Abstract
Atm-deficient mice (Atm−/−) recapitulate many aspects of the ataxia telangiectasia (AT) syndrome, including the susceptibility to tumors of lymphoid origin. To investigate the mechanism of tumorigenesis, we have examined a panel of 8 thymic lymphomas from Atm−/− mice. AllAtm−/− tumors are of thymic lymphoblastoid origin, display an immature CD3− and CD4+/CD8+ phenotype, and arise coincident with V(D)J recombination. Cytogenetically, all tumors are diploid or near diploid but exhibit multiple chromosome aberrations with an average of 4 abnormal chromosomes per tumor. All the tumors revealed chromosome 14 rearrangements precisely at the T-cell receptorα/δ(Tcrα/δ) locus, suggesting the involvement of V(D)J recombination in these translocations. In addition, 11.5% ofAtm−/− peripheral T cells showed chromosome 14 translocations, suggesting that rearrangements at theTcrα/δ locus occur early during tumor development in the absence of ATM. However, additional genetic aberrations are required for tumorigenesis. For example, translocations involving chromosome 12, often with chromosome 14 (more than 60%), and partial or complete trisomy of chromosome 15, with copy number increases of the c-myc oncogene were frequently observed. These observations suggest that ATM is required for normal rearrangement of the Tcrα/δ locus but not for V(D)J recombination at other loci. The mechanisms that lead to tumorigenesis may be due to the involvement of ATM in monitoring double-stranded DNA breaks.
Introduction
AT is an autosomal recessive disease with a pleiotropic phenotype involving the nervous, immune, and reproductive systems.1 The primary features include progressive cerebellar ataxia, difficulties with speech and abnormal eye movements, oculocutaneous telangiectasia, immunodeficiencies, and susceptibility to cancer. Other characteristics of the disease include growth retardation, chromosomal instability, hypersensitivity to ionizing radiation, hypogonadism and infertility, thymic dysplasia, and elevated serum alpha-fetoprotein (reviewed by Segwick and Boder,2Hosking et al,3 Waldmann,4 and Gatti5).
Individuals with AT are predisposed to leukemia and lymphomas.6 Thirty-eight percent of patients with AT have a malignancy develop during their lifetime,7 the majority of which are hematologic tumors of the lymphoid lineage in the absence of tumors of myeloid origin. Leukemias in patients with AT are usually of T-cell origin, whereas lymphomas originate primarily from B cells, Moreover, there is an estimated 4- to 5-fold increased frequency of T-cell tumors compared with B-cell tumors in these patients. The majority of T-cell tumors in patients with AT are T-ALL and T-cell lymphoma, but in older patients with AT, the expansion of T-cell clones with karyotypic abnormalities has been followed to the point to which they develop into T-PLL.8 The characteristic translocations present in these leukemias and lymphomas contain breakpoints that cluster in the T-cell receptor (TCR) genes, immunoglobulin genes, and at least 2 putative oncogenes,TCL1 and MTCP, which are both involved in T-cell lymphomagenesis (reviewed by Taylor et al8). Interestingly, patients with AT also display immunologic defects during T-cell development. These results suggest that abnormalities in T-cell development result in translocations that arise during V(D)J recombination, and that these translocations are important for the development of lymphoid malignancies. However, no direct evidence of the involvement of T-cell developmental defects during V(D)J recombination in human tumors exists, and the mechanism of tumorigenesis remains unknown. In addition, human AT tumors are rare, making it difficult to study tumorigenesis.
The identification of the murine homologue, Atm, allowed for the generation of several mouse models, andAtm−/− mice recapitulate many features of the phenotype of patients with AT.9-12 For example,Atm−/− mice display defects in mitotic cells in response to ionizing radiation,13,14 in meiotic cells during the process of homologous recombination,15-17 and evidence of oxidative damage to the central nervous system (CNS).17 These studies are consistent with a role forATM in managing DNA double-stranded breaks (DSBs), either through a role in cell cycle regulation or directly in DNA repair (reviewed by Rotman and Shiloh18 and Brown et al19). Nearly all Atm−/− mice succumb to aggressive T-cell lymphoblastic lymphomas between 3 to 6 months of age.10 As in humans with AT,Atm−/− mice display T-cell developmental defects, consistent with the hypothesis that defects in T-cell development are related to the propensity for lymphoreticular malignancies in humans and mice. The ability to produceAtm−/− mice and collect multiple tumor samples allowed us to compare the pattern of cytogenetic aberrations in these tumors and study the molecular mechanisms of lymphomagenesis inAtm−/− mice. Our findings indicate that lymphomagenesis in Atm−/− mice proceeds via multiple chromosomal aberrations that begin to occur during V(D)J recombination. Translocations present in these tumors suggest that loss of Atm results in the inability of T cells to manage double-stranded DNA (dsDNA) breaks, leading to chromosome aberrations and tumors.
Materials and methods
Preparation of metaphase chromosomes
Eight Atm−/− tumor samples designated AT-1, AT-4, AT-5, AT-7, AT-10, AT-11, AT-12, and AT-13 were derived from primary tumors of thymic origin as previously described.10 Primary cultures were grown in RPMI medium (Life Technologies, Bethesda, MD), supplemented with 10% heat-inactivated fetal calf serum (FCS) and 20 U/mL of human interleukin-2 (Boehringer Mannheim, Mannheim, Germany). Metaphase chromosomes for molecular cytogenetic analyses were prepared from early passages (passage 2 to 6) as described previously.10Splenocyte chromosomes were prepared from spleens cultured for 48 hours in RPMI + 20% fetal bovine serum (FBS) with the addition of 6 μg/mL concanavilin A to preferentially stimulate T cells or 25 μg/mL lipopolysaccharide to stimulate B cells. After an additional 30 minutes incubation in Colcemid (Gibco, Bethesda, MD), cells were harvested and metaphase chromosomes prepared as for the tumor cells.
Spectral karyotyping and in-situ hybridization with locus specific bacterial artificial chromosome probes
Spectral karyotyping (SKY) of tumor metaphases was performed as previously described.20 21 SKY probes were prepared from flow-sorted mouse chromosomes (kindly provided by J. Wienberg and M. Ferguson-Smith, Department of Pathology, Cambridge, UK) by degenerate oligonucleotide-primed polymerase chain reaction (PCR), incorporating haptenized or fluorochrome-conjugated nucleotides. To identify bacterial artificial chromosome (BAC) probes for in situ hybridization, PCR primers were prepared to target sequences published in the GenBank database. Using these primers, locus-specific BAC clones were isolated from Down-to-the-Well PCR pools using procedures described by the manufacturer (Genome Systems, St Louis, MO). The clone addresses for the isolated BACs are as follows: IghCα-141E18; IghV-225N21; c-Myc-51A4; Tcl1-226E11; TcrCα-232F19; TcrCδ-216F1; TcrVδ3/Vα6-46G9; TcrβC-164G11; and TcrVβ17a-23N16. Labeled BAC probes were prepared by nick-translation of the respective BAC DNA with either digoxigenin-12-dUTP, biotin-16-dUTP (Boehringer Mannheim), or Spectrum Orange-dUTP (Vysis, Downers Grove, IL). The 50 ng of labeled BAC DNA was precipitated in the presence of 5 μg of mouse Cot1 DNA (Life Technologies, Gaithersburg, MD) and resuspended in 50% formamide, 10% dextran sulfate, 2xSSC. After denaturation (5 minutes, 80°C), hybridization with BAC probes was carried out using an excess of Cot1 DNA to metaphase chromosomes on glass slides for 1 to 2 days. Biotin- and digoxigenin-labeled probes were detected with Cy5 and FITC conjugates. Samples were counterstained with DAPI and embedded in an antifade reagent containing para-phenylenediamine (Sigma Chemical, St Louis, MO).
Results
Chromosomal rearrangements are characteristic of Atm−/− mouse lymphomas
We performed comprehensive molecular cytogenetic analyses on 8 thymic lymphoblastic lymphomas from Atm−/−mice using SKY and FISH with locus-specific probes. Additionally, histopathology and flow cytometric analyses with antibodies to CD3, CD4, and CD8 cell surface markers were performed on tumors AT-4, AT-7, AT-10, AT-11, AT-12, and AT-13 to identify the stage of T-cell maturation that coincides with the emergence of malignant clones. Histologically, all tumors consisted of monomorphic lymphoblastic cells, and flow cytometry with cell surface markers revealed that they were CD3−, CD4+, and CD8+, indicating that all tumors were of immature T-cell origin. (Barlow et al10 and data not shown) The chromosomal aberrations are summarized in Table 1 and an example of SKY analysis is shown in Figure 1.Atm−/− lymphomas were diploid with a mean chromosome number of 40. Rarely were multiple karyotypes found in different cells of a tumor (less than 5% among all tumors), indicating that the tumors were derived from a predominant clone and remained clonal in culture. As shown in Table 1, thymic lymphomas exhibited chromosomal abnormalities with an average of slightly more than 4 chromosomal aberrations per tumor. The most frequent type of aberration was an unbalanced rearrangement involving 2 or more chromosomes with net deletion or duplication of genetic material. In tumor AT-4 (Figure1), the rearrangement of one copy of chromosome 12 with chromosome 14 leads to the partial deletion of chromosome 12 and partial loss of chromosome 14 material. Furthermore, AT-4 displayed a duplication and translocation of chromosome 15 material to chromosome 14, an aberration also found in several other tumors (AT-1, AT-5, AT-13; see Table 1). Two additional unbalanced translocations found in AT-4, T(14;X), and T(X;11) resulted in a partial gain of chromosome 11 and loss of chromosome 14. Whole chromosome numerical changes were less common, and there were more gains5 than losses.1 Notably, a whole gain of chromosome 15 was found in AT-7, AT-11, and AT-12 (Table 1). Other abnormalities leading to net gains and losses of genetic material were deletions and duplications of individual chromosomes, dicentric chromosomes, and a Robertsonian translocation. Balanced translocations were not observed.
Chromosomal abnormalities detected by SKY and FISH in Atm-deficient mouse thymic lymphomas
| Tumor . | Chromosomal aberrations* . | No. gains/losses† . | FISH‡ . | |
|---|---|---|---|---|
| TcrCα/Vα 14D . | IgH 12F . | |||
| AT-1 | T(12;14)1-153 T(14;Dp15)1-155 T(14;Dp3) Is(6;14) | 2/1 | Not done | Not done |
| AT-4 | T(12;14) T(14;Dp15) T(14;X) T(X;Dp11) | 2/2 | LR | LD |
| AT-5 | T(12;14) T(14;Dp15) T(14;DpX) | 2/2 | LR | LD |
| AT-7 | T(12;14) T(Is(1;14);16)1-154 Del(14)# +15 | 1/3 | LR | LD |
| AT-10 | T(12;10) Dp(14) T(X;Dp15) T(17;Dp1) Del(16) −11 | 3/4 | LR | LD |
| AT-11 | T(12;Dp6) Dic(14;14)1-160 Dp(14) Dic(15;Dp15) Rb(16.Dp16)1-164 +15 | 6/1 | LR | LD |
| AT-12 | T(12;9) T(9;Dp15) Del(14) Ace(14)1-161 +10 +13 +15 | 4/3 | LR | LD |
| AT-13 | T(12;14) T(Is(15;14);Dp15) Del(14) | 1/2 | LR | LD |
| Tumor . | Chromosomal aberrations* . | No. gains/losses† . | FISH‡ . | |
|---|---|---|---|---|
| TcrCα/Vα 14D . | IgH 12F . | |||
| AT-1 | T(12;14)1-153 T(14;Dp15)1-155 T(14;Dp3) Is(6;14) | 2/1 | Not done | Not done |
| AT-4 | T(12;14) T(14;Dp15) T(14;X) T(X;Dp11) | 2/2 | LR | LD |
| AT-5 | T(12;14) T(14;Dp15) T(14;DpX) | 2/2 | LR | LD |
| AT-7 | T(12;14) T(Is(1;14);16)1-154 Del(14)# +15 | 1/3 | LR | LD |
| AT-10 | T(12;10) Dp(14) T(X;Dp15) T(17;Dp1) Del(16) −11 | 3/4 | LR | LD |
| AT-11 | T(12;Dp6) Dic(14;14)1-160 Dp(14) Dic(15;Dp15) Rb(16.Dp16)1-164 +15 | 6/1 | LR | LD |
| AT-12 | T(12;9) T(9;Dp15) Del(14) Ace(14)1-161 +10 +13 +15 | 4/3 | LR | LD |
| AT-13 | T(12;14) T(Is(15;14);Dp15) Del(14) | 1/2 | LR | LD |
SKY = spectral karyotyping; FISH = fluorescence in situ hybridization; LR = locus rearranged; LD = locus deleted.
Chromosome rearrangements were detected by SKY analysis of at least 6 metaphase cells per tumor. Aberrations listed were present in at least 5 of the 6 metaphases. Aberrations involving chromosomes 12, 14, and 15 are shown in bold.
The number of partial and whole chromosome gains or losses identified by SKY and FISH.
The results of FISH analysis with bacterial artificial chromosome (BAC) clones to mouse IgH and TcrCα/Vδ loci. As described in the text, all tumors exhibited rearrangement within the Tcr locus on both alleles of chromosome 14 and deletion of the IgH locus on the translocated allele of chromosome 12.
T: translocation. The chromosome donating the centromere is listed first.
Dp: duplication of a chromosome segment.
Is: insertion. The inserted segment is listed first.
#Del: interstitial deletion of a chromosome segment.
Dic: a dicentric chromosome.
Rb: Robertsonian translocation.
Ace: acentric fragment of a chromosome.
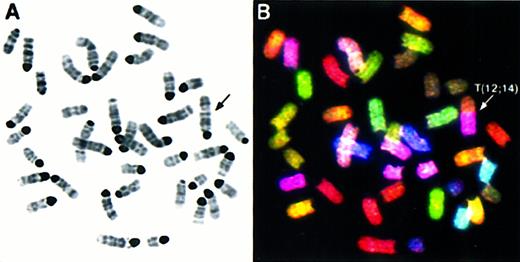
Spectral karyotyping analysis of thymic lymphoma AT-4 reveals multiple chromosomal aberrations.
SKY analysis of a representative metaphase from thymic lymphoma AT-4. The inverted DAPI-stained image is shown in (A), and the RGB display image in (B), with arrows indicating the chromosomal aberrations. The full karyotype is shown in (C), with each chromosome in its spectra-based classification color flanked by the DAPI and RGB images (for details see “Materials and methods”). The karyotype of tumor AT-4 is 39, XY, T(12;14), T(14;15); T(14;X), T(X;11).
Spectral karyotyping analysis of thymic lymphoma AT-4 reveals multiple chromosomal aberrations.
SKY analysis of a representative metaphase from thymic lymphoma AT-4. The inverted DAPI-stained image is shown in (A), and the RGB display image in (B), with arrows indicating the chromosomal aberrations. The full karyotype is shown in (C), with each chromosome in its spectra-based classification color flanked by the DAPI and RGB images (for details see “Materials and methods”). The karyotype of tumor AT-4 is 39, XY, T(12;14), T(14;15); T(14;X), T(X;11).
Mouse chromosomes 12, 14, and 15 are frequently altered in Atm−/− lymphomas
Hallmark features of Atm−/− lymphomas were intrachromosomal and interchromosomal rearrangements of chromosome 14, translocations involving chromosome 12, and gains of chromosome 15 (Table 1). In all tumors, chromosome 14 was aberrant. Notably, 15 of the 16 chromosome 14 homologues in the 8 tumors were rearranged (Table1). Frequently, chromosome 14 was involved in translocations with chromosome 15 (AT-1, AT-4, AT-5, and AT-13) and/or chromosome 12 (AT-1, AT-4, AT-5, AT-7, and AT-13) or had undergone an intrachromosomal rearrangement, leading to deletion or duplication of material relative to the diploid karyotype (AT-10, AT-11, AT-12, AT-13; Table 1). Tumor AT-4 has both the t(14;15) and the t(12;14) characteristic of manyAtm−/− tumors (Figure 1).
Mouse chromosomes 12 and 14 are homologous to human chromosome 14, which is frequently rearranged in hematologic malignancies from patients with AT. Translocations involving chromosome 12 were found in all tumors as unbalanced translocations. Therefore, chromosome 12 material was always deleted. Chromosome 12 to 14 translocations were found in 50% of the tumors. Other chromosome 12 translocation partners included chromosome 6 (AT-11), 9 (AT-12), and 10 (AT-10).
Chromosome 15 aberrations were found in 100% ofAtm−/− tumors (Table 1), all of which resulted in a gain of chromosome 15 material. As noted above, in 50% of the tumors chromosome 15 was involved in translocations with chromosome 14. Whole chromosomal gains of 15 were found in 3 tumors (37%).
The Tcrα/δ locus on chromosome 14 is disrupted in Atm−/− lymphomas
Mouse chromosome 14D1-D2 harbors the Tcrα/δlocus. The SKY results therefore suggested that the recurrent rearrangements observed in Atm−/− lymphomas recapitulated the cytogenetic events described in human lymphomas, where translocations and inversions involving the Tcrα/δlocus on chromosome 14 are frequently observed.8Furthermore, FACS analysis had shown that tumors appeared to arise from CD4/CD8 double positive T-cell precursors. Double positive T cells are produced after rearrangement of Tcrβ but before productive rearrangement of the Tcrα/δ gene. To examine whether the observed chromosome 14 abnormalities involved the Tcr loci, we prepared BAC probes from a mouse SV129 library (described in “Materials and methods”).
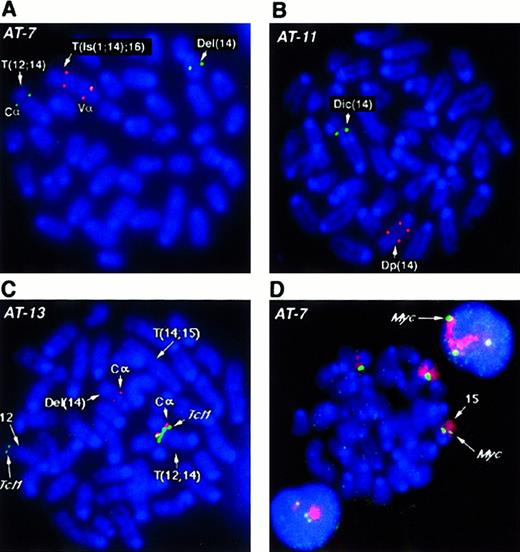
Several BAC probes to different regions of the more than 1 MBTcrα/δ locus were identified. TcrCα andTcrVδ3/Vα6 probes flank the locus, while the constant region TcrCδ maps within the gene. Results obtained withTcrCδ probes were essentially the same as those withTcrCα, therefore only those with the latter are shown. With the exception of AT-1, we analyzed all tumors forTcrα/δ gene rearrangements by fluorescence in situ hybridization (FISH) and all analyzed cases showed disruption of this locus. For example, in tumor AT-7 (Figure2A), both alleles of chromosome 14 have undergone rearrangement and translocation with breakpoints within theTcrVα/δ locus. One allele lost the TcrVαregion with a deletion of chromosomal material also detectable in the SKY analysis. In the second allele, a complex rearrangement resulted in duplication of the region containing TcrVα, an insertion of chromosome 1 material, and translocation of this rearranged chromosome 14 to chromosome 16 (T(Is(1;14);16)). TcrCα is in the portion of 14 translocated to chromosome 12. Tumor AT-11 (Figure2B) lost the TcrVα signal on the dicentric chromosome 14 allele, in addition to an intrachromosomal rearrangement, leading to separation or duplication of the second TcrVα region and loss of the TcrCα (Figure 2B). Other tumors exhibited similar rearrangements of both alleles of the Tcrα/δlocus, suggesting that rearrangements at this locus during V(D)J recombination are critical for tumorigenesis.
FISH analysis of
Tcrα/δ, Tcl1 and c-myc genes inAtm−/− lymphomas. (A, B) FISH analysis of the Tcrα/δ locus on mouse chromosome 14 with BAC probes to constant region alpha (TcrCα) and variable region alpha (TcrVα). TcrCα signals are shown in green and the TcrVα signal in red for all panels A-D as depicted in D. Chromosomes and DNA are counterstained with DAPI (blue). Structural aberrations were identified by SKY (not shown). At least 10 nuclei were examined and the presence of the same pattern of FISH signals was confirmed in the majority of nuclei. Representative metaphase nuclei are shown for tumors AT-7 (A) and AT-11 (B). (C) FISH analysis of AT-13 with BAC probes to the T-cell receptor constant region TcrCα (red) and the Tcl1 locus (green).TcrCα on chromosome 14 is translocated into the vicinity of the Tcl1 gene on chromosome 12 (T12;14). TheTcl1 locus appears to be amplified compared with the second FISH signal on a normal chromosome 12 allele. One allele of chromosome 14 with the second TcrCα has undergone deletion of theTcrVα region (Del(14); Table 1). The second allele of chromosome 14, T(14;15), has lost the TcrCα region and therefore shows no hybridization signal. Chromosomes are counterstained with DAPI (blue). (D) FISH analysis of chromosome 15 and thec-myc locus in thymic lymphoma AT-7. The AT-7 lymphoma has gained an extra copy of chromosome 15 (chromosome painted in red), along with an additional copy of the c-myc allele (green). Note the 3 c-myc FISH signals captured in the interphase nuclei above and below the metaphase plate. Chromosomes and DNA are counterstained with DAPI (blue).
FISH analysis of
Tcrα/δ, Tcl1 and c-myc genes inAtm−/− lymphomas. (A, B) FISH analysis of the Tcrα/δ locus on mouse chromosome 14 with BAC probes to constant region alpha (TcrCα) and variable region alpha (TcrVα). TcrCα signals are shown in green and the TcrVα signal in red for all panels A-D as depicted in D. Chromosomes and DNA are counterstained with DAPI (blue). Structural aberrations were identified by SKY (not shown). At least 10 nuclei were examined and the presence of the same pattern of FISH signals was confirmed in the majority of nuclei. Representative metaphase nuclei are shown for tumors AT-7 (A) and AT-11 (B). (C) FISH analysis of AT-13 with BAC probes to the T-cell receptor constant region TcrCα (red) and the Tcl1 locus (green).TcrCα on chromosome 14 is translocated into the vicinity of the Tcl1 gene on chromosome 12 (T12;14). TheTcl1 locus appears to be amplified compared with the second FISH signal on a normal chromosome 12 allele. One allele of chromosome 14 with the second TcrCα has undergone deletion of theTcrVα region (Del(14); Table 1). The second allele of chromosome 14, T(14;15), has lost the TcrCα region and therefore shows no hybridization signal. Chromosomes are counterstained with DAPI (blue). (D) FISH analysis of chromosome 15 and thec-myc locus in thymic lymphoma AT-7. The AT-7 lymphoma has gained an extra copy of chromosome 15 (chromosome painted in red), along with an additional copy of the c-myc allele (green). Note the 3 c-myc FISH signals captured in the interphase nuclei above and below the metaphase plate. Chromosomes and DNA are counterstained with DAPI (blue).
If V(D)J recombination was generally disrupted inAtm−/− mice, rearrangement of other TCR loci would be expected in tumors. However, only one tumor (AT-1), displayed abnormalities of a chromosome containing another TCR locus (Tcrβ), which maps to mouse chromosome 6. AT-1 had an insertion of chromosome 6 material into chromosome 14. However, molecular cytogenetic techniques using the BAC probes, as described previously, uncovered no evidence of rearrangement of this gene. Although it remains a possibility that smaller rearrangements do occur but are not detected by SKY or FISH with BAC probes, these results suggest that V(D)J recombination in general is not impaired by the loss of Atm.
Tcl1-Tcrα/δ gene fusions are not frequently observed in Atm−/− lymphomas
SKY consistently showed translocations involving chromosome 12 and gains of chromosome 15 in thymic lymphomas fromAtm−/− mice. Mouse chromosome 12 harbors theTcl1 oncogene, and chromosome band 15D2-D3, the c-myc oncogene. Human TCL1 and c-MYChave been implicated in T-cell leukemias from patients with AT, as well as other types of leukemias and lymphomas. To analyze the genomic rearrangements more precisely, we prepared BAC probes to theIgh and Tcl1 loci on mouse chromosome 12 and to the c-myc locus on chromosome 15.
SKY revealed T(12;14) translocations in 5 tumors analyzed in this study. These tumors were analyzed for possible fusion of theTcrα/δ locus on chromosome 14 to the Tcl1oncogene on chromosome 12. Surprisingly, we found only one tumor (AT-13) in which the Tcrα/δ locus and theTcl1 locus co-localized to the breakpoint. On the T(12;14) chromosome of AT-13 TcrCα and Tcl1 BAC probes hybridize together at the breakpoint (Figure 2C). The Del(14) chromosome has undergone rearrangement leading to loss of theTcrVα locus and the second chromosome 14 has lost theTcrCα region and undergone duplication of theTcrVα region. In all other cases (AT-1, AT-4, AT-5, and AT-7), the Tcl1 locus was deleted, whereas chromosome 14 had retained the TcrCα element on the distal side of the breakpoint. We also examined Tcl1 in the tumors without a T(12;14). In AT-10 Tcl1 was deleted, whereas in AT-11 and AT-12, Tcl1 was present on the translocation chromosome. In addition, Tcl1 was not localized to the breakpoints of the T(12;6) and T(12;9) in AT-11 and AT-12, respectively.
Igh is located telomeric to Tcl1 on chromosome 12. We further analyzed each tumor with Igh BAC probes to confirm that the loss of Tcl1 was not restricted to this particular locus but was due to a larger loss of the distal region of chromosome 12. As expected, all T(12;14) translocation chromosomes including AT-13 showed loss of the Igh locus. The tumors with other translocations involving chromosome 12 had also lost theIgH locus on the translocated chromosome. In all tumors,Igh and Tcl1 loci on the second chromosome 12 allele appeared unaffected. It therefore appears that Tcl1genomic rearrangements, although present in rare cases, are not required for tumorigenesis in Atm−/− mice. In addition, Northern blot analysis for Tcl1 expression showed that no tumor cell line expressed Tcl1, including AT-13 (data not shown).
SKY analysis also revealed partial or whole chromosome gains of chromosome 15 in all the tumors. We therefore analyzed the panel of tumors for the presence of additional copies of the c-myclocus using a BAC clone isolated from the mouse library. We found an additional copy of c-myc in every tumor that had either a partial gain (AT-1, AT-4, AT-5, AT-10, and AT-13) or a whole chromosome 15 gain (AT-7, AT-11, and AT-12) (Table 1). For example, 3 copies of c-myc were detected in AT-7 cells with the c-mycBAC probe (Figure 2D). To determine whether c-myc was overexpressed, we analyzed expression levels of c-myc RNA inAtm−/− tumors compared with the level in normal mouse thymus tissue. Analysis of total RNA from several tumors did not show a dramatic increase of c-myc message in most samples. However, in tumor AT-7, we did record a modest but reproducible 1.2-fold increase in c-myc messenger RNA (mRNA) levels (data not shown).
T cells from Atm−/− mice show chromosomal aberrations
Because T-cell lymphomas fromAtm−/− mice invariably display chromosome 14 rearrangements at the Tcrα/δ locus, we wanted to determine whether this was an early event in tumorigenesis. Therefore, we separately cultured splenic T and B cells from 2-month-oldAtm−/−mice without any evidence of tumor, and analyzed the cells for karyotypic abnormalities by SKY. In the T-cell population, 3 of 10 metaphases contained structural chromosomal rearrangements, and 2 of these were translocations involving chromosome 14. An example is shown in Figure 3 of a spleen cell with a T(12;14) similar to the T(12;14) observed in tumor AT-4 (Figure 1). One rearrangement, not involving 14, was found in the B cells (data not shown). To further examine the prevalence of chromosome 14 translocations in premalignant cells, we performed FISH with a chromosome 14 painting probe on a larger number of cells. Table2 shows the number of chromosome 14 structural abnormalities found in ATM versus wild-type spleen cells. Twenty-three of 200 (11.5%) of the cells contained translocations, deletions, or large duplications of chromosome 14 (likely an underestimate as inversions and small duplications could not be detected by these methods). Only 1 of 200 or 0.5% of the wild-type spleen cells had a chromosome 14 rearrangement. Therefore, the spleens of Atm−/− mice contain a significant number of cells with nonclonal chromosome aberrations of a single allele of chromosome 14, similar to the clonal aberrations found on both alleles of chromosome 14 in the tumors. By flow cytometry, the spleens ofAtm−/− mice did contain single-positive mature T cells (although a reduced number10). The high incidence of chromosome 14 abnormalities (implicitly rearrangements of the Tcrα/δ locus) indicates that abnormal rearrangement of both alleles of chromosome 14 is likely to be an early step in tumorigenesis. Similarly, in patients with AT, up to 10% of stimulated lymphocytes display stable translocations and inversions resembling the clonal populations found in subsequent malignancies.22
SKY analysis of
Atm−/−spleen cells reveals chromosome 14 rearrangements. Inverted DAPI-stained (A) and RGB display images (B) of a representative metaphase from anAtm−/− mouse splenic T cell. The karyotype is 40, XX, T(12;14). Band analysis indicates that the breakpoint of the translocation is in band 14D. There are 2 normal copies of chromosome 14, so the 14 material in the T(12;14) is a duplication of bands D-E.
SKY analysis of
Atm−/−spleen cells reveals chromosome 14 rearrangements. Inverted DAPI-stained (A) and RGB display images (B) of a representative metaphase from anAtm−/− mouse splenic T cell. The karyotype is 40, XX, T(12;14). Band analysis indicates that the breakpoint of the translocation is in band 14D. There are 2 normal copies of chromosome 14, so the 14 material in the T(12;14) is a duplication of bands D-E.
Number of chromosome 14 structural abnormalities in ATM versus wild-type spleen cells
A painting probe for chromosome 14 was hybridized to metaphase cells from either a wild-type mouse spleen or non-malignant Atm−/− mouse spleen. Structural abnormalities include translocations and large duplications or deletions visible by fluorescence in situ hybridization analysis.
Metaphases from 2 Atm−/− spleens were pooled.
Metaphases from one wild-type (Atm+/+) spleen.
Spleen cells were cultured separately with either concanavilin A or LPS (see “Materials and methods”) to stimulate T or B cells, respectively.
Discussion
We have analyzed lymphomas in Atm−/− mice by FACS, SKY, and FISH with probes for the Tcrα/δ locus on mouse chromosome 14. The results indicate that the tumors are of T-cell origin, and reveal chromosomal rearrangements that invariably involve the Tcrα/δ locus. Although multiple chromosomal aberrations are observed, the breakpoints cluster in chromosomes 12, 14, and 15 (Figure 4). The results recapitulate those seen in lymphomas and leukemias from patients with AT, which frequently bear rearrangements in Tcr andTcl1 loci. Similarly, c-MYC amplification on human chromosome 8q24 (mouse chromosome 15), has been shown to occur in T-prolymphocytic leukemia (T-PLL). In addition, other studies have established the association of somatic inactivation of ATMin the pathogenesis of sporadic T-PLL, suggesting that ATMacts as a tumor suppressor in certain types of leukemias.23-25 Importantly, as shown in the Oxford comparative homology map,26 93% (66 of 71) of the known homologues from human chromosome 14, the chromosome most frequently affected in AT leukemias and T-PLL either map to mouse chromosome 12 (41 of 66) or chromosome 14 (22 of 66), indicating a high degree of structural conservation of human chromosome 14 across species. It may be that defective Tcrα/δ rearrangements increase the propensity to develop lymphomas. However, the fact that chromosome 12 aberrations are found in all tumors suggests that faulty Tcrrearrangement is not sufficient for tumorigenesis. In addition, we did not find high levels of overexpression of the Tcl1 or c-myc oncogenes in our tumors. Therefore, the genes that are either abnormally activated or lost as a result of these translocations remain to be identified. Recently, genomic analysis of the mouse and human Tcl1 locus has revealed several tightly clustered, homologous genes in the breakpoint cluster regions observed in human T-cell neoplasias,27 suggesting that some of the non-Tcl1 chromosome 12 tumor breakpoints inAtm−/− mice might be found in these genes. Therefore, the potential involvement of these genes in tumorigenesis inAtm−/− mice will be investigated.
Karyogram of breakpoints and band duplications in
Atm−/− lymphomas determined by SKY and FISH analysis. Breakpoints and regions of band duplication (from the chromosome aberrations listed in Table 1) were estimated by aligning the DAPI-banded, RGB display, and classified images of each chromosome. All aberrations for which the breakpoints and regions of band duplication could be determined are shown; the question marks indicate uncertainty in the placement of the breakpoint. Filled circles indicate breakpoints and lines indicate band duplication regions. Each tumor is identified by its number in Table 1, for example AT-1 = 1.
Karyogram of breakpoints and band duplications in
Atm−/− lymphomas determined by SKY and FISH analysis. Breakpoints and regions of band duplication (from the chromosome aberrations listed in Table 1) were estimated by aligning the DAPI-banded, RGB display, and classified images of each chromosome. All aberrations for which the breakpoints and regions of band duplication could be determined are shown; the question marks indicate uncertainty in the placement of the breakpoint. Filled circles indicate breakpoints and lines indicate band duplication regions. Each tumor is identified by its number in Table 1, for example AT-1 = 1.
In addition to comparisons of the specific genes involved in the chromosome aberrations observed in mouse and human AT tumors, the results of our tumor analyses help to explain the relationship between the decrease in production of mature T cells and the predisposition to development of T-cell lymphomas in the absence of ATM. T-cell development proceeds through various stages that require the orderly progression of Tcr locus rearrangements (reviewed by Fischer and Malissen28). First, the Tcrβ locus is rearranged, followed by the Tcrα/δ locus. Immature precursor cells that express little or no CD4 or CD8, termed double-negative (DN) cells, migrate to the thymus. Tcrβrearrangement initiates in DN cells and differentiation to the subsequent developmental stage normally proceeds once a functionalTcrβ chain is expressed. Signaling mediated by the presence of these chains results in the rapid proliferation and differentiation to the CD4/CD8 double-positive (DP) stage where mostTcrα rearrangements are thought to occur. Productive rearrangement of a Tcrα allele allows expression of the mature TCR/CD3 complex that is required for further maturation to single positive (SP) CD4+ or CD8+ thymocytes. Patients with AT have reduced numbers of mature T cells. In addition, we and others have found that Atm−/− mice have abnormalities in progression from the DP to the SP stage and have reduced numbers of T cells expressing a normal Tcrα.Tumors from Atm−/− mice arise in DP cells at the time of Tcrα/δ V(D)J recombination, at the same T-cell stage where normal T-cell development is defective in these mice. Thus, these results link the T-cell developmental defects with tumorigenesis. In addition, translocations at this locus were observed in normal splenic T cells, suggesting that such translocations are an early event in the progression of lymphomas.
A recent study demonstrated that in the absence of RAG1-mediated V(D)J recombination, Rag1−/−Atm−/−double mutant mice did not develop tumors.29 However, inRag2−/−Atm−/−mice,30 the onset of tumorigenesis was substantially delayed but not eliminated. Tumors fromRag2−/−Atm−/− mice displayed chromosomal translocations, but not at the site ofTcrα/δ V(D)J recombination. Taken together, these results are most consistent with a role for ATM not specifically inTcrα rearrangement as part of a DNA repair pathway, but as part of a broader mechanism in signaling to a cell that dsDNA breaks are present, perhaps as a consequence of the ATM cell cycle checkpoint function. It is possible that the need for ATM is particularly critical in cells undergoing rapid proliferation when they are in the process of undergoing Tcrα/δ V(D)J recombination.
We propose the following model for tumorigenesis in AT, based on these results and the study of Petiniot et al.30 In the absence of ATM, successful V(D)J recombination occurs in some cells, despite the absence of normal signaling of dsDNA breaks. This leads to the production of a small number of normal functional T cells that migrate to peripheral lymphoid organs. In other cases, ATM-deficient T cells mature and differentiate, despite the presence of dsDNA breaks at theTcrα/δ locus because of cell cycle checkpoint defects in this rapidly proliferating population of DP T cells. Most of these cells undergo cell cycle arrest or apoptosis, because the presence of even a single unrepaired dsDNA break is sufficient to induce arrest,31 providing an explanation for the reduced T-cell numbers displayed by ATM-deficient mice and humans. In a subset of these cells, however, the dsDNA breaks are “repaired” by undergoing translocations with other chromosomes. Some of these translocations may provide a growth advantage as a consequence of placing an active T-cell enhancer in the Tcrα/δ locus in the proximity of a growth promoting oncogene, and clonal expansion of these cells occurs. Some of these cells will then undergo normal rearrangement of the second allele of the Tcrα/δ locus, permitting further differentiation to mature single positive T cells, which then migrate to the periphery and represent the more than 11% of cells with a single chromosome 14 rearrangement that we observed there. However, a subset of cells undergoes a translocation of the secondTcrα/δ locus. If this again results in activation of a growth promoting oncogene, clonal expansion results. On average, 2 or more rearrangements occur as a result of the inability of cells lacking ATM to signal the presence of dsDNA breaks that occur during DNA synthesis. If these additional aberrations result in activation of growth promoting genes, or the inactivation of growth suppressors, then a thymic lymphoma will occur. The frequency of these events is high enough that nearly all mice succumb to lymphomas within 6 months of age.
It is unclear why no Tcrβ translocations were observed in any of the tumors studied, whereas all tumors, except AT-10, displayed 2 Tcrα/δ rearrangements. During normal T-cell development, dsDNA breaks are generated in the Tcrβ locus as well as the Tcrα/δ locus as an early step in the process of rearrangement of these loci. Successful rearrangement of theTcrβ locus has been shown to be a critical stage in T-cell development, and aberrant rearrangements of this locus in ATM-deficient cells would therefore be expected to lead to maturational arrest, with failure to clonally expand or to progress to the DP stage of thymocyte development.32 Under these conditions, the occurrence and selection of subsequent events necessary for malignant transformation of T cells may be unlikely. In contrast, failure ofTcrα/δ rearrangement does not prevent clonal expansion or differentiation to the DP stage, hence the presence of aberrantTcrα/δ rearrangements in the thymic lymphomas ofAtm−/− mice, and the failure to detect aberrant Tcrβ translocations.
In any case, the involvement of the Tcrα/δ locus in translocations does not appear to be due solely to a role of ATM in V(D)J recombination. Rather, it may be the result of at least 3 factors. First, the Tcrα/δ locus can undergo multiple attempts at successful rearrangement. Second, V(D)J recombination produces dsDNA breaks, and cells deficient in ATM have an inability to handle such breaks efficiently. Finally, translocations of aTcr locus to a growth promoting gene will result in its activation in T cells, leading to a higher probability of thymic lymphomagenesis.
Acknowledgments
We thank Johannes Wienberg and Malcolm A. Ferguson-Smith (Department of Pathology, Cambridge, UK) for providing flow-sorted mouse chromosomes, and Danny Wangsa, Veronique Bruniquel, and Joseph Cheng for technical assistance.
M.L., Z.W., and C.B. contributed equally to this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Zoë Weaver, Genetics Department, Division of Clinical Sciences, National Cancer Institute, NIH, 9 Memorial Dr, Bldg 9, Rm 1N-105, MSC 0913, Bethesda, MD 20892; e-mail:weaverz@mail.nih.gov.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal