . | Thymus . |
|---|---|---|---|---|---|---|---|
| 13.2 kb | 3 | 88 | 92.6 | 147.7 | 472.4 | 163.9 | 257.7 |
| Δ6-18b | 9 | 25 | 30.5 | 35 | 47.7 | 106.5 | 52.7 |
| 12 | 28 | 114.6 | 53.1 | 61.1 | 62.2 | 60.2 | |
| 13 | 7 | 106.2 | 141.5 | 240.2 | 165.5 | 138.7 | |
| 0.5-kb HSabc | 1 | 2 | 8.1 | 264.1 | 366.9 | 85 | 25.9 |
| 4 | 7 | 37.3 | 55.5 | 137 | 76.2 | 28.8 | |
| 7 | 40 | 15.8 | 25.2 | 89.3 | 44 | 250.6 | |
| 8 | 13 | — | 66.9 | 271.8 | 65 | 32 | |
| 10 | 5 | — | 13.3 | 313 | 19.8 | 21.6 | |
| 12 | 9 | 51.4 | 100 | 106.7 | 62 | — | |
| 21 | 1 | 56.2 | 96.6 | 198.8 | 377 | — | |
| 23 | 1 | 9.9 | 42.7 | 798.2 | 88.9 | 130.1 | |
| 25 | 2 | 11.4 | 12.4 | 156.6 | 12.9 | — |
| Construct . | Animal . | Copy no. . | Bone marrow . | Spleen . | Brain . | Lung . | Thymus . |
|---|---|---|---|---|---|---|---|
| 13.2 kb | 3 | 88 | 92.6 | 147.7 | 472.4 | 163.9 | 257.7 |
| Δ6-18b | 9 | 25 | 30.5 | 35 | 47.7 | 106.5 | 52.7 |
| 12 | 28 | 114.6 | 53.1 | 61.1 | 62.2 | 60.2 | |
| 13 | 7 | 106.2 | 141.5 | 240.2 | 165.5 | 138.7 | |
| 0.5-kb HSabc | 1 | 2 | 8.1 | 264.1 | 366.9 | 85 | 25.9 |
| 4 | 7 | 37.3 | 55.5 | 137 | 76.2 | 28.8 | |
| 7 | 40 | 15.8 | 25.2 | 89.3 | 44 | 250.6 | |
| 8 | 13 | — | 66.9 | 271.8 | 65 | 32 | |
| 10 | 5 | — | 13.3 | 313 | 19.8 | 21.6 | |
| 12 | 9 | 51.4 | 100 | 106.7 | 62 | — | |
| 21 | 1 | 56.2 | 96.6 | 198.8 | 377 | — | |
| 23 | 1 | 9.9 | 42.7 | 798.2 | 88.9 | 130.1 | |
| 25 | 2 | 11.4 | 12.4 | 156.6 | 12.9 | — |
Transgene copy number was quantified by Southern blot hybridization performed in triplicate. Murine c-fes RNA levels were given the arbitrary value 2, corresponding to the expression of both endogenous genes. Transgenic human c-fesRNA expression was compared with murine c-fes RNA levels and corrected for differences in gene copy number. For example, transgenic no. 3 (88 copies) would be expected to express 44 times as much humanc-fes RNA as murine c-fes RNA and listed as 100% of the expected value (2 copies). Thus, the value of humanc-fes expression is the volume of the human 273–base pair (bp) protected fragment (minus background) divided by the volume of the murine 288-bp protected fragment (minus background) and compared with the expected abundance on the basis of the copy number of each transgene. The value of human c-fes expression for all splenic samples is based on a single RPA autoradiogram for direct comparison.
HS indicates hypersensitive site; — indicates data were not determined.
Myeloid-specific expression of a heterologous gene (encoding the EGFP) in transgenic mice by means of the minimalc-fes expression cassette
To assess the usefulness of the minimal c-fesexpression cassette to drive expression of a heterologous gene in the myeloid compartment in transgenic mice, we removed the c-fescDNA from the 0.5-kb HSabc construct, introduced a new polylinker, and inserted the gene encoding the EGFP. The function of this construct was evaluated by transient transfection of the bone-marrow–derived myeloid cell line FDC-P1 and subsequent fluorescence microscopy and flow cytometry. Transfected FDC-P1 cells expressed high levels of green fluorescent protein (data not shown). After microinjection of the EGFP construct, we obtained 8 transgenic founders. F1 animals from all 8 lines were analyzed for EGFP expression by flow cytometry. Surprisingly, EGFP expression was detected in only 3 of the transgenic lines. Apparently, the integration-site–independent expression observed with the 0.5-kb HSabc construct was lost when a heterologous gene was substituted for the c-fes cDNA, which may indicate that the LCR acts in conjunction with elements in the cDNA sequence. The apparent loss of integration-site independence is also observed when other genes are expressed in mice by means of this cassette (as evaluated by RNase protection; data not shown).
To determine the cell-type specificity of EGFP transgene expression, we analyzed cells from bone marrow, spleen, and thymus by flow cytometry. The cells were incubated with myeloid (Gr-1 and Mac-1) or B-lymphoid (B220) lineage markers prior to analysis, and expression of these markers was evaluated in combination with EGFP expression. Representative data from the transgenic line with highest EGFP expression are depicted in Figure 7. Expression of EGFP is highest in bone marrow (Figure 7A), lower but significant in spleen (Figure 7B), and essentially absent in thymus (Figure 7C). B220+ cells did not express EGFP, indicating that the transgene is not expressed in the B-cell lineage. In constrast, 50% of Gr-1+ (granulocytes) or Mac-1+ (granulocytes, monocytes, macrophages) bone marrow cells were EGFP+. Similarly, 30% to 50% of splenic myeloid cells expressed the transgene. These percentages of Gr-1+ and Mac-1+ bone marrow cells may represent the total number of cells actually expressing endogenousc-fes. Thioglycollate mobilization of peritoneal cells also revealed EGFP expression in macrophages (not shown). The absence or very low level of EGFP expression in the thymus demonstrates that thec-fes expression cassette is not active in T cells. A myeloid restricted-expression pattern was also observed in 2 additional transgenic c-fes EGFP lines, demonstrating the consistency of this minimal c-fes expression cassette (data not shown).
The c-fes expression cassette directs myeloid specific expression of a heterologous gene in hematopoietic tissues.
Representative data from a flow cytometry analysis of hematopoietic cells from a transgenic mouse expressing a c-fes EGFP construct (tg) and a nontransgenic littermate (neg). (A) Bone marrow. Of the cells, 33% express the transgene. Approximately 50% of the Gr-1+ cells coexpress EGFP. The same is true for the Mac-1+. The B-cells (B220+) are EGFP−. (B) Spleen. Approximately 5% of the cells express the transgene, and expression is observed only in Gr-1+ or in Mac-1+ cells. (C) Thymus. Expression of EGFP is absent or very low, indicating that the c-fes expression cassette is inactive in T cells. In the double histograms, the numbers indicate percentages of cells present in the given quadrant. Cell-density scaling parameters are identical for all double histograms from the same tissue, but differ slightly between bone marrow and spleen. There were 50 000 counts from each sample analyzed.
The c-fes expression cassette directs myeloid specific expression of a heterologous gene in hematopoietic tissues.
Representative data from a flow cytometry analysis of hematopoietic cells from a transgenic mouse expressing a c-fes EGFP construct (tg) and a nontransgenic littermate (neg). (A) Bone marrow. Of the cells, 33% express the transgene. Approximately 50% of the Gr-1+ cells coexpress EGFP. The same is true for the Mac-1+. The B-cells (B220+) are EGFP−. (B) Spleen. Approximately 5% of the cells express the transgene, and expression is observed only in Gr-1+ or in Mac-1+ cells. (C) Thymus. Expression of EGFP is absent or very low, indicating that the c-fes expression cassette is inactive in T cells. In the double histograms, the numbers indicate percentages of cells present in the given quadrant. Cell-density scaling parameters are identical for all double histograms from the same tissue, but differ slightly between bone marrow and spleen. There were 50 000 counts from each sample analyzed.
Discussion
LCRs confer integration-site–independent, copy-number–dependent expression at high levels on transgenes. Previous experiments strongly suggested that the 13.2-kb human c-fes gene included such a dominant, myeloid-specific LCR element. Therefore, the relatively shortc-fes genomic locus must include all necessarycis-acting DNA elements for high levels of myeloid-cell–specific expression. Surprisingly, this 13.2-kb genomic fragment contains only 446 bp of 5′ and 1.4 kb of 3′ flanking sequences, respectively. To locate where cis-acting sequences reside within the 13.2-kb DNA element, we have studied the expression of various c-fes constructs in a large number of transgenic mice. Two series of transgenic constructs convincingly demonstrate that the c-fes LCR lies within introns 1 and 3. These sequences direct integration-site–independent and copy-number–dependent expression in transgenic mice in conjunction with the myeloid-specific c-fes promoter. The deletion series diagrammed in Figure 2 established that sequences between +3767 and +10738 and downstream of +11377 are unnecessary for LCR activity. Furthermore, the 0.5-kb HSabc minilocus construct confirms that DNA sequences between +28 and +2523 in conjunction with the 446-bp promoter are sufficient for LCR regulation of thec-fes cDNA. All minilocus integration events express the transgene, demonstrating integration-site–independent expression. Furthermore, ribonuclease protection analyses of splenic RNA obtained from the 8 animals containing this minilocus support copy-number–dependent expression in a tissue-specific manner. RNase protection analyses revealed that expression of the 0.5-kb HSabc construct in the spleen is comparable to bone marrow expression. When used to express a heterologous gene (EGFP), this minimal construct directs myeloid-specific expression in hematopoietic tissues in transgenic mice.
LCR characteristics are probably mediated by higher-order chromatin structure.17,22,40-42 The colocalization of DNase I HSs to LCR elements in a number of genes, such as β-globin,17CD2,22 and LAP,31 supports this hypothesis. Higher-order chromatin structures include nuclear-matrix–associated regions within the LCR sequences,43-45 suggesting that the LCR regulates chromatin structure by forming a chromosomal loop.46,47 The 13.2-kb human c-fesgenomic DNA contains 3 tissue-specific DNase I HSs,30 2 of which reside within intron 3. All 3 HSs are present within the 0.5-kb HSabc minilocus, which maintains locus control activity.
Similar expression cassettes have previously been used for analysis of the PML-RARα oncoprotein in acute promyelocytic leukemia. PML-RARα expression regulated by a constitutive, housekeeping promoter such as β-actin results in the death of transgenic fetuses.48 Therefore, a myeloid-cell–specific, human cathepsin G cassette (based on the experiments of Grisolano et al49) was designed to control PML-RARα production in transgenic animals.48,50 The CD11b promoter also directs myeloid-specific gene transcription in transgenic mice.51 52 However, both the cathepsin G and CD11b cassettes fail to yield high levels of transgene expression. These examples illustrate the need for a myeloid-cell–specific expression cassette that provides high levels of expression in macrophages and neutrophils. We believe that the c-fes construct provides such a reagent.
In contrast to the 0.5-kb HSabc construct where integration-site–independent expression was observed, only 3 out of 8 established lines expressed EGFP, as evaluated by flow cytometry. One likely explanation is that sequences in the c-fes cDNA are important for the integration-site–independent expression. Another explanation for the lack of expression for some EGFP lines is that F1 animals and not transgenic founders were analyzed. Perhaps some EGFP transgenes were methylated upon germ-line transmission. Finally, RPAs may be more sensitive than flow cytometry. The myeloid- restricted expression pattern of the cassette is, however, maintained after substitution of the c-fes cDNA, and we still observe a copy-number dependency for expression levels (data not shown). The usefulness of the c-fes cassette will be further enhanced by precisely defining the pattern of expression during embryonic development, together with an exact description of the expressing cell types in the adult mouse. We are currently investigating these issues.
The identification of the c-fes minimal expression cassette also permits myeloid-cell–specific knockouts using the cre/lox system (reviewed in Marth53). An obvious use for such a strategy is to overcome embryonic lethality of particular mutants. Even if a null allele is not lethal, it may be desirable to investigate the cell-specific deletion of certain proteins in an otherwise normal cellular environment. Cell-intrinsic vs cell-extrinsic questions can be addressed in such a cell-type–specific chimeric animal. Thec-fes cassette can be used in such a deletion strategy to study the role of many genes: for example, the C/EBPα transcription factor. C/EBPα is expressed in adipose, hepatic, and myeloid cells.54,55 C/EBPα null mice die shortly after birth owing to faulty glucose metabolism.56,57 These mice also display a significant defect in granulocyte numbers,58liver architecture, and lung development.57 C/EBPα regulates critical myeloid genes, such as those encoding GM-CSF receptor α59 and granulocyte colony-stimulating factor receptor.60 A more complete investigation of macrophage and neutrophil functional defects is not possible owing to an inability to produce viable mice. A c-fes/cre–mediated myeloid-cell–specific knockout of the C/EBPα factor would greatly assist in analyzing the granulocyte defects in the absence of this factor.
In summary, using a significant number54 of transgenic mice, we have successfully located the myeloid-specific LCR within the human c-fes locus and determined that it resides in intron 1 and intron 3. This DNA element, encompassing approximately 2.5 kb ofc-fes genomic sequences, is necessary and sufficient for conferring integration-site–independent, copy-number–dependent expression on a cDNA construct in conjunction with the c-fespromoter. We have used this minimal expression cassette to drive transcription of the gene encoding EGFP in a myeloid-specific manner. Apparently, integration-site–independent expression is lost when thec-fes cDNA is substituted for a heterologous gene. However, the cassette is still capable of directing copy-number–dependent and myeloid-specific expression in transgenic mice. It thus provides a unique tool for expression of nonmyeloid genes or oncogenes in the monocytic and neutrophilic hematopoietic lineages. Furthermore, the human c-fes construct will be highly useful for conducting tissue-specific, myeloid-lineage gene targeting experiments.
Supported by National Institutes of Health grant R01 HL52094; Howard Hughes Medical Institute; The Danish Medical Research Council; and The Karen Elise Jensen Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
M. Celeste Simon, Howard Hughes Medical Institute, Abramson Cancer Research Institute, University of Pennsylvania Cancer Center, Biomedical Research Bldg II/III, Rm 456, 421 Curie Blvd, Philadelphia, PA 19104; e-mail: celeste2@mail.med.upenn.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal