Abstract
Congenital afibrinogenemia is a rare autosomal recessive disorder characterized by a hemorrhagic diathesis of variable severity. Although more than 100 families with this disorder have been described, genetic defects have been characterized in few cases. An investigation of a young propositus, offspring of a consanguineous marriage, with undetectable levels of functional and quantitative fibrinogen, was conducted. Sequence analysis of the fibrinogen genes showed a homozygous G-to-A mutation at the fifth nucleotide (nt 2395) of the third intervening sequence (IVS) of the γ-chain gene. Her first-degree relatives, who had approximately half the normal fibrinogen values and showed concordance between functional and immunologic levels, were heterozygtes. The G-to-A change predicts the disappearance of a donor splice site. After transfection with a construct, containing either the wild-type or the mutated sequence, cells with the mutant construct showed an aberrant messenger RNA (mRNA), consistent with skipping of exon 3, but not the expected mRNA. Sequencing of the abnormal mRNA showed the complete absence of exon 3. Skipping of exon 3 predicts the deletion of amino acid sequence from residue 16 to residue 75 and shifting of reading frame at amino acid 76 with a premature stop codon within exon 4 at position 77. Thus, the truncated γ-chain gene product would not interact with other chains to form the mature fibrinogen molecule. The current findings show that mutations within highly conserved IVS regions of fibrinogen genes could affect the efficiency of normal splicing, giving rise to congenital afibrinogenemia.
Introduction
Congenital afibrinogenemia is a rare autosomal recessive disorder characterized by the complete absence of detectable fibrinogen and an infinite prolongation of functional assays of clot formation. Patients with congenital afibrinogenemia have a lifelong hemorrhagic diathesis of variable severity, although abnormal bleedings do not occur more frequently than in hemophilias A and B.1Since the first case reported in 1920,2 approximately 150 cases have been described.3
Fibrinogen is a complex dimeric protein of 340 kd, composed of 3 pairs of nonidentical polypeptides designated as Aα-, Bβ-, and γ-chains.4 Human fibrinogen chains Aα, Bβ, and γ contain 610, 461, and 411 amino acid residues, respectively,4 and are interconnected by 29 disulphide bridges.5 The covalent structure of the human protein was first elucidated by studies on protein sequence,6 and then confirmed and extended by DNA sequence analysis.7 The DNA sequences encoding the human fibrinogen chains occur as closely linked single-copy genes lying within a 50-kilobase (kb) segment of DNA in the distal third of the long arm of chromosome 4, region q28.8,9 The Bβ gene is in opposite transcriptional orientation with respect to the other 2 genes.10 The tight clustering of the 3 genes encoding this protein, the amino acid and nucleotide sequence homologies among these genes, and certain similarities of exon-intron domains at the 5′ ends suggest that these genes have arisen by duplication and divergence from an ancestral entity.10 Each gene is separately transcribed and translated in the liver, and the nascent chains are discharged into the endoplasmic reticulum,11 where chain assembly into dimeric fibrinogen occurs in a stepwise fashion.12 The control of fibrinogen gene expression seems to depend to a large extent on the interaction of certain factors with specific nucleotide sequences predominantly located in the 5′-flanking region of the genes.13
In congenital afibrinogenemia a failure of synthesis, secretion, or intracellular transport has been supposed. However, to date, only few defects in the fibrinogen genes have been identified: an 11-kb deletion of the fibrinogen Aα-chain gene14 and 2 missense mutations in the fibrinogen Bβ-chain (Leu353Arg and Gly400Asp), leading to impaired fibrinogen secretion.15
We now present the molecular basis of a novel form of congenital afibrinogenemia in a young female patient. Genetic characterization demonstrated that the patient was homozygous for a G-to-A transition at the fifth nucleotide (nt 2395) of the third intervening sequence (IVS) of the γ-chain gene. This mutation altered the pattern of RNA processing.
Materials and methods
Informed consent was obtained from the patient and other members of the family, after approval of the local Human Ethics Committee. The studies were carried out according to the Principles of the Declaration of Helsinki.
Materials
Reagents were of an analytic grade or the best available commercial grade.
Fibrinogen measurement
Functional plasma fibrinogen was assayed by the Clauss clotting method. Both the reagent and apparatus (CoA Data, 2000) were from Boehringer-Mannheim, Milan, Italy. Plasma fibrinogen was quantitatively evaluated using the NOR Partigen Fibrinogen kit (Dade Behring, Marburg, Germany).
Fibrinogen analysis
Plasma was prepared from citrate anticoagulated whole blood and fibrinogen purified from 800 μL of plasma by precipitation with 225 μL of saturated ammonium sulfate. The pellet was washed 3 times with 25% saturated ammonium sulfate, and the fibrinogen was dissolved in 200 μL of an 8 mol/L urea solution, 0.1 mol/L Tris-HCl pH 8.0, 20 mmol/L dithiothreitol, and reduced for 4 hours at 37°C. Five microliters of sample buffer were added to 10 μL of reduced fibrinogen and 5 μL of this mix were loaded onto a 12% w/v polyacrylamide gel (PhastGel, Pharmacia, Uppsala, Sweden) and allowed to run at 30 mA for 6 hours (PhastSystem, Pharmacia). Then, the bands were silver stained.
DNA analysis
Isolation of DNA and polymerase chain reaction (PCR) analysis were performed according to standard procedures.16 For DNA extraction, peripheral blood leukocytes were separated by sedimentation and incubated overnight at 37°C in digestion buffer (100 mmol/L NaCl, 10 mmol/L Tris-HCl, 25 mmol/L EDTA, 1% w/v SDS) containing 0.1 mg/mL of proteinase K. The nucleic acid was isolated by phenol/chloroform extraction and ethanol precipitation. Amplifications of all coding regions of fibrinogen chain genes and intron/exon boundaries were achieved using sense and antisense oligonucleotides designed on the basis of known sequences of fibrinogen gene loci (GenBank accession numbers M64982, M64983, and M10014). Oligonucleotide custom synthesis service was from Life Technologies (Paisley, UK). PCR was carried out on 50 μL volume samples, in a Perkin Elmer-Cetus thermal cycler (Perkin-Elmer Cetus, Norwalk, CT). Each sample contained 0.1 μg of genomic DNA, 10 pmoles of each primer, 125 μmol/L of dNTP, 5 mmol/L Tris HCl pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 1 unit Taq polymerase. The solution was overlaid with 50 μL of mineral oil and, after an initial denaturation step (3 minutes at 95°C), it was put through 30 cycles each consisting of 1 minute at 95°C, 1 minute at 56°C to 60°C and 2 minutes at 72°C. Thereafter, 5 μL volumes of the amplification products were separated in a 2% w/v agarose-gel electrophoresis in TAE buffer (40 mmol/L Tris-Acetate,1 mmol/L EDTA pH 7.7), containing 0.5 μg/m ethidium bromide, and visualized under ultraviolet (UV) light. SSCP analyses were performed as described.17 Briefly, 3 μL of each amplification were added to 5 μL of formamide, containing loading dye, heated at 95°C for 5 minutes, chilled on ice, and immediately loaded onto a 20 cm × 60 cm × 0.4 mm 20% w/v acrylamide/TBE gel, with a acrylamide/bisacrylamide ratio of 29:1, without any other additive. The gel was run at 20 W for 16 hours at room temperature and single-stranded DNA bands were detected by using a silver staining method. Any difference was observed when the run was performed at 4°C or at 15°C. Amplified DNA fragments showing abnormal SSCP patterns were purified and subjected to direct cycle sequence analysis using the Taq dye-deoxy terminator method and an ABI PRISM 310 Genetic Analyzer sequencer (PE Biosystems, Foster City, CA) according to the manufacturer's instructions.
RNA analysis
A 766-base pair (bp) DNA fragment, spanning from 92-bp upstream exon 2 to 43-bp downstream exon 4, was amplified. Primer sequences were (sense: 1821-1832) 5′-GTGTGCTCTTCACAAAACGTTG-3′ and (antisense: 2646-2625) 5′-ACTAAATCAGTCTT GCAGAGCA-3′. PCR was carried out on 50 μL volume samples, in a 480 Perkin Elmer-Cetus termal cycler (Perkin-Elmer Cetus, Norwalk, CT). Each sample contained 0.2 μg of genomic DNA, 25 pmoles of each primer, 200 μmol/L of dNTP, 5 mmol/L Tris HCl pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 1 units Taq polymerase. The solution was overlaid with 50 μL of mineral oil and, after an initial denaturation step (3 minutes at 95°C), it was put through 35 cycles each consisting of 1 minute at 94°C, 1.5 minutes at 60°C, and 1.5 minutes at 72°C. A final step at 72°C for 10 minutes to ensure a 3′ adenylated PCR product was added. Before cloning, PCR product was purified from a 1.8% agarose gel by Concert Rapid Gel Extraction System (Life Technologies) according to manufacturer's instructions. The purified PCR product was cloned using Eucariotic TOPO TA Cloning Kit Version C (Invitrogen, Groningen, NL) according to manufacturer‘s instructions. Plasmid DNA was purified by QIAprep Spin Miniprep Kit (Quiagen, Valencia, CA) according to manufacturer’s instructions. Positive clones were identified by digesting plasmid DNA from 10 random colonies withBstXI, according to pcDNA3.1/V5/His-TOPO restriction map (Invitrogen). Briefly, 5 μL of purified plasmid DNA was digested with 1 μL of BstXI (Promega, Madison, WI) in 20 μL of reaction volume. Positive clones were sequenced in an ABI PRISM 310 Genetic Analyzer (PE Biosystems) to identify clones showing the correct 5′→3′ orientation. Clones that showed the correct 5′→3′ orientation were transfected in HEK 293 cells. Briefly, human kidney HEK293 cells were grown in 10% fetal bovine serum (FBS)/MEM-alfa medium (Life Technologies). One day before the transfection, cells were seed in 6-well plates (400 000 cells per well). The transfection was performed by calcium phosphate method as previously described.18 Total RNA was purified by TRIzol Reagents (Life Technologies) according to the manufacturer's instructions. Reverse transcriptase was made by Reverse Transcription System (Promega). Briefly, 2 μl of total RNA was treated with 1 unit of RQ1 DNase (Promega) with 40 units of RNasin (Promega) in a total volume of 5 μL. A 2.5 μL aliquot was transcribed at 42°C for 30 minutes. Before the transcription, the RNA aliquot was keep at 60°C for 15 minutes with 25 pmoles of the reverse primer to permit annealing. A 5 μL aliquot of complementary DNA (cDNA) was amplified by PCR as previously described and 5 μL volumes of the amplification products were separated in a 1.8% agarose-gel electrophoresis in TAE buffer (40 mmol/L Tris-Acetate,1 mmol/L EDTA pH 7.7), containing 0.5 μg/mL ethidium bromide, and visualized under UV light. The abnormal messenger RNA (mRNA) splicing product was purified and subjected to direct cycle sequence analysis using the Taq dye-deoxy terminator method and an ABI PRISM 310 Genetic Analyzer sequencer (PE Biosystems) according to the manufacturer's instructions.
Results
Case presentation
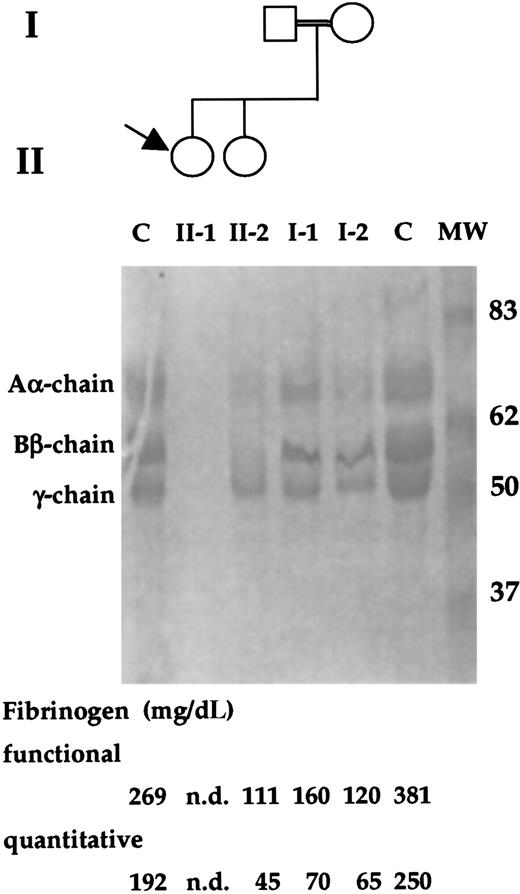
The patient was a 6-year-old white girl whose parents were first cousins (Figure 1). When she was 1 year old, a diagnosis of afibrinogenemia was made because of a posttraumatic and life-threatening bleeding that required hospitalization and blood tranfusions. After that, she had mild bleeding, experiencing posttraumatic muscle hematomas. A very prolonged aPTT and PT, and undetectable fibrinogen levels were observed. As to her family history, no bleeding episode was reported. In the propositus, undetectable levels of both functional (normal range: 160-400 mg/dL) and quantitative (normal range: 160-400 mg/dL) fibrinogen were observed. Both parents of the propositus, as well as her sister, had low levels of functional and quantitative fibrinogen (Figure 1). In addition, single-dimension SDS-PAGE electrophoresis revealed the complete absence of fibrinogen chains in the proband and reduced amounts in family members (Figure 1).
Family study of the index patient and relatives.
The propositus is indicated by an arrow. The double line indicates a consanguineous marriage. PAGE analysis of the reduced fibrinogen of family members and 2 control (C) subjects are shown. The molecular weights (kd) of the size marker (MW) are reported. In the lower part, functional and immunologic fibrinogen values of family members and control subjects are shown. nd, not detected.
Family study of the index patient and relatives.
The propositus is indicated by an arrow. The double line indicates a consanguineous marriage. PAGE analysis of the reduced fibrinogen of family members and 2 control (C) subjects are shown. The molecular weights (kd) of the size marker (MW) are reported. In the lower part, functional and immunologic fibrinogen values of family members and control subjects are shown. nd, not detected.
Genetic characterization
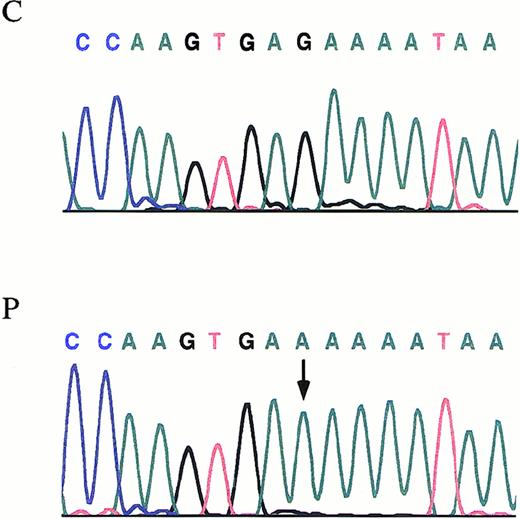
Fragments covering the entire coding region of Aα-, Bβ-, and γ-chain genes were amplified from patient's and her relatives' genomic DNA. Then, PCR products were subjected to SSCP analysis. All the amplified segments from Aα- and Bβ-chain genes in the kindred proved to be identical to those obtained in the controls. As for the γ-chain gene, all amplified segments were identical to those obtained in the controls, except for the segment spanning the exon 3 and intron/exon boundary regions. Direct DNA sequence analysis of the PCR product from the patient showed a homozygous G-to-A transversion at the fifth nucleotide (nt 2395) after the termination of the exon 3 (Figure2). The same mutation was found, in a heterozygous form in samples from both parents and sister. The G-to-A transition is within the consensus sequence of the donor splice site, a region believed to be critical for accurate mRNA splicing. In 72 healthy subjects (144 chromosomes), screening for the G-to-A transition failed to identify this molecular change. Using the Neural Network Promoter Prediction Tool program (http://www.fruitfly.org/seq_tools/splice.html)19 to predict changes induced by mutations in mRNA splicing, we found that the mutation causes the disappearance of a donor splice site. In the patient, further sequencing of all coding regions of fibrinogen chain genes and intron/exon boundaries did not detect any additional mutation.
The γ-chain fibrinogen IVS-3 nt 2395.
Electropherograms showing the mutation identified in the propositus with afibrinogenemia. An arrow indicates the G-to-A transition. P, propositus. C, control.
The γ-chain fibrinogen IVS-3 nt 2395.
Electropherograms showing the mutation identified in the propositus with afibrinogenemia. An arrow indicates the G-to-A transition. P, propositus. C, control.
In vitro messenger RNA splicing
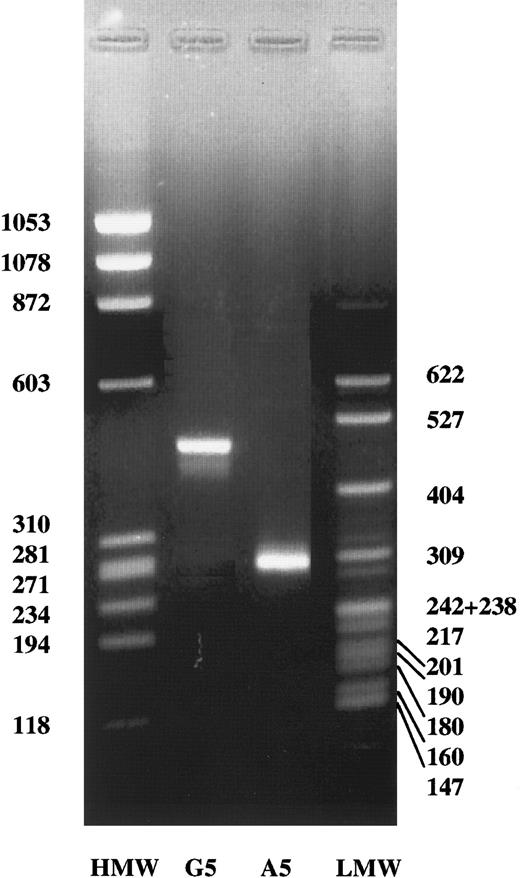
To determine whether the G2395A transvertion affects the processing of the γ-chain gene primary transcript, we transfected HEK 293 cells with both normal and mutant constructs, 766-bp spanning from 92- bp upstream exon 2 to 43-bp downstream exon 4 (Figure3A). The results of this analysis are shown in Figure 4. Under these conditions, HEK 293 cells containing the normal γ-chain construct displayed a band of approximately 450 bp, which corresponded fairly well to the expected splicing product of 458 bp (Figure 3B). In contrast, HEK 293 cells transfected with the mutant construct showed a band, shorter than that observed in cells transfected with the normal construct, of approximately 280 bp (Figure 4). This result suggested that the splicing product observed was consistent with an aberrant mRNA, resulting from exon 3 skipping, and expected to be 274 bp (Figure3C). The skipping of exon 3 predicts the deletion of amino acid sequence from residues 16 to 75 and the shift of the reading frame at amino acid 76 with a premature stop codon within exon 4 at position 77. To confirm that the G-to-A transition at the γ-chain, 2395 nucleotide causes an abnormal mRNA processing, resulting in skipping of the exon 3, the abnormal band observed in HEK 293 cells transfected with the mutant construct was isolated and then sequenced. The complete absence of exon 3 was observed (Figure 5), thus confirming that the A2395 construct hardly affects mRNA processing.
Experimental design.
(A) Schematic representation of the construct transfected in human kidney HEK 293 cells. The position of primers is reported. (B) Predicted splicing (top) and mRNA product (bottom) in cells transfected with the normal construct. (C) Predicted splicing (top) and mRNA product (bottom) in cells transfected with the construct containing the A2395 mutation. Dashed boxes refer to exons. Closed loops indicate spliced RNA sequences.
Experimental design.
(A) Schematic representation of the construct transfected in human kidney HEK 293 cells. The position of primers is reported. (B) Predicted splicing (top) and mRNA product (bottom) in cells transfected with the normal construct. (C) Predicted splicing (top) and mRNA product (bottom) in cells transfected with the construct containing the A2395 mutation. Dashed boxes refer to exons. Closed loops indicate spliced RNA sequences.
Detection of mRNA splicing products.
In vitro splicing of HEK 293 cells transfected with the normal construct (G2395) or with the construct containing the mutation (A2395). HMW, ΦX174 HaeIII digest DNA molecular weight markers. LMW, pBR322 MspI digest DNA molecular weight markers. The molecular weights are indicated.
Detection of mRNA splicing products.
In vitro splicing of HEK 293 cells transfected with the normal construct (G2395) or with the construct containing the mutation (A2395). HMW, ΦX174 HaeIII digest DNA molecular weight markers. LMW, pBR322 MspI digest DNA molecular weight markers. The molecular weights are indicated.
Identification of the abnormal mRNA.
Sequencing of the abnormal mRNA product obtained in HEK 293 cells transfected with the mutated construct. The arrow indicates the lack of the exon 3.
Identification of the abnormal mRNA.
Sequencing of the abnormal mRNA product obtained in HEK 293 cells transfected with the mutated construct. The arrow indicates the lack of the exon 3.
Discussion
The central event in the coagulation of vertebrate blood is the thrombin-catalyzed conversion of fibrinogen into fibrin. The architecture of the fibrinogen molecule has been adapted over the course of several hundred millions of years of evolution. In addition, fibrinogen is also involved in platelet aggregation, endothelial cell injury, blood rheology, and cell proliferation. 20,21
During the past decades a large number of variant fibrinogens have been reported in more than 300 families and about 90 inherited variants have been identified in association with fibrinogen dysfunction.3 These aberrant fibrinogens have been very useful in the continuous effort to correlate structure and function. The complete absence of fibrinogen from the bloodstream may be predicted by many types of abnormal fibrinogen-chain gene structures and/or proteins. However, despite about 150 patients with congenital afibrinogenemia described, only very few cases have been characterized.14,15 Previously identified genetic basis of congenital afibrinogenemia includes homozygous truncation of the fibrinogen Aα-chain gene14 and 2 homozygous missense mutations in the Bβ-chain gene, Leu353Arg and Gly400Asp,15 resulting in impaired assembly and release of the fibrinogen molecule.
We now demonstrate the functional consequences of an additional case of congenital afibrinogenemia. We have investigated a young propositus, offspring of a consanguineous marriage, who presented with undetectable levels of functional and quantitative fibrinogen. Her first-degree relatives had approximately half the normal fibrinogen values and showed a good concordance between functional and immunologic levels.
The molecular basis of this condition is a G-to-A transition at position 2395 in IVS-3 of the γ-chain gene. Most of the mammalian genes are interrupted by introns, which are removed from mRNA precursors by the splicing machinery. At the 5′ and 3′ ends of each intron, dinucleotides GT and AG, respectively, are invariably present.22,23 Flanking these invariable dinucleotides are sequences that are fairly well conserved. Mutations in these sequences have been described that reduce, to various degrees, the efficiency of normal splicing, giving rise to abnormal mRNAs and producing affected phenotypes.24 Within the 5′ splice site consensus sequence (GTAAGT), the fifth nucleotide was a G in 84% of 5′ splice sites from about 400 vertebrate genes.24 Six of the nine 5′ splice sites of the γ-chain gene, including IVS-3 (GTGAGA), contain a G at the fifth position.
The analysis of mRNA isolated from human kidney HEK cells, transfected with the construct containing the A2395 transition, showed an abnormally processed transcript. The abnormal mRNA originated from the skipping of exon 3 is 184 nucleotides shorter than normal. If the aberrant mRNA were translated, the corresponding peptide would have an extremely altered amino acid sequence. Skipping of exon 3 predicts the deletion of the amino acid sequence from residue 16 to 75 and the shift of the reading frame at amino acid 76 with a premature stop codon within exon 4 at position 77. Certainly, it would not be a functional peptide. A peptide originating from its translation would have only 16 amino acids. As a consequence of drastic alterations in its structure, the truncated γ-chain gene product would not contain the coiled-coil and terminal domains and would thus be unable to interact with other fibrinogen chains to form the mature fibrinogen molecule.
Aberrant mRNA transcripts, which could be the result of the activation of cryptic splice sites or of a read-through of the intervening sequence were not observed. Several point mutations within the intron 5′ splice site consensus region and their effects on gene expression have been reported in a number of human disorders. Aberrant mRNA transcripts arising from mutations within the first few nucleotides have been observed in several patients with β-thalassemia. Abnormally processed transcripts corresponded to mRNA products spliced at cryptic splice sites or to exon skipping.25-28 This suggests, in comparison with results presented in this report, that different nucleotides within the consensus sequence of the donor splice site may play different roles in selection of splice sites and in the kinetics of splicing.
In conclusion, in addition to gross gene deletion and impaired secretion, we have documented a new mechanism of congenital afibrinogenemia, due to abnormal mRNA processing, that affects the γ-chain gene. The current data clearly support the concept of a close link in the processing of the 3 fibrinogen gene products and that the correct expression of each chain allows for the synthesis of mature fibrinogen molecules.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Maurizio Margaglione, Unità di Aterosclerosi e Trombosi, IRCCS “Casa Sollievo della Sofferenza,” viale Cappuccini, San Giovanni Rotondo (FG) 71013, Italy; e-mail:ate.tro@operapadrepio.it.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal