Abstract
To understand the regulation of CC chemokine receptor 3 (CCR3) expression, its gene structure and promoter have been characterized. The CCR3 gene contains 4 exons that give rise to multiple messenger RNA (mRNA) species by alternative splicing. Exon 1 is present in all transcripts, whereas exon 2 or 3 is present at low frequency (< 10%). Exon 4 contains the open reading frame and 11 bp of the 5′ untranslated region. Northern analysis revealed 4 species of CCR3 mRNA. Direct sequencing revealed that the first 1 kb of the promoter and exon 1 contained only one mutation in 19 individuals, indicating that the CCR3 promoter and exon 1 are conserved between individuals. The first 1.6 kb of the 5′ flanking region of exon 1 contained promoter elements including a TATA box and motifs for myeloid transcription factors and had strong promoter activity in eosinophilic, lymphoid, myeloid, and respiratory epithelial cell lines. Deletion analysis revealed differential regulation of the CCR3 promoter in eosinophilic and epithelial cells suggesting the presence of lineage-specific elements. Interestingly, exon 1 enhanced the activity of the promoter and this effect was especially prominent in eosinophilic cells. Thus, the humanCCR3 gene has a complex 5′ exon structure, a conserved promoter with strong activity in multiple cell types, and a functional 5′ untranslated exon.
Introduction
The eotaxin receptor, CCR3, is the major chemokine receptor expressed on eosinophils.1-4 In addition, it is expressed by other cell types such as basophils and a subpopulation of Th2 lymphocytes.5-10 Recently, CCR3 has been shown to be up-regulated on neutrophils and monocytoid U937 cells by interferons in vitro and to be expressed by endothelial cells, epithelial cells, and mast cells.11-16 The relevance of these findings and the function of CCR3 on noneosinophils remain to be elucidated because administration of the CCR3-specific ligand eotaxin in vivo induces only eosinophil accumulation.17-19 Recent studies in mice demonstrated CCR3 expression on eosinophils in vivo20 and polarized Th2 cells in vitro.21 Additionally, mice deficient in eotaxin have a major impairment in eosinophil trafficking.22 23 Taken together, these studies imply a critical role for CCR3 in the orchestration of allergic inflammation.
Because CCR3 is a relatively eosinophil-selective marker, analysis of the signals that induce its expression may give insight into the molecular mechanisms for the commitment of myeloid progenitors to the eosinophil lineage. It is generally believed that specific combinations of transcription factors determine the lineage fate of hematopoietic progenitors.24 Although no eosinophil-specific transcription factors have been reported, eosinophil commitment appears to be regulated by GATA-1, PU-1, and C/EBP proteins.25-29Consistent with this, DNA binding sites for these transcription factors are found in several eosinophil-selective promoters, such as the promoter for major basic protein (MBP), interleukin-5 receptor alpha (IL-5Rα) chain, and Charcot-Leyden crystal (CLC) protein.
To date, the complete messenger RNA (mRNA) and genomic organization of only a limited number of chemokine receptors has been described (eg, CXCR1, CXCR2, CCR2, and CCR5).30-33 These studies have shown that the 5′ untranslated region (5′-UTR) can be complex and contain up to 11 exons as in the CXCR2 gene. As a result, alternative splicing and transcription directed by multiple promoters can give rise to variable mRNA isoforms. The function of these 5′ untranslated exons has not been examined except for a single study focused on CCR2, demonstrating a transcriptional role for exon 1.33 Furthermore, the promoter can be highly polymorphic as in the case of CCR5 where a common haplotype, CCR5P1, confers rapid progression of human immunodeficiency virus infection.34-37
Taken together, these studies highlight the importance of further elucidating the gene structure, genetic polymorphisms, and transcriptional regulation of CCR3.
Materials and methods
Cell culture and isolation
The AML14.3D10 cell line (kindly provided by C. C. Paul, Dayton VA Medical Center, Dayton, OH),38 L1.2 (kindly provided by Paul Ponath, LeukoSite, Cambridge, MA), Jurkat, and U937 (ATCC, Rockville, MD) cells were grown in RPMI 1640 (Gibco BRL, Gaithersburg, MD) containing 10% fetal calf serum (FCS, Gibco BRL), 50 μmol/L 2-mercaptoethanol (Sigma, St Louis, MO), 0.1 mmol/L nonessential amino acids (Gibco BRL), 1 mmol/L sodium pyruvate (Sigma) and penicillin-streptomycin (Gibco BRL). The AML14.3D10 cells were further differentiated with butyric acid and IL-5 as previously described.39 The A549 cell line (ATCC) was grown in Dulbecco modified Eagle medium (DMEM; Gibco BRL) supplemented with 10% FCS and penicillin-streptomycin. Eosinophils were isolated by anti-CD16 negative selection from granulocyte preparations of healthy or atopic volunteers as described previously.40
RNA preparation and Northern blot analysis
Total RNA was prepared using the Trizol reagent (Gibco BRL) according to the manufacturer's instructions. RNA was electrophoresed in an agarose-formaldehyde gel, transferred to Gene Screen transfer membranes (NEN, Boston, MA) in 10 × standard sodium citrate (SSC) and cross-linked by UV radiation. The chemokine receptor probe was labeled with 32P using the Klenow reaction with random priming. The open reading frame (ORF) encoding for CCR3 was amplified by polymerase chain reaction (PCR) from human genomic DNA,41 subcloned into pCR2.1 (Invitrogen, Carlsbad, CA), and the CCR3 fragment liberated by EcoRI digestion. Blots were hybridized under standard conditions and washed under high stringency (0.1 × SSC, 1% sodium dodecyl sulfate [SDS] at 65°C). Oligonucleotide probes were labeled with 32P using T4-kinase (Gibco BRL) as per the manufacturer's instructions. Membranes were hybridized with the probe in oligo hybridization buffer, (6 × SSC, 5 × Denhardt solution, 50 mmol/L sodium phosphate buffer [pH 6.8], 0.1 mg/mL herring sperm DNA and 1% SDS) overnight at 42°C. Following 3 washes at room temperature and 1 at 42° in 5 × SSC, 0.1% SDS, membranes were exposed to film.
5′-Rapid amplification of cDNA ends
The template for 5′-rapid amplification of cDNA ends (RACE) was total RNA (1 μg) isolated from human eosinophils and butyric acid/IL-5-differentiated AML14.3D10 cells.39 The Marathon complementary DNA (cDNA) Amplification Kit (Clontech, Palo Alto, CA) was used for 5′-RACE according to the manufacturer's instructions. The sequences of the gene- specific primers were: primary 5′-TCC GGG CTC GAA GGG CAA ACA CA-3′ and nested 5′-CCC AAG AGG CCC ACA GTG AAC AC3-′. The 5′-RACE products were subcloned into pCR2.1 and sequenced by the DNA Core Facility, University of Cincinnati.
Genomic DNA analysis
Human CCR3 genomic clones were isolated by screening a phage P1 library by PCR (DMPC-HFF#1; Genome Systems, St Louis, MO). PCR primers were chosen from the CCR3 ORF sequence and the exon 1 sequence identified by 5′-RACE. The clone identified by the ORF primers (#427-G2)42 and the clone identified by the exon 1 primers (#350-B3) were used for Southern blot analysis and sequencing of theBamHI/HindIII and BglII fragment, respectively.
Reporter gene constructs
The human CCR3 promoter construct was made as follows: a 1.6-kb sequence proximal to transcription initiation site at position −1544 to +60 of exon 1 was amplified by PCR, cloned into pEGFP-1 (Clontech), and subcloned into pGL3.basic (Promega, Madison, WI) via theBglII and BamHI sites. This construct is referred to as the CCR3-1.6pGL3 construct. The CCR3-1.6pGL3 construct without exon 1 was made by digesting the CCR3-1.6pGL3 construct withKpnI and ligating the insert into pGL3.basic vector linearized with KpnI and is referred to as CCR3-1.6(-exon1)pGL3. The exon 1 construct was engineered by re-ligating the CCR3-1.6pGL3 construct digested with KpnI following removal of the insert and is referred to as CCR3-exon1pGL3. Deletion constructs (referred to as CCR3-0.892pGL3, CCR3-0.257pGL3, CCR3-0.222pGL3, and CCR3-0.102pGL3) were amplified by PCR, cloned into pEGFP-1 or pCR2.1, and subcloned into pGL3.basic.
Transient transfection of cell lines and reporter gene expression measurements
Hematopoietic cells (AML14.3D10, L1.2, and Jurkat) were transfected by electroporation as previously described26(with kind guidance from Dr Steven Ackerman, University of Illinois, Chicago, IL). Briefly, 1.5 × 107 cells were electroporated in RPMI with 0.3 to 15 μg plasmid DNA containing the reporter construct and an appropriate amount (5 μg for L1.2 cells and 10 μg for AML and Jurkat) of control construct (pcDNA3.βGal) at 960 μF and 350 V for AML, 300 V for Jurkat, and 250 V for L1.2. Cells were incubated for 7 hours in RPMI with 10% FCS (10 mL per electroporation) and lysates were made using 0.25 mL reporter lysis buffer (Promega) per electroporation. In other experiments, cells were transfected with Effectene (Qiagen, Valencia, CA) following the manufacturer's instructions. Promoter activity results were comparable between experiments performed using the 2 methods. A549 and U937 cells were transfected using Effectene and lysed 24 hours after transfection. The pGL3.SV40 (Promega) and the promoterless pGL3.basic vectors were used as positive and negative controls, respectively. The luciferase assay was performed per manufacturer's instructions (Promega) using 20 μL of the cell lysate and data were recorded with a Monolight 3010 luminometer (Analytical Luminescence Laboratory, Ann Arbor, MI) as relative light units (RLU). β-Galactosidase activity (from 50 μL cell lysate) was measured using ONPG (o-nitrophenyl β-d-galacto-pyranoside; Sigma) as a substrate in sodium phosphate buffer for 2 hours at 37°C. The reaction was stopped by addition of sodium carbonate and optical density (OD) was measured at 405 nm. All data were normalized by dividing RLU (luciferase assay) by OD (β-galactosidase assay). When the deletion constructs were compared to wild-type in the AML14.3D10 cell line, transfection efficiency was normalized by co-transfecting with the renilla luciferase vector, pRL.SV40 (Promega) and the firefly and renilla luciferase activities were determined according to manufacturer's instructions (Dual Luciferase Reporter Assay System; Promega).
Polymorphism detection
The human CCR3 promoter and exon 1 were screened for polymorphisms by sequencing 3 overlapping segments amplified by PCR from genomic DNA from 19 individuals (15 with severe allergic asthma and 4 normal controls). The diagnosis of asthma was made based on symptoms and a 12% or greater increase in forced expiratory volume in 1 second (FEV1) after a bronchodilator or after a 2-week trial of oral corticosteroids.43 44 It was classified as severe based on the FEV1 being below 60% and allergic based on a positive skin prick test (≥ 3 mm wheal with erythema) to one or more antigens tested (environmental antigens indigenous to the Ohio valley). Normal controls were nonallergic, nonasthmatic volunteers with a negative skin prick test to all allergens tested (excluding histamine). Informed consent was obtained from all participants in these studies. The PCR reactions were performed with approximately 0.3 μg genomic DNA, 0.5 μmol/L each primer, 0.2 mmol/L dNTPs (Roche, Indianapolis, IN), 1.25 U Taq Polymerase (Roche) in a total volume of 50 μL. Primer pairs were as follows: P1 5′-(TGTAAAACGACGGCCAGT CCC AAG GGA CAC ATC AGC) and 5′-(CAGGAAACAGCTATGACC CCC GGC AAA GGA ATA AAC T); P2 5′-(TGTAAAACGACGGCCAGT AAC CTT TGC AGC CAC ATT TTG) and 5′-(CAGGAAACAGCTATGACC GCT GCT TTA GGG GCT CTC CAC); P3 5′-(TGTAAAACGACGGCCAGT CCC CCA CCA CTA AAA ATG AGC) and 5′-(CAGGAAACAGCTATGACC CCT GGA AAA GCG ACA CCT ACC). All primers had the M13 primer sequence tagged (underlined). PCR products (420-575 bp in length) were purified (Qiagen PCR Purification Kit) and sequencing was performed on the ABI sequencer (DNA Core Facility, University of Cincinnati) using dye-primer chemistry to facilitate detection of heterozygosity. Data were analyzed using DNA Star software (DNA Star, Madison, WI).
Results
Northern blot analysis of CCR3
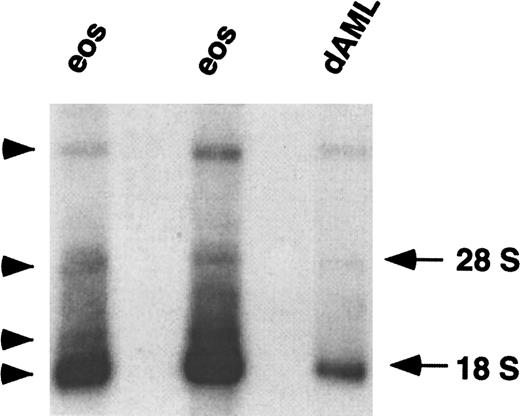
The promoters of chemoattractant receptor genes are often separated from the ORF by one or more large introns. The first evidence that this was the case for CCR3 was derived from analysis of CCR3 mRNA expression. Northern blot analysis using a CCR3 ORF probe revealed multiple hybridizing mRNA bands. The main CCR3 mRNA migrated at ∼1.8 kb and 3 weaker species migrated at ∼2, ∼4, and ∼20 kb (Figure1). The presence of multiple bands may indicate cross-hybridization with related gene products, detection of unspliced heterogeneous nuclear RNA, or the presence of multiple mature CCR3 transcripts that could arise either by alternative splicing or use of different transcription initiation or polyadenylation sites. Therefore, to characterize the CCR3 promoter, we were first interested in determining the complete sequence of the 5′-UTR.
Northern blot analysis of CCR3 mRNA.
Total RNA (4 μg) from the eosinophils (eos) of 2 individuals and from butyric acid/IL-5 differentiated AML14.3D10 cells (dAML) was hybridized with a radiolabeled full-length CCR3 ORF probe under high stringency conditions. Autoradiography was performed for 72 hours. The locations of 28 S and 18 S RNA are shown on the right and hybridized bands are labeled with an arrowhead.
Northern blot analysis of CCR3 mRNA.
Total RNA (4 μg) from the eosinophils (eos) of 2 individuals and from butyric acid/IL-5 differentiated AML14.3D10 cells (dAML) was hybridized with a radiolabeled full-length CCR3 ORF probe under high stringency conditions. Autoradiography was performed for 72 hours. The locations of 28 S and 18 S RNA are shown on the right and hybridized bands are labeled with an arrowhead.
5′-RACE of CCR3 mRNA
To characterize the CCR3 promoter, we identified the 5′ sequence of the mature mRNA. To accomplish this, we performed 5′-RACE using RNA isolated from eosinophils and butyric acid/IL-5 differentiated AML14.3D10 cells. Products were subsequently subcloned and 12 clones were selected for sequencing by choosing clones with a range of insert sizes. Alignment of the 5′-UTR sequence of 7 clones originally derived from eosinophil RNA revealed a complex organization (Figure2). All clones had 11 bases upstream from the ATG that were identical to the genomic sequence. Additionally, all clones contained up to 93 bases of the 5′-UTR sequence that is labeled as exon 1 in Figure 2. The truncated forms (with fewer than 93 bases) may arise from premature termination of cDNA synthesis by the reverse transcriptase in vitro or may indicate the presence of multiple transcription start sites in vivo. One 5′-RACE product had 69 bp between the 2 sequences (clone EO12). Another clone had 89 bp between the 2 segments (clone EO9). These data indicate that there are three 5′ exons alternatively spliced into the final mRNA. Exon 1 is present in all transcripts, whereas either exon 2 or 3 is present in a small subset of mRNA species. To verify the occurrence of exons 2 and 3, we screened all cloned 5′-RACE products for their expression. Using oligonucleotide probes for each of the exons (EO9: 5′-TCA CTG GCT CCC TCA TTC CG-3′ and EO12: 5′-CTG CTG TGG ATT GGA TTA TG3-′), a low frequency of clones (< 10%) containing exons 2 and 3 were identified (data not shown).
Alignment of 5′-RACE products.
5′-RACE was performed on RNA isolated from purified eosinophils. Separate clones were sequenced and are labeled as EO with an assigned number. Alignment of the 5′-UTR is shown in capital letters and coding sequence as small letters. Upstream ATGs are boxed. The positions of exons 1 through 4 are indicated; — indicates a gap.
Alignment of 5′-RACE products.
5′-RACE was performed on RNA isolated from purified eosinophils. Separate clones were sequenced and are labeled as EO with an assigned number. Alignment of the 5′-UTR is shown in capital letters and coding sequence as small letters. Upstream ATGs are boxed. The positions of exons 1 through 4 are indicated; — indicates a gap.
Genomic organization of the CCR3 gene
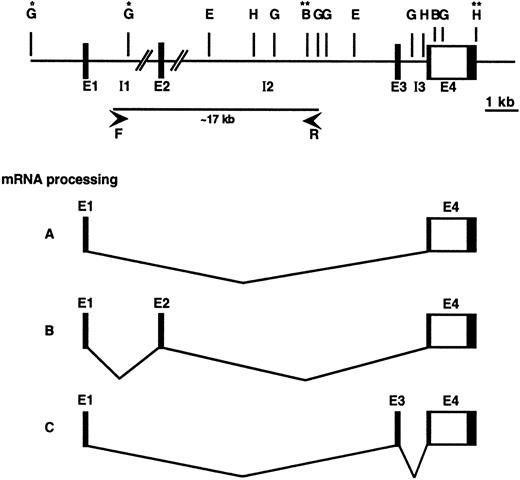
To determine the intron/exon structure of the CCR3 gene in more detail, genomic clones containing CCR3 were isolated and characterized. A genomic library was screened using PCR primers specific for the entire CCR3 ORF and exon 1. One of the clones contained the ORF and was used for Southern blot analysis and restriction map analysis. Two overlapping segments (3.8 kbBamHI and 1.7 kb HindIII fragments) were fully sequenced and shown to contain the entire ORF (located on exon 4) as well as 3591 bp of 5′ sequence and 445 bp of 3′ sequence. Analysis of this sequence revealed that it also contains the 69 additional bases found in 5′-RACE clone EO12, designated exon 3. Another genomic clone was identified by the exon 1 primers, and a 2.9 kb BglII fragment was fully sequenced and found to contain the entire exon 1. Analysis of the intron/exon junctions revealed that they conformed to the splice donor consensus sequence. Southern blotting with the exon 2 sequence (the 89 additional base pairs found in 5′-RACE clone EO9) indicated this exon is located in the ∼8 kb BglII- Eco RI segment. To analyze the length of the sequence intervening exons 1 and 3, long-range PCR analysis was performed using primers within the sequenced regions of introns 1 and 2, respectively (Figure3). This analysis resulted in a specific band of ∼17 kb (data not shown). The proposed genomic organization is shown in Figure 3 and the sequence of the 5′ BglII fragment is shown in Figure 4.
Genomic organization and mRNA processing of theCCR3 gene.
A schematic diagram of the organization of the CCR3 gene is shown. The translated DNA area is depicted as an open rectangle and untranslated DNA is shown as shaded rectangles. Restriction enzymes are labeled as follows: E, EcoRI; G, BglII; B,BamHI; and H, HindIII. Exons are labeled as E1 through E4; introns as I1 through I3. The precise position of exon 2 has not been determined and this is indicated by the pair of slashed lines. DNA fragments flanked by single asterisk (*) and double asterisk (**) were fully sequenced (Genbank accession numbers AF237380,AF237381, and U51241) and the BglII sequence is shown in Figure 4. Primers used for long-range PCR are labeled as F and R flanking the ∼17 kb PCR product. Below are the corresponding mRNA species: A, majority of mRNA species contain only exons 1 and 4; B and C show usage of exon 1 with exons 2 or 3, respectively.
Genomic organization and mRNA processing of theCCR3 gene.
A schematic diagram of the organization of the CCR3 gene is shown. The translated DNA area is depicted as an open rectangle and untranslated DNA is shown as shaded rectangles. Restriction enzymes are labeled as follows: E, EcoRI; G, BglII; B,BamHI; and H, HindIII. Exons are labeled as E1 through E4; introns as I1 through I3. The precise position of exon 2 has not been determined and this is indicated by the pair of slashed lines. DNA fragments flanked by single asterisk (*) and double asterisk (**) were fully sequenced (Genbank accession numbers AF237380,AF237381, and U51241) and the BglII sequence is shown in Figure 4. Primers used for long-range PCR are labeled as F and R flanking the ∼17 kb PCR product. Below are the corresponding mRNA species: A, majority of mRNA species contain only exons 1 and 4; B and C show usage of exon 1 with exons 2 or 3, respectively.
Sequence of the CCR3 promoter.
The 2.7-kb 5′ BglII fragment (Figure 3) was fully sequenced. The exon 1 is bolded. The splice donor consensus sequence is double-underlined. Numbering is based on assigning +1 to the first base of the longest 5′-RACE product. Underlined are restriction enzyme sites (BglII, KpnI); overlined are consensus transcription factor binding sites; boxed are pyrimidine-rich sequences; the shaded box depicts the area of high homology with CCR2 that contains the Alu repeat and the light gray box marks the area of homology with hIL-5Rα. The asterisk depicts the site of the identified polymorphism.
Sequence of the CCR3 promoter.
The 2.7-kb 5′ BglII fragment (Figure 3) was fully sequenced. The exon 1 is bolded. The splice donor consensus sequence is double-underlined. Numbering is based on assigning +1 to the first base of the longest 5′-RACE product. Underlined are restriction enzyme sites (BglII, KpnI); overlined are consensus transcription factor binding sites; boxed are pyrimidine-rich sequences; the shaded box depicts the area of high homology with CCR2 that contains the Alu repeat and the light gray box marks the area of homology with hIL-5Rα. The asterisk depicts the site of the identified polymorphism.
Analysis of the CCR3 promoter sequences
The human CCR3 promoter contained 2 putative TATA-boxes: one from position −298 to −294 and the other from position −108 to −103 proximal to the first base of the longest 5′-RACE product. There are several pyrimidine (CT)-rich segments in the promoter region. For example, regions from −1361 to −1300 and from −1282 to −1224 have more than 90% C+T over more than 50 nucleotides (boxed in Figure 4). Interestingly, it has been reported that CT-rich segments are present in genes abundantly expressed in myeloid cells, that is, fMLP-receptor and myeloperoxidase.45 46 The promoter sequence was analyzed using the publicly available TFSEARCH engine (http://pdap1.trc.rwcp.or.jp/research/db/TFSEARCH.html) and found to contain consensus DNA-binding sites for several transcription factors (ie, GATA-1, AML1, C/EBP, etc). In addition to those shown in Figure 4, other transcription factor motifs found included AP-1, NFκB, Oct-1, CdxA, CREB, and STAT-x.
The promoter sequence was compared to other chemokine receptor promoter sequences using BestFit (SeqWeb, Genetics Computer Group, Madison, WI). Comparison with CCR2 (accession number AF068 265)33 and CCR5 (accession numbers AF082 742 and AF017 632)31,36revealed 40% overall identity. However, there were areas of up to 72% identity between the human CCR3 and CCR2 promoters spanning over 200 bp (shaded area in Figure 4). Further examination revealed that this sequence represents an Alu family repeat.47 Comparison to eosinophil-selective promoters (eg, hCLC, accession number L01 665; human eosinophil peroxidase (hEPO), accession number M29 904, hIL-5Rα, accession number U18 373, and hMBP, accession number M34 462)25,26,48 49 revealed 36% to 39% overall identity. There was a 31-bp segment with 67% homology between the CCR3 and IL-5Rα promoter (shaded box in Figure 4).
Genetic polymorphisms in the CCR3 noncoding sequences
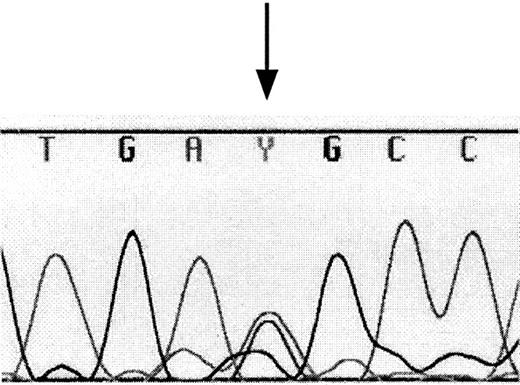
Several chemokine receptor promoters have been shown to be highly polymorphic and these polymorphisms are sometimes correlated with disease processes. Hence, we hypothesized that polymorphisms may exist in the 5′-UTR and promoter of CCR3 and that these polymorphisms may have consequences on diseases such as asthma in which CCR3-expressing cells have a major role. We therefore screened genomic DNA from 19 individuals (15 with severe asthma and 4 normal controls) for polymorphisms in the first 1 kb of the promoter sequence and the entire exon 1 using dye-primer sequencing. Surprisingly, we found only one heterozygous polymorphism. DNA from one normal control individual had equal representation of cytosine and thymine bases in position −37 (Figure 5 and asterisk on Figure 4), whereas all other individuals had cytosine in that position. To control for PCR-introduced mutations, we repeated the same analysis using a separate PCR-amplified DNA and obtained the same results. Interestingly, this polymorphism lies in a putative CREB binding site. This indicates that the CCR3 promoter region and exon 1 are conserved between individuals.
Detection of the promoter polymorphism.
PCR-amplified DNA from 19 individuals was subjected to sequence analysis using dye-primer sequencing. A heterozygous polymorphism was detected in the DNA from one individual. Equal amounts of thymine and cytosine peaks indicate heterozygosity at position −37 of theCCR3 gene (as indicated by the arrow). Y indicates thymine and cytosine.
Detection of the promoter polymorphism.
PCR-amplified DNA from 19 individuals was subjected to sequence analysis using dye-primer sequencing. A heterozygous polymorphism was detected in the DNA from one individual. Equal amounts of thymine and cytosine peaks indicate heterozygosity at position −37 of theCCR3 gene (as indicated by the arrow). Y indicates thymine and cytosine.
Characterization of the functional CCR3 promoter
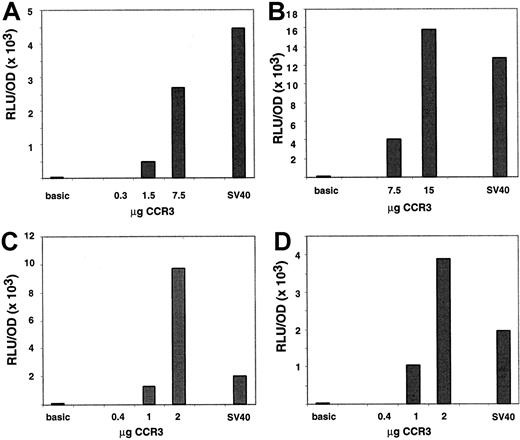
To prove that the 5′ genomic region has promoter activity, 1.6 kb of 5′ DNA containing 60 bp of exon 1 was cloned into a firefly luciferase reporter vector, referred to as CCR3-1.6pGL3. The ability of the promoter construct to promote the expression of the reporter gene was first tested in AML14.3D10 cells, an eosinophilic myelocyte cell line. To correct for differences in transfection efficiency, promoter constructs were co-transfected with pcDNA3.βGal. The eosinophilic AML14.3D10 cells were transfected either by electroporation as previously reported (Figure 6A-B) or by a new method using the Effectene kit (Figure 6C,D). Interestingly, expression of transfected proteins peaked at different times between the 2 methods. With electroporation, expression of luciferase was ∼30-fold higher 7 hours after the transfection as compared to 24 hours. This kinetic pattern is similar to that reported previously.26 Conversely, with Effectene expression peaked at 24 hours (∼8-fold higher than at 7 hours, data not shown). When CCR3 promoter activity was measured at the optimal time point for each of the methods, strong expression of the luciferase gene was observed and a dose response was seen with increased DNA concentrations. Of note, the activity of the CCR3 promoter was comparable to that of the SV40 promoter used as a positive control. Because of variability between experiments, 2 representative experiments for each of the methods are shown in Figure 6. When compared to the basic promoterless vector, the CCR3 promoter activity was 45-fold higher at 7.5 μg and 130-fold at 15 μg when transfection was performed by electroporation. For comparison, SV40 promoter activity was 100-fold higher than the basic promoter at 15 μg. When transfection was performed with Effectene, the CCR3 promoter activity was 23- and 120-fold over the basic vector at 1 and 2 μg, respectively. In the same experiments, SV40 activity was 40-fold above the promoterless vector at 1 μg.
Characterization of the functional CCR3 promoter in AML14.3D10 cells.
Cells (AML14.3D10) were transiently transfected with a reporter plasmid containing 1.6 kb of human CCR3, SV40, or no promoter ligated to the luciferase reporter gene. Cells were cotransfected with the pCDNA3.βGal plasmid and data are normalized to β-galactosidase activity. In panels A and B, 2 separate experiments are shown where cells were transfected by electroporation, whereas in panels C and D cells were transfected with Effectene. A dose response of the CCR3 promoter activity is depicted as well as the value of the negative control, promoterless vector (basic), and the positive control (SV40) promoter. The control vectors were used at 15 μg in panels A and B and 1 μg in panels C and D. On the y-axis, data are presented as RLU (luciferase activity)/OD (β-galactosidase activity).
Characterization of the functional CCR3 promoter in AML14.3D10 cells.
Cells (AML14.3D10) were transiently transfected with a reporter plasmid containing 1.6 kb of human CCR3, SV40, or no promoter ligated to the luciferase reporter gene. Cells were cotransfected with the pCDNA3.βGal plasmid and data are normalized to β-galactosidase activity. In panels A and B, 2 separate experiments are shown where cells were transfected by electroporation, whereas in panels C and D cells were transfected with Effectene. A dose response of the CCR3 promoter activity is depicted as well as the value of the negative control, promoterless vector (basic), and the positive control (SV40) promoter. The control vectors were used at 15 μg in panels A and B and 1 μg in panels C and D. On the y-axis, data are presented as RLU (luciferase activity)/OD (β-galactosidase activity).
To assess whether the CCR3 promoter is specific for eosinophilic cells in vitro, we tested the activity of the CCR3 promoter in the L1.2 (mouse B cell), Jurkat (human T cell leukemia), U937 (human myelomonocytic cells), and A549 (human bronchial epithelial) cell lines. Activity in those cell lines was above the promoterless vector and dose dependent (Figure 7). For instance, in a representative experiment (n = 2-4 for each cell line), when 1 μg of DNA was transfected using Effectene in the U937 cells the CCR3 promoter activity was 30-fold and the SV40 promoter activity was 500-fold over the promoterless vector. In the L1.2 cells, the CCR3 promoter activity was 4-fold and SV40 promoter activity was 37-fold over the promoterless vector. In the Jurkat cells, the CCR3 promoter activity was 13-fold and SV40 promoter activity was 70-fold over the promoterless vector. In the A549 cells, the CCR3 promoter activity was 56-fold and SV40 promoter activity was 2000-fold over the promoterless vector.
Activity of the CCR3 promoter in noneosinophilic cell lines.
Cells were transiently transfected using the Effectene method with a reporter plasmid containing 1.6 kb of human CCR3 promoter or no promoter ligated to the luciferase gene. Cells were cotransfected with the pcDNA3.βGal plasmid and the data are normalized to β-galactosidase activity. Cell lines shown are A, U937; B, L1.2; C, Jurkat; and D, A549. A dose response of CCR3 promoter activity is depicted as well as the value of the promoterless vector (basic). The control vector was used at 1 μg in A through C and 0.4 μg in D. On the y-axis, data are presented as RLU (luciferase activity)/OD (β-galactosidase activity). Data are presented for a representative experiment (n = 2-4 for each cell line).
Activity of the CCR3 promoter in noneosinophilic cell lines.
Cells were transiently transfected using the Effectene method with a reporter plasmid containing 1.6 kb of human CCR3 promoter or no promoter ligated to the luciferase gene. Cells were cotransfected with the pcDNA3.βGal plasmid and the data are normalized to β-galactosidase activity. Cell lines shown are A, U937; B, L1.2; C, Jurkat; and D, A549. A dose response of CCR3 promoter activity is depicted as well as the value of the promoterless vector (basic). The control vector was used at 1 μg in A through C and 0.4 μg in D. On the y-axis, data are presented as RLU (luciferase activity)/OD (β-galactosidase activity). Data are presented for a representative experiment (n = 2-4 for each cell line).
Deletion analysis of the CCR3 promoter
To more precisely localize the regulatory regions of the promoter, a series of deletion mutants were generated and tested for promoter activity (Figure 8). Four deletions of the CCR3-1.6pGL3 construct were prepared in the pGL3 vector. These constructs included the promoter elements starting at positions −892, −257, −222, −102 (referred to as CCR3-0.892pGL3, CCR3-0.257pGL3, CCR3-0.222pGL3, and CCR3-0.102pGL3, respectively). These constructs were tested for promoter activity by transiently transfecting the A549 cell line. The A549 cells were initially chosen because the CCR3-1.6pGL3 construct displayed strong promoter activity in this cell line and transfection efficiency was highest in A549 compared with the other cell lines (Figure 7D and data not shown). Interestingly, deletion of nucleotides 5′ of bp 102 did not diminish promoter activity compared to the full length vector, CCR3-1.6pGL3 (Figure 8A). Similar levels of relative promoter activity were seen at 2 lower doses (0.1 and 0.3 μg) of the deletion constructs (data not shown). These data suggest that the optimal promoter activity is located within the first 102 bp of the region 5′ to exon 1.
Activity of CCR3 promoter deletion constructs.
A schematic representation of the deletion constructs cloned into the pGL3 luciferase vector is shown in the left panel. The promoter sequence is shown as a line and exon 1 is depicted as an open box. The position of the KpnI restriction site and the deletion construct end positions are shown with arrowheads. In the right panel, cells were transfected with the reporter plasmid indicated. In A and B, A549 cells were cotransfected with the pcDNA3.βGal plasmid and the data are normalized to β-galactosidase activity. On the x-axis, data are presented as RLU (luciferase activity)/OD (β-galactosidase activity). In C, AML14.3D10 cells were cotransfected with the pRL.SV40 and the data are normalized to the activity of the renilla luciferase. On the x-axis, data are presented as RLU1 (firefly luciferase activity)/RLU2 (renilla luciferase activity). Data are presented for a representative experiment (n = 3). Each point was performed in triplicate and is expressed as mean ± SD.
Activity of CCR3 promoter deletion constructs.
A schematic representation of the deletion constructs cloned into the pGL3 luciferase vector is shown in the left panel. The promoter sequence is shown as a line and exon 1 is depicted as an open box. The position of the KpnI restriction site and the deletion construct end positions are shown with arrowheads. In the right panel, cells were transfected with the reporter plasmid indicated. In A and B, A549 cells were cotransfected with the pcDNA3.βGal plasmid and the data are normalized to β-galactosidase activity. On the x-axis, data are presented as RLU (luciferase activity)/OD (β-galactosidase activity). In C, AML14.3D10 cells were cotransfected with the pRL.SV40 and the data are normalized to the activity of the renilla luciferase. On the x-axis, data are presented as RLU1 (firefly luciferase activity)/RLU2 (renilla luciferase activity). Data are presented for a representative experiment (n = 3). Each point was performed in triplicate and is expressed as mean ± SD.
We were next interested in delineating the role of exon 1 because the function of untranslated exons in chemokine receptor genes has not been extensively examined. Additionally, the finding that exon 1 was conserved between individuals suggested that this exon might have a functional role. To investigate this, we examined the promoter activity of a construct containing full promoter elements without exon 1 (referred to as CCR3-1.6(-exon1)pGL3) and a construct containing exon 1 alone (referred to as CCR3-exon1pGL3). Interestingly, in the absence of exon 1, the promoter activity was reduced by 60% ± 12% (mean ± SD, n = 3) as shown in Figure 8B. Additionally, exon 1 alone exhibited minimal promoter activity. These data indicate that exon 1 enhances the expression of CCR3, but does not act as a promoter element by itself.
After establishing the regulatory regions of the CCR3 promoter that were operational in epithelial cells, we were next interested in determining if these same regions were involved in eosinophils. We hypothesized that eosinophilic cells would use distinct promoter regions because CCR3 is relatively eosinophil selective in vivo. To test this hypothesis, we examined the activity of the critical promoter constructs identified in respiratory epithelial cells (Figure 8C). We initially examined the CCR3-0.102pGL3 construct because this region has full promoter activity in epithelial cells. Interestingly, in eosinophilic cells, the activity of this construct was reduced by 86.1% ± 11.6% (mean ± SD, n = 3) compared to the full length construct (CCR3-1.6pGL3). Taken together, these results establish that the major promoter elements in eosinophilic cell are present 5′ of bp −102. Lastly, we examined the role of exon 1 in eosinophilic cells. The activity of the CCR3 construct without exon 1 was reduced by 85% ± 17.7% (mean ± SD, n = 4). Thus, similar to respiratory cells, maximal CCR3 transcription is dependent on exon 1, and this effect appears to be particularly important in eosinophilic cells.
Discussion
In this study we have demonstrated that the human CCR3gene has a complex 5′ exon structure and a highly conserved promoter with activity in eosinophilic cells. We further report that theCCR3 gene spans ∼23 kb and consists of 4 exons. Although exons 1 and 4 are used in all transcripts, exon 2 or 3 is used in only a minority of transcripts. Moreover, the presence of exon 1 enhances the promoter activity of the endogenous promoter, and this effect is especially prominent in eosinophilic cells.
Although the full function of the 5′ untranslated exons in CCR3 remains to be elucidated, it is interesting to consider that these exons may have regulatory functions. For example, 5′-UTR exons may have a transcriptional role because they can bind transcription factors as is the case for the CCR2 promoter. Using gel mobility shift assays and mutation analysis, it was shown that binding of C/EBP to exon 1 is critical for transcription of CCR2.33 We have obtained evidence for the importance of exon 1 for CCR3 transcription by demonstrating diminished promoter activity when exon 1 is deleted from the promoter construct compared to the CCR3-1.6pGL3 construct (Figure8). Interestingly, although exon 1 does not contain a C/EBP motif, there are motifs for GATA-1. It still remains possible that the 5′ exon may alter the thermodynamic stability of CCR3 mRNA, which has not been examined in the present experiments.
Expression of CCR-3 protein may also be regulated at the level of translation. In previous studies, we have shown that CCR3 is regulated by post-translational events in eosinophilic cells such as receptor internalization and degradation.40 The identification of the complex genomic structure of CCR3 gives further insight into possible mechanisms of CCR3 regulation. It is interesting to speculate that the 5′-UTR may regulate CCR3 expression. Factors in the 5′UTR that promote efficient translation include: (1) the Kozak sequence immediately surrounding the start codon, (2) the absence of upstream ATG codons, and (3) short 5′-UTR (most vertebrate mRNAs have a 5′-UTR of 20–100 nucleotides).50 The Kozak sequence of CCR3 is relatively weak, with only the purine in position −3 conserved. Additionally, transcripts containing sequence from either exon 2 or 3 have 2 upstream ATG codons (Figure 2). The ATG codons derived from exon 2 are followed immediately by termination codons; thus they may give rise to short peptides leading to reduced protein output; this type of mechanism has been reported for the β-adrenergic receptor.51 52 However, these 2 ATGs are not surrounded by a consensus Kozak sequence. On the contrary, the 2 ATGs derived from usage of exon 3 are surrounded by a Kozak sequence comparable to the one used by the major ORF (purine at position −3) and are in frame with the major ORF. This suggests that there may be translation of 2 additional proteins 15 and 21 amino acids longer than the one originally identified.
In this report, we have identified strong CCR3 promoter activity in the eosinophilic AML14.3D10 cell line. In addition, dose-dependent activity was detected in other myeloid (U937), lymphoid (L1.2 and Jurkat), and nonhematopoietic (A549) cell lines. This suggests that additional mechanisms besides transcription may be used to regulate the ultimate level of CCR3 protein in various cell types because most of these cells do not express CCR3 constitutively. For instance, the CCR3 promoter may be active in noneosinophilic cell types, but the transcribed mRNA may be preferentially spliced to include exon 2 and/or 3 and therefore translation may be inefficient. This in turn may result in lack of protein production in these cell types. It remains possible that the activity of the CCR3 promoter in transfected cells may not accurately reflect the CCR3 promoter activity in vivo. This may be particularly important for eosinophilic cell lines because these cells are difficult to transfect compared to other hematopoietic cells. Additionally, stronger eosinophil selectivity might be observed with different promoter constructs (eg, longer than 1.6 kb). However, we have repeatedly observed the activity of the CCR3 promoter to be comparable or stronger than that of the SV40 promoter in eosinophilic cells. In contrast, the SV40 promoter activity was always significantly stronger in noneosinophilic cell lines. We cannot, however, rule out the possibility that this is due to different, cell-specific requirements of the SV40 promoter itself. Several eosinophil-selective gene product promoters have been studied so far. For instance, the IL-5Rα, EPO, MBP, and CLC promoters are all active in nonhematopoietic cell lines tested even though their expression in vivo is relatively limited to the eosinophilic lineage.25,26,28,48 Of those, only the IL-5Rα and the MBP promoter have demonstrated some eosinophil-lineage specificity in vitro. It is interesting that CCR3 has recently been reported to be expressed by respiratory epithelial cells13 and we have found promoter activity in this cell type (A549). Our data, taken together with this preliminary study,13 suggests that CCR3 may indeed be involved in airway epithelial biology. A further clarification of this role may be determined by analysis of mice genetically deficient in CCR3. Future studies will be required to determine the significance and regulation of CCR3 in noneosinophilic cell types.
To begin addressing the issue of cell-specific elements, we performed CCR3 promoter deletion construct analysis in respiratory epithelial and eosinophilic cells (Figure 8). Even though the full length construct was fully active in a broad array of cells, deletion construct analysis suggested the presence of eosinophil-specific regulatory elements. For instance, the promoter construct containing −102 bp of the CCR3 promoter had activity comparable to the full length 1.6 kb construct in A549 cells. However, in eosinophilic cells the promoter construct containing −102 bp had reduced activity. This suggests the presence of eosinophil-specific elements that bind to the region upstream of −102 and act as an amplifier of promoter activity. Additionally, the presence of exon 1 was important for transcription in both cell types, but this effect is especially prominent in eosinophilic cells.
Finally, our studies elucidate the genetic stability of the CCR3 promoter. Several recent reports have described at least 12 single nucleotide polymorphisms (SNP) within the 5′ upstream regulatory region of the human CCR5 gene.35-37 Furthermore, a common 10-SNP haplotype, called CCR5P1, has been shown to confer rapid clinical progression of acquired immunodeficiency syndrome. We have previously reported 4 polymorphisms (0.005–0.13 allele frequency) in the ORF of CCR3.41 Taken together, we therefore hypothesized that polymorphisms may exist in the regulatory region of the CCR3 gene. Surprisingly, after analysis of the first 1 kb of CCR3 promoter and exon 1, we found only one individual who had a heterozygous mutation. These results indicate that, in contrast to the coding region, the promoter and exon 1 of CCR3 are conserved between individuals. The conservation of exon 1 supports our results showing an important functional role for this exon. Future studies will need to be conducted to explore the frequency of this polymorphism in the population and any possible correlation with disease phenotypes.
In summary, we report that (1) the CCR3 gene has a complex structure containing 4 exons 2 of which are alternatively used in mRNA composition; (2) the CCR3 promoter and exon 1 are conserved between individuals; (3) the 5′-flanking region of exon 1 has strong promoter activity in both hematopoietic and nonhematopoietic cells in vitro; (4) the CCR3 promoter activity is differentially regulated in eosinophilic and respiratory epithelial cells; and (5) the exon 1 is involved in the regulation of CCR3 expression. These studies provide the molecular framework to analyze the regulation of CCR3 transcriptional activation and tissue expression.
Acknowledgments
The authors wish to thank Drs Jonathan Bernstein and Gurjit Hershey for DNA samples, and Drs Steve Ackerman, Cindy Bachurski, Jeff Whitsett, and Simon Hogan for helpful discussions and critical reading of the manuscript.
Division of Pulmonary Medicine, Allergy and Clinical Immunology, Department of Pediatrics, Children's Hospital Medical Center, Cincinnati, OH; Department of Immunology and Rheumatology, Merck Research Laboratories, Rahway, NJ.
Submitted April 18, 2000; accepted June 5, 2000.
N.Z. is a Parker B. Francis fellow in pulmonary research. This work was supported in part by the AAAAI President's Grant-in-Aid to N.Z.
N.Z. and B.L.D. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Marc E. Rothenberg, Division of Pulmonary Medicine, Allergy and Clinical Immunology, Department of Pediatrics, Children's Hospital Medical Center, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: rothenberg@chmcc.org.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal