Abstract
Duffy antigen/receptor for chemokines (DARC) is a promiscuous receptor for chemokines that is required for Plasmodium vivax infection of erythroid cells. This receptor is expressed by subsets of endothelial, as well as erythroid cells. Selection for protection from malaria infection resulted in an erythroid-specific defect, suggesting that DARC may play a critical role in endothelial biology. Mice with targeted disruption of this gene were generated, and the function of DARC in inflammation was explored. RNA from spleens of homozygous mutant mice lacked DARC transcripts, which were abundant in wild-type (+/+) and heterozygote (+/−) mice. DARC−/− mice lacked developmental abnormalities and were healthy at 1 year. Whereas hematologic parameters were within normal ranges, erythrocytes from nullizygous mice lacked CXC and CC chemokine-binding activity. Challenge with lipopolysaccharide resulted in significantly increased inflammatory infiltrates in lung and liver of nullizygous mice. These results suggest that DARC modulates the intensity of inflammatory reactions as a sink for chemokines.
Introduction
The Duffy blood group was first described as an alloantigen in a multiply transfused hemophiliac.1 The finding that expression of this blood group protein was significantly under-represented in individuals of African ancestry, particularly in regions where the population was resistant to infection byPlasmodium vivax,2 led to the recognition that the Duffy-negative erythroid phenotype was protective for infection byP vivax.3
A second major advancement of insight into the physiologic role of the Duffy blood group antigen was the discovery that it was identical to a promiscuous receptor for chemokines expressed on the surface of erythrocytes.4 Chemokines promote the directed migration of specific leukocyte subsets.5 The chemokine family can be separated into branches based on the configuration of the 2 amino-proximal, of 4 positionally conserved, cysteine residues. In the CXC branch they are separated by a single residue, whereas they are juxtaposed in the CC branch. In general, CXC and CC chemokines promote the formation of acute and chronic inflammatory infiltrates, respectively. The receptors that transduce the signals of chemokines are members of the serpentine receptor family, and separate subsets of receptors mediate signaling by CXC and CC chemokines.6 The Duffy antigen was shown to be identical to the erythroid chemokine receptor, which has the unique capacity to bind members of both CXC and CC branches.7 In contrast to other receptors, the binding of ligands to Duffy antigen/receptor for chemokines (DARC) does not induce signal transduction,8 complicating the interpretation of its function.
The genetic mechanism for the erythroid Duffy-negative phenotype9 preserved the expression of this receptor in endothelium,10 inviting speculation that it plays an important role in the pathophysiology of inflammation.11In the current studies, the physiological role of DARC was studied in mice rendered deficient of this receptor by gene targeting. These mice showed an exaggerated inflammatory response to intraperitoneal administration of bacterial lipopolysaccharide (LPS), indicating that DARC may be a regulatory sink for chemokines.
Materials and methods
Targeted disruption of the mouse DARC gene
A 9-kilobase (kb) EcoRI fragment containing the DARC locus was cloned from an isogenic mouse genomic library. This fragment was used to generate the targeting construct shown in Figure1A, which contained 4.3-kb and 1.2-kb arms of homology in the 5′ and 3′ regions, and a neo gene in opposite transcriptional orientation that replaced 90 base pairs (bp) of the DARC open reading frame. Correctly targeted embryonic stem (ES) cell lines were injected into C57BL/6 blastocysts and subsequently implanted into CD1 host pseudopregnant females to obtain 10 chimeric pups. Three of these chimeras were able to readily transmit the DARC mutation through the germline when mated with C57BL/6 mice, resulting in F1 heterozygotes. F1 heterozygotes were intercrossed to obtain F2 homozygous DARC-deficient mice, which served as the experimental animals (along with the wild-type littermate controls) in these studies. All animal experiments were approved by institutional committees and were in compliance with National Institutes of Health guidelines.
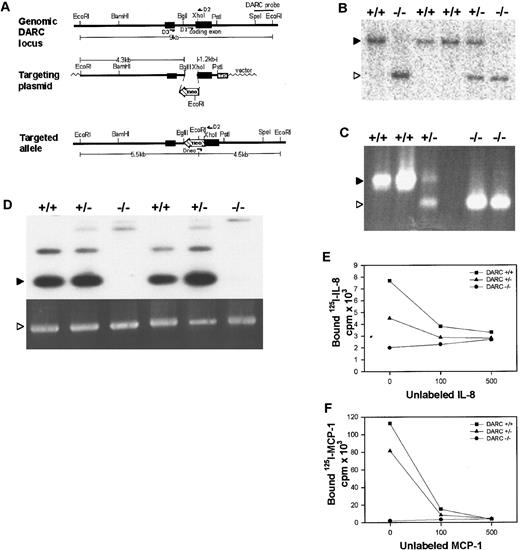
Targeted disruption of the mouse DARC gene.
(A) mDARC targeting strategy. Top line: Endogenous DARC gene locus showing coding exon I and exon II in a 9-kb EcoRI fragment. A 3′ 1-kb SpeI-EcoR1 located outside the targeting construct was used as a probe for Southern blot analysis. Primers D1 and D2 were used for PCR identification of the wild-type DARC gene, and primers D2 and D3 were used to detect DARC cDNA by RT-PCR analysis. Middle line: The targeting plasmid, a 1.1-kbneo gene, was inserted to replace theBglII-XhoI fragment. This eliminates a part of the intron and 90 bp of exon II. A 5′ 4.3-kbEcoRI-BglII fragment was used as the 5′ arm of homology and a 3′ 1.2-kb XhoI-PstI fragment containing most of the coding exon was used for the 3′ homology. A TK gene was inserted for negative selection of ES cells. The OSDUPDEL vector was used to create the targeting construct. Bottom line: The correctly targeted allele. There is a unique EcoRI site introduced with the neo gene that results in a reduction of the germline 9-kb EcoRI fragment to 4.5 kb when the DARC probe is used for Southern blot analysis. Alternatively, the correctly targeted allele can be detected by PCR analysis by using primers D2 and Dneo−. (B) Southern blot showing correct targeting at the DARC locus. Genomic DNA prepared from the tails of F2+/+, F2+/−, and F2−/− mice were digested with EcoRI and probed with the DARC probe, detecting bands of 9.0 kb (▸, wild-type) and 4.5 kb (▹, targeted). (C) PCR gel showing correct targeting at the DARC locus. Genomic tail DNA from F2+/+, F2+/−, and F2 −/− mice was analyzed by PCR analysis with the use of primers D1, D2, and Dneo, resulting in a 400-bp wild-type product (▸) and a 300-bp targeted product (▹). (D) RT-PCR analysis of DARC-deficient mice. Total RNA was isolated and cDNA was obtained from the spleens of F2 DARC+/+, F2 DARC +/−, and F2 DARC −/− mice. There was no observable difference in the expression of DARC mRNA (▸) between +/+ and+/− mice; however, we did observe an alternate splicing product that is produced in the spleen at 800 bp. GAPDH mRNA (▹) served as a control. There is a faint 1.4-kb band that results from a nonfunctional message that is produced across the neoinsert. This RT-PCR analysis indicates that DARC−/− mice have no normal DARC mRNA in their spleens. (E) Absence of IL-8 binding to DARC-deficient RBCs. For a competitive binding assay, RBCs were isolated from F2 DARC+/+, +/−, and−/− mice and incubated with 0.5 nmol/L of125I-labeled IL-8 and varying amounts of unlabeled IL-8 (0, 100, or 500 nmol/L). Each data point represents the average of duplicate samples. Similar results were obtained in 3 independent experiments. (F) Absence of MCP-1 binding to DARC-deficient RBCs. Competitive binding assays with 125I-MCP-1 were performed as described above. Similar results were obtained in 2 independent experiments.
Targeted disruption of the mouse DARC gene.
(A) mDARC targeting strategy. Top line: Endogenous DARC gene locus showing coding exon I and exon II in a 9-kb EcoRI fragment. A 3′ 1-kb SpeI-EcoR1 located outside the targeting construct was used as a probe for Southern blot analysis. Primers D1 and D2 were used for PCR identification of the wild-type DARC gene, and primers D2 and D3 were used to detect DARC cDNA by RT-PCR analysis. Middle line: The targeting plasmid, a 1.1-kbneo gene, was inserted to replace theBglII-XhoI fragment. This eliminates a part of the intron and 90 bp of exon II. A 5′ 4.3-kbEcoRI-BglII fragment was used as the 5′ arm of homology and a 3′ 1.2-kb XhoI-PstI fragment containing most of the coding exon was used for the 3′ homology. A TK gene was inserted for negative selection of ES cells. The OSDUPDEL vector was used to create the targeting construct. Bottom line: The correctly targeted allele. There is a unique EcoRI site introduced with the neo gene that results in a reduction of the germline 9-kb EcoRI fragment to 4.5 kb when the DARC probe is used for Southern blot analysis. Alternatively, the correctly targeted allele can be detected by PCR analysis by using primers D2 and Dneo−. (B) Southern blot showing correct targeting at the DARC locus. Genomic DNA prepared from the tails of F2+/+, F2+/−, and F2−/− mice were digested with EcoRI and probed with the DARC probe, detecting bands of 9.0 kb (▸, wild-type) and 4.5 kb (▹, targeted). (C) PCR gel showing correct targeting at the DARC locus. Genomic tail DNA from F2+/+, F2+/−, and F2 −/− mice was analyzed by PCR analysis with the use of primers D1, D2, and Dneo, resulting in a 400-bp wild-type product (▸) and a 300-bp targeted product (▹). (D) RT-PCR analysis of DARC-deficient mice. Total RNA was isolated and cDNA was obtained from the spleens of F2 DARC+/+, F2 DARC +/−, and F2 DARC −/− mice. There was no observable difference in the expression of DARC mRNA (▸) between +/+ and+/− mice; however, we did observe an alternate splicing product that is produced in the spleen at 800 bp. GAPDH mRNA (▹) served as a control. There is a faint 1.4-kb band that results from a nonfunctional message that is produced across the neoinsert. This RT-PCR analysis indicates that DARC−/− mice have no normal DARC mRNA in their spleens. (E) Absence of IL-8 binding to DARC-deficient RBCs. For a competitive binding assay, RBCs were isolated from F2 DARC+/+, +/−, and−/− mice and incubated with 0.5 nmol/L of125I-labeled IL-8 and varying amounts of unlabeled IL-8 (0, 100, or 500 nmol/L). Each data point represents the average of duplicate samples. Similar results were obtained in 3 independent experiments. (F) Absence of MCP-1 binding to DARC-deficient RBCs. Competitive binding assays with 125I-MCP-1 were performed as described above. Similar results were obtained in 2 independent experiments.
Genotype analysis
The genotypes of all ES cells and mice were determined by Southern blot or polymerase chain reaction (PCR) analysis. Insertion ofneo introduces a unique EcoRI site, reducing the 9-kb EcoRI fragment to 4.5 kb in the correctly targeted allele by Southern blotting with the use of a 3′ DARC probe. PCR was initially used to screen ES cells and to genotype F2 mice. The wild-type DARC allele was detected by amplification of a 400-bp PCR product, using D1 and D2 primers that flank the DARC coding exon. Primer D1 (5′-GCTAGATGTCCTGACTGTCC) is a sequence that is deleted in the correctly targeted allele. Primer D2 (5′-CCAGTAGCCCAGGTTGCATA) is complementary to sequences in exon 2. Primer Dneo(5′-TATGGCGCGCCATCGATCTC-3′) is complementary to a sequence within the inserted neo gene. D2 and Dneo were used to amplify a 300-bp fragment from the targeted DARC gene.
RT-PCR analysis
Total RNA for RT-PCR analysis was obtained from spleens of F2 mice by using TRIzol Reagent (Gibco-BRL). Complementary DNA (cDNA) was synthesized from RNA templates with a poly-T primer and M-MLV RT (Gibco-BRL). DARC cDNA was amplified by PCR, using primers D2 and D3 (5′-GCCCTGAGCCTGCAGTGCCAT-3′) in exon 1, resulting in a 400-bp product.
Red blood cell chemokine binding assay
Red blood cells (RBCs), obtained from F2 mice, were incubated with 0.5 nmol/L [125I]-labeled interleukin-8 (IL-8) and MCP-1 (DuPont NEN) and saturating amounts of unlabeled ligands. The incubation was terminated by centrifugation through an oil mixture. Erythrocyte pellets were counted in a gamma counter.
LPS challenge
LPS (Escherichia coli, 0111:B4; Sigma Chemical, St Louis, MO) was administered to wild-type and nullizygous mice by intraperitoneal injection (30 mg/kg). Control mice (+/+) received phosphate-buffered saline (PBS). Mice were killed 2 hours after injection, and tissues were sampled for myeloperoxidase (MPO) determination12 and histopathology.
Results
Chimeras generated from correctly targeted ES cells transmitted the mutation through the germline when mated with C57BL/6J mice. ES cell lines and F1 heterozygotes were genotyped by Southern blot analysis (Figure 1B). F2 mice were genotyped by PCR analysis (Figure 1C).
The nullizygous mice exhibited normal growth, development, fertility, and were healthy at 1 year. No significant differences between DARC+/+ and DARC−/− mice were detected in their standard hematological parameters (data not shown).
Normal DARC messenger RNA (mRNA) expression was completely abolished in the −/− mice as measured by RT-PCR analysis from splenic total RNA (Figure 1D), indicating the absence of DARC expression by endothelial cells. The effect of the DARC gene disruption on expression of the erythrocyte chemokine receptor was examined, using [125I]-labeled IL-8 and MCP-1. The RBCs from the DARC−/− mice showed a marked reduction in specific binding relative to the cells from control mice (Figure 1E,F), indicating that they lack expression of DARC on erythroid cells.
Histologic examination of tissues from animals treated with LPS revealed a mild granulocytic infiltrate in lung and liver of+/+ mice (Figure 2A,D). In contrast, nullizygous mice had intense granulocytic infiltrates in lung (Figure 2B) and scattered granulocytes in hepatic sinusoids, with foci of microabscess formation (Figure 2E). Organ neutrophil accumulation was assessed indirectly by measuring tissue MPO content. Lung and liver from −/− mice had significantly higher MPO levels than+/+ mice (Figure 2C,F). Mice receiving PBS injections lacked evidence of a significant infiltrate. There was no difference between +/+ and −/− mice in LPS-induced leukocyte recruitment into the peritoneal cavity (data not shown).
Organ leukocyte accumulation is augmented in DARC nullizygous mice.
Mild leukocytic infiltrates were observed in lung and liver sections from DARC+/+ mice (panels A and D, respectively). Accumulation of leukocytes was markedly increased in lungs and livers from DARC−/− mice (panels B and E, respectively). Neutrophil accumulation, as measured by tissue MPO content, was significantly greater in DARC−/− mice in lung (panel C) and liver (panel F), compared with DARC+/+ mice (*P < .05). All sections were stained with hematoxylin and eosin; original magnification: A-E, 4×; insets, 20×. For analysis of tissue MPO content, bars represent mean ± SEM with n = 5 per group.
Organ leukocyte accumulation is augmented in DARC nullizygous mice.
Mild leukocytic infiltrates were observed in lung and liver sections from DARC+/+ mice (panels A and D, respectively). Accumulation of leukocytes was markedly increased in lungs and livers from DARC−/− mice (panels B and E, respectively). Neutrophil accumulation, as measured by tissue MPO content, was significantly greater in DARC−/− mice in lung (panel C) and liver (panel F), compared with DARC+/+ mice (*P < .05). All sections were stained with hematoxylin and eosin; original magnification: A-E, 4×; insets, 20×. For analysis of tissue MPO content, bars represent mean ± SEM with n = 5 per group.
Discussion
The exaggerated inflammatory response observed in nullizygous mice suggests that DARC may serve as a chemokine sink and that normal expression of DARC regulates leukocyte trafficking during inflammation.
Note added in proof.
Following the resubmission of our manuscript, Luo et al13reported that DARC knockout mice have lower MPD values than wild-type controls in lung and intestine 24 hours after intraperitoneal injection of 10 mg/kg LPS. This represents a lower dose and a longer time interval than employed in the current study.
Acknowledgments
The authors thank Annette Staton, Kimberly Kluckman, Silvia Hiller, and Tianyvan Zhang for technical assistance and Drs Oliver Smithies, Don Cook, and Terence J. Hadley for helpful discussions. This work represents partial fulfillment of requirements for a PhD in Biochemistry and Molecular Biology (Zx.-W.).
Supported in part by the Agnes Brown Duggan Endowment, the Humana Fund for Excellence, and by National Institutes of Health grants DK56029 (A.B.L.) and HL42630 (N.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stephen C. Peiper, J.G. Brown Cancer Center, 529 South Jackson St, Louisville, KY 40202; e-mail:scp@bcc.louisville.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal