Abstract
Histone deacetylase (HDAC) inhibitors can induce transcriptional activation of a number of genes and induce cellular differentiation as histone acetylation levels increase. Although these inhibitors induce apoptosis in several cell lines, the precise mechanism by which they do so remains obscure. This study shows that HDAC inhibitors, sodium butyrate and trichostatin A (TSA), abrogate interleukin (IL)-2–mediated gene expression in IL-2–dependent cells. The HDAC inhibitors readily induced apoptosis in IL-2–dependent ILT-Mat cells and BAF-B03 transfectants expressing the IL-2 receptor βc chain, whereas they induced far less apoptosis in cytokine-independent K562 cells. However, these inhibitors similarly increased acetylation levels of histones in both cells. Although histone hyperacetylation is believed to lead to transcriptional activation, the results showed an abrogation of IL-2–mediated induction of c-myc,bag-1, and LC-PTP gene expression. This observed abrogation of gene expression occurred prior to phosphatidylserine externalization, a process that occurs in early apoptotic cells. Considering the biologic role played by IL-2–mediated gene expression in cell survival, these data suggest that its abrogation may contribute to the apoptotic process induced by HDAC inhibitors.
Introduction
Transcriptional regulation is a major event in cell differentiation, proliferation, and apoptosis. Transcriptional activation of a set of genes determines cell destination and for this reason transcription is tightly regulated by a variety of factors. One of its regulatory mechanisms involved in the process is an alteration in the tertiary structure of DNA, which affects transcription by modulating an accessibility of transcription factors to their target DNA segments.1 Nucleosomal integrity is regulated by the acetylation status of the core histones. In a hypoacetylated state, nucleosomes are tightly compacted and thus are nonpermissive for transcription. On the other hand, nucleosomes are relaxed by acetylation of the core histones, with the result being permissiveness to transcription.2 The acetylation status of the histones is governed by the balance of the activities of histone acetyl transferase (HAT) and histone deacetylase (HDAC).
Recently, HDAC inhibitors, sodium butyrate and trichostatin A (TSA), have been found to arrest growth and apoptosis in several types of cancer cells, including colon cancer, T-cell lymphoma, and erythroleukemic cells.3-5 Given that apoptosis is a crucial factor for cancer progression,6,7 these HDAC inhibitors are promising reagents for cancer therapy as effective inducers of apoptosis. However, the molecular mechanisms underlying the HDAC inhibitor-induced apoptosis remain to be elucidated. Several reports have suggested that HDAC inhibitor-treated cells undergo apoptosis as a terminal event in differentiation and maturation, based on the fact that inhibition of HDAC leads to histone hyperacetylation and transcriptional activation of several genes related to differentiation.8-10 In contrast, butyrate blocks the primary mechanisms involved in signal transduction, such as the release of Ca++ from intracellular stores and activation of serine/threonine kinases, and down-regulates expression of the c-myc gene.11 12 These data raise the possibility that inhibition of the cell survival signals may contribute to HDAC inhibitor-induced apoptosis.
We thus investigated the effects of HDAC inhibitors on cytokine-mediated cell survival signals. We first studied the apoptotic effect of HDAC inhibitors using a variety of hematopoietic cells and found that interleukin (IL)-2–dependent cells demonstrated the highest sensitivity to HDAC inhibitors and readily went into apoptosis in response to the inhibitors. Interestingly, HDAC inhibitors increased histone acetylation at similar levels in both IL-2–dependent and IL-2–independent cells, despite their different sensitivity to the inhibitors. This suggests that histone hyperacetylation alone does not account for their apoptotic effect. We thus focused on the biologic effects of HDAC inhibitors on IL-2–mediated signals, which are known to mediate cell survival signals in IL-2–dependent cells. In this report, we observed that sodium butyrate and TSA strongly suppressed IL-2–mediated gene expression prior to induction of apoptosis in 2 IL-2–dependent hematopoietic cell lines, ILT-Mat and BAF-B03 transfectant F7. Because they did not affect the IL-2–mediated tyrosine phosphorylation of SHP-2, Jak1, and STAT5, HDAC inhibitors appear to suppress IL-2–mediated gene expression selectively. These findings suggest that abrogation of IL-2–mediated gene expression may contribute to HDAC inhibitor-induced apoptosis in hematopoietic cells.
Materials and methods
Reagents and cell culture
Human IL-2–dependent leukemia cell line, ILT-Mat, and murine IL-2–dependent hematopoietic cell line BAF-B03 derived transfectant F7, expressing the human IL-2 receptor βc chain, were maintained in RPMI-1640 medium (GIBCO, Grand Island, NY) supplemented with 10% fetal calf serum (FCS) and 400 U/mL of IL-2 (kindly provided by Shionogi Chemical Pharmacy, Osaka, Japan). Human erythroleukemia cell line K562 was maintained in RPMI-1640 medium supplemented with 10% FCS. Sodium butyrate and actinomycin D were purchased from Sigma (St Louis, MO). TSA was from WAKO Pure Chemicals (Osaka, Japan).
DNA isolation and agarose gel electrophoresis
Cells were cultured with sodium butyrate or TSA for the indicated periods and their apoptotic states were evaluated with DNA fragmentation. After washing with phosphate-buffered saline (PBS), their low molecular weight genomic DNA was extracted from ILT-Mat cells with 0.5% Triton X-100, 10 mmol/L EDTA, and 10 mmol/L Tris, pH 7.4 at 4°C for 10 minutes. The DNA was treated with 400 μg/mL of RNase A for 1 hour at 37°C, incubated with 400 μg/mL of proteinase K for 1 hour at 37°C, ethanol precipitated and analyzed on 1.8% agarose gels.13 F7 cells were incubated in the lysis buffer containing 1% sodium dodecyl sulfate (SDS) and 500 μg/mL of proteinase K, and their genomic DNA was extracted from with phenol/chloroform/isoamyl alcohol and then treated with 100 μg/mL of RNase A for 1 hour at 37°C. The gels were stained with 1 μg/mL of ethidium bromide and photographed under UV light.
Histone preparation and gel fractionation
Histones were prepared by a modification of a previously described method.14 Cells (5 × 106) were exposed to sodium butyrate for 18 hours and washed twice in PBS. Cells were lysed by Dounce homogenization in 1 mL ice-cold lysis buffer (10 mmol/L Tris pH 8, 50 mmol/L NaCl, 1% Triton X-100, 10 mmol/L MgCl2, 8.6% sucrose, and 0.5 mmol/L dithiothreitol), and nuclei were collected by centrifugation for 5 minutes at 6000 rpm in a microcentrifuge. Sulfuric acid was added to a concentration of 0.4 N and the resultant supernatant was collected. Histones were precipitated by addition of 10 volumes of acetone and incubation at −20°C overnight. Histone acetylation was evaluated by fractionating histones on acid/urea/acrylamide gels. Gels were fixed and stained in 0.25% Coomassie blue/10% acetic acid/40% methanol.
Annexin V detection
Translocation of the membrane phospholipid phosphatidylserine from the inner face of the plasma membrane to the cell surface was detected by binding of annexin V according to the manufacturer's protocol15 (ApoAlert Annexin V Apoptosis Kit, Clontech, Palo Alto, CA). ILT-Mat cells were treated with 200 nmol/L of TSA for the indicated hours and 10 000 cells were analyzed by FACS.
Northern blot analysis
Total RNAs were extracted from the treated cell lines by the acid guanidium thiocyanate/phenol/chloroform method. Ten micrograms of the total RNAs was separated by 1% agarose formaldehyde gel electrophoresis and transferred onto nitrocellulose filters. The filters were hybridized with 32P-labeled complementary DNA (cDNA) fragments as probes: the full-length of the human c-myc,16 the 0.6-kb 5′-end ofLC-PTP17 and the full-length ofbag-118 cDNA fragments. The hybridization was performed under the condition of 50% formamide, 2 × standard sodium citrate (SSC) (1 × SSC is 150 mmo/L NaCl/15 mmol/L trisodium citrate), 2.5 × Denhart's solution, 0.1% SDS, 2.5% dextran sulfate, and denatured salmon testes DNA (100 μg/mL). The filters were washed with 2 × SSC at 50°C.
Western blot analysis
The ILT-Mat cells were washed with cold PBS and lysed in 100 μL of a buffer containing 100 mmol/L NaCl, 2 mmol/L EDTA, 10 mmol/L sodium orthovanadate, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 1% NP-40, and 50 mmol/L Tris (pH 7.2). Protein concentrations of the lysates were analyzed by the Protein Assay kit (BioRad, Melville, NY) and each lysate was immunoprecipitated with anti-SHP-2,19anti-Jak1 (Transduction Laboratory, Lexington, KY), or anti-STAT5A (R&D Systems, Minneapolis, MN) antibody. The immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), followed by electrophoretic transfer onto Immobilon (Millipore, Bedford, MA). The blots were incubated with blocking buffer containing 3% bovine serum albumin (BSA), 10 mmol/L Tris (pH 8.2), 140 mmol/L NaCl, and 0.01% NaN3. Then, they were incubated with either 1 μg/mL of anti-SHP-2, anti-Jak1, anti-STAT5A, or antiphosphotyrosine antibody 4G10 (Upstate Biotechnology, Lake Placid, NY) for 2 hours in washing solution 150 mmol/L NaCl, 10 mmol/L Tris (pH 7.5) and 0.01% Tween20 with 2% FCS, and washed several times in washing solution, followed by an additional 1 hour of incubation with peroxidase-conjugated antirabbit IgG or antimouse IgG antibody (Amersham Life Sciences, Arlington Heights, IL). The blots were developed by a standard ECL method.
Results
Sodium butyrate induces apoptosis in ILT-Mat cells
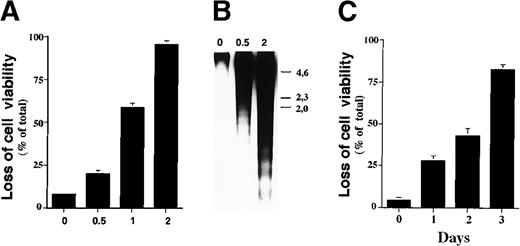
Sodium butyrate inhibits proliferation of cancer cell lines and induces expression of genes associated with maturation in a given cell lineage, with the ultimate result being apoptosis.20-22 To investigate the mechanism by which sodium butyrate induces apoptosis, we first examined its effect on cell viability using a variety of hematopoietic cell lines. In our experiments, we found that sensitivity to sodium butyrate varied among the cell lines examined (data not shown), with the human IL-2–dependent leukemia cell line, ILT-Mat, showing the highest sensitivity to the inhibitor. When ILT-Mat cells were cultured with sodium butyrate for 48 hours, cell viability was substantially reduced in a dose-dependent manner. Exposure of more than 0.5 mmol/L sodium butyrate significantly reduced cell viability of ILT-Mat cells (Figure 1A). Confirmation of the induction of apoptosis can be attained by internucleosomal fragmentation of the DNA. Electrophoresis of the low molecular weight DNA prepared from butyrate-treated ILT-Mat cells showed substantial DNA fragmentation, with the amount increasing in a dose-dependent manner (Figure 1B). We confirmed cellular morphologic changes in the butyrate-treated cells, which exhibited a shrunken cell shape characteristic of apoptosis (data not shown). We then examined the time course for butyrate-induced apoptosis. ILT-Mat cells treated with 1 mmol/L sodium butyrate gradually reduced cell viability and at day 3 of the treatment approximately 80% of the cells exhibited cell death (Figure 1C). All of these findings indicated that sodium butyrate strongly induced apoptosis in ILT-Mat cells and that a concentration as low as 0.5 mmol/L butyrate was sufficient for induction of apoptosis.
Sodium butyrate induces apoptosis in ILT-Mat cells.
(A) Dose response of the loss of cell viability by sodium butyrate. Human IL-2–dependent adult T-cell leukemia (ATL) ILT-Mat cells were treated with sodium butyrate at the indicated concentrations (mmol/L) for 48 hours. Cell viability was assessed by trypan blue exclusion assay. The results, 7.2 ± 0.6(0), 21.0 ± 2.4 (0.5), 55.3 ± 3.7(1), and 93.3 ± 2.9%(2) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) DNA fragmentation in ILT-Mat cells. The proliferating ILT-Mat cells were incubated with sodium butyrate at the indicated concentrations (mmol/L) for 48 hours. Their low molecular weight genomic DNA was extracted and analyzed on 1.2% agarose gels, which were stained with 1 μg/mL of ethidium bromide. The size marker (λ phage/Hind III-digested DNA) is indicated. (C) Time course of apoptosis in response to sodium butyrate. ILT-Mat cells were treated with sodium butyrate at 1 mmol/L for the indicated days. Cell viability was assessed by trypan blue exclusion assay. The results, 4.3 ± 1.5(0), 27.7 ± 3.2(1), 42.7 ± 4.7(2), and 82.3 ± 2.9%(3) of total cells, are averages of triplicate samples from 3 experiments ± SD.
Sodium butyrate induces apoptosis in ILT-Mat cells.
(A) Dose response of the loss of cell viability by sodium butyrate. Human IL-2–dependent adult T-cell leukemia (ATL) ILT-Mat cells were treated with sodium butyrate at the indicated concentrations (mmol/L) for 48 hours. Cell viability was assessed by trypan blue exclusion assay. The results, 7.2 ± 0.6(0), 21.0 ± 2.4 (0.5), 55.3 ± 3.7(1), and 93.3 ± 2.9%(2) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) DNA fragmentation in ILT-Mat cells. The proliferating ILT-Mat cells were incubated with sodium butyrate at the indicated concentrations (mmol/L) for 48 hours. Their low molecular weight genomic DNA was extracted and analyzed on 1.2% agarose gels, which were stained with 1 μg/mL of ethidium bromide. The size marker (λ phage/Hind III-digested DNA) is indicated. (C) Time course of apoptosis in response to sodium butyrate. ILT-Mat cells were treated with sodium butyrate at 1 mmol/L for the indicated days. Cell viability was assessed by trypan blue exclusion assay. The results, 4.3 ± 1.5(0), 27.7 ± 3.2(1), 42.7 ± 4.7(2), and 82.3 ± 2.9%(3) of total cells, are averages of triplicate samples from 3 experiments ± SD.
TSA induces apoptosis in ILT-Mat cells
Sodium butyrate has pleiotropic effects aside from inhibition of HDAC. Thus, we next investigated the apoptotic effect of a specific HDAC inhibitor, TSA.23 Exposure of more than 100 nmol/L TSA for 48 hours markedly reduced viability of ILT-Mat cells (Figure 2A). TSA-treated cells showed a substantial amount of DNA fragmentation, and the amount increased in a dose-dependent manner (Figure 2B). When ILT-Mat cells were incubated with 100 nmol/L TSA for 2 days, the treated cells exhibited a substantial amount of DNA fragmentation. These findings indicate that these 2 HDAC inhibitors strongly induce apoptosis in ILT-Mat cells.
TSA induces apoptosis in ILT-Mat.
(A) Dose response of cell death in ILT-Mat cells by TSA. ILT-Mat cells were treated with TSA at the indicated concentrations (nmol/L) for 48 hours and investigated by trypan blue dye exclusion assay. The results, 4.3 ± 1.5(0), 24.6 ± 2.1(50), 51.7 ± 6.7(100), and 83.0 ± 7.6%(200) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) DNA fragmentation in ILT-Mat cells. Proliferating ILT-Mat cells were incubated with TSA at the indicated concentrations (nmol/L) for 48 hours. Low molecular DNAs were extracted from the treated and untreated cells and loaded onto 1.2% agarose gel with the size marker (λ phage/Hind III-digested DNA).
TSA induces apoptosis in ILT-Mat.
(A) Dose response of cell death in ILT-Mat cells by TSA. ILT-Mat cells were treated with TSA at the indicated concentrations (nmol/L) for 48 hours and investigated by trypan blue dye exclusion assay. The results, 4.3 ± 1.5(0), 24.6 ± 2.1(50), 51.7 ± 6.7(100), and 83.0 ± 7.6%(200) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) DNA fragmentation in ILT-Mat cells. Proliferating ILT-Mat cells were incubated with TSA at the indicated concentrations (nmol/L) for 48 hours. Low molecular DNAs were extracted from the treated and untreated cells and loaded onto 1.2% agarose gel with the size marker (λ phage/Hind III-digested DNA).
Sodium butyrate and TSA did not induce apoptosis substantially in K562 cells
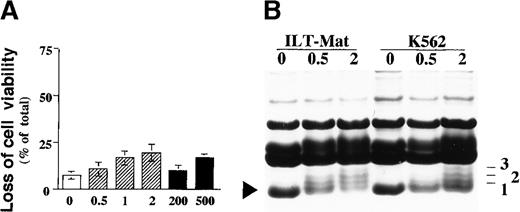
Although sodium butyrate is a nontoxic natural product and is normally found in the large intestine, the possibility remains that a high concentration of sodium butyrate and the synthetic compound TSA may represent nonspecific toxicity to these cells. To evaluate whether HDAC inhibitors induce apoptosis through their nonspecific toxic effects, we examined further the apoptotic effects of these inhibitors in human erythroleukemia K562 cells, which grow independently of IL-2. Treatment with 2 mmol/L sodium butyrate or 500 nmol/L TSA for 2 days did not induce cell death substantially in K562 cells (Figure3A), indicating that HDAC inhibitor-induced apoptosis in ILT-Mat cells was not due to their nonspecific toxic effects at the concentration used in the present study.
Effect of HDAC inhibitors on K562 cells and histone hyperacetylation in ILT-Mat and K562 cells in response to butyrate.
(A) K562 cells were cultured for 48 hours without (open column) or with sodium butyrate (shaded column) or TSA (closed column) at the indicated concentrations (mmol/L or nmol/L, respectively). Cell viability was assessed by trypan blue dye exclusion assay. The results, 7.3 ± 0.8(0), 10.6 ± 3.8 (0.5), 16.6 ± 3.6(1), 19.4 ± 4.4(2), 8.4 ± 1.0(200), and 16.2 ± 0.6%(500) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) Histone acetylation in ILT-Mat and K562 cells after butyrate treatment was evaluated by fractionating histones on acid/urea/acrylamide gels. Cells (5 × 106) were exposed to sodium butyrate at the indicated concentration (mmol/L) for 18 hours. Histones were extracted and loaded onto SDS-PAGE gels that were fixed and stained in 0.25% Coomassie blue/10% acetic acid/40% methanol. The lowest band in each lane is unacetylated H4, and the numbers to the right indicate the number of acetylated lysines in H4.
Effect of HDAC inhibitors on K562 cells and histone hyperacetylation in ILT-Mat and K562 cells in response to butyrate.
(A) K562 cells were cultured for 48 hours without (open column) or with sodium butyrate (shaded column) or TSA (closed column) at the indicated concentrations (mmol/L or nmol/L, respectively). Cell viability was assessed by trypan blue dye exclusion assay. The results, 7.3 ± 0.8(0), 10.6 ± 3.8 (0.5), 16.6 ± 3.6(1), 19.4 ± 4.4(2), 8.4 ± 1.0(200), and 16.2 ± 0.6%(500) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) Histone acetylation in ILT-Mat and K562 cells after butyrate treatment was evaluated by fractionating histones on acid/urea/acrylamide gels. Cells (5 × 106) were exposed to sodium butyrate at the indicated concentration (mmol/L) for 18 hours. Histones were extracted and loaded onto SDS-PAGE gels that were fixed and stained in 0.25% Coomassie blue/10% acetic acid/40% methanol. The lowest band in each lane is unacetylated H4, and the numbers to the right indicate the number of acetylated lysines in H4.
Hyperacetylation of histones was similarly observed in ILT-Mat and K562 cells
To explore the correlation between HDAC inhibitor-mediated apoptosis and histone hyperacetylation, we investigated the level of histone acetylation in ILT-Mat and K562 cells after the treatment with sodium butyrate. Isolated histones were fractionated on acid/urea/polyacrylamide gels, and visualized by staining with Coomassie blue. The H4 band at the bottom of each lane represents unacetylated H4, which was predominantly found in untreated cells. In ILT-Mat cells treated with 0.5 mmol/L butyrate, there was a modest increase in H4 acetylation, and treatment with 2 mmol/L butyrate resulted in a clear accumulation of diacetylated and triacetylated H4 (Figure 3B). In contrast to the fact that K562 cells did not show a significant loss of viability in response to treatment with 0.5 mmol/L butyrate, the treatment clearly increased the accumulation of acetylated H4 (Figure 3). A similar effect was found in TSA-treated cells (data not shown). The above findings strongly suggest that histone hyperacetylation is not directly linked to HDAC inhibitor-induced apoptosis.
Sodium butyrate and TSA block IL-2–mediated gene expression
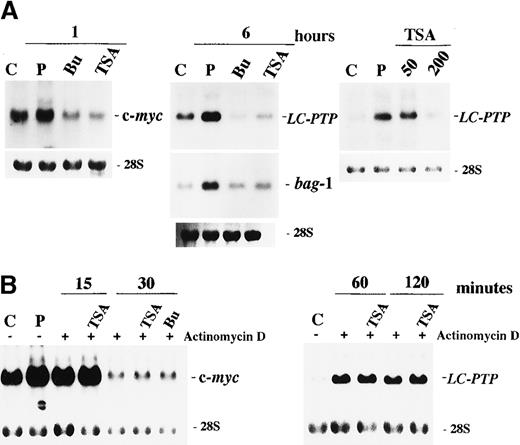
The preferential effect of HDAC inhibitors on apoptosis in IL-2–dependent ILT-Mat cells led us to consider that the inhibitors may affect IL-2–mediated signaling. Numerous studies have investigated intracellular signal transductions mediated through the IL-2 receptor (IL-2R). There are at least 2 functional regions in the IL-2Rβc chain. The acidic (A) region is required for induction of c-fos /c-jun gene expressions.24 The serine-rich (S) region is a crucial site for association with Syk25 and Jak1,26 and is essential for the induction of c-myc gene expression and for subsequent cellular proliferation.27 To explore the mechanism(s) underlying HDAC inhibitor-induced apoptosis, we investigated their effects on IL-2–mediated gene expression. We initially investigated whether HDAC inhibitors affected the S region-mediated IL-2 signal via an induction of c-myc gene expression. When cells were deprived of IL-2 for 24 hours and then restimulated with IL-2 for 1 hour, a substantial increase in c-myc messenger RNA (mRNA) levels was observed in control cells. In contrast, in cells treated with either butyrate or TSA for 30 minutes before IL-2 stimulation, no induction of c-myc mRNA expression was observed (Figure4A).
Effect of butyrate and TSA on IL-2-mediated gene expression in ILT-Mat cells.
(A) Butyrate and TSA abrogated the IL-2-mediated induction of c-myc, LC-PTP, and bag-1 genes. ILT-Mat cells were treated with 2 mmol/L butyrate (Bu) or 200 nmol/L TSA 0.5 hours prior to stimulation with IL-2. Following IL-2 stimulation for 1 or 6 hours, RNAs were extracted from the cells. Dose dependency for suppression of LC-PTP mRNA induction was analyzed (right panel). Cells were treated with 50 to 200 nmol/L TSA 0.5 hours prior to stimulation with IL-2 and incubated for 6 hours. (B) Butyrate and TSA did not affect stability of c-myc and LC-PTP (right panel) mRNAs. ILT-Mat cells were stimulated with IL-2 for 1 or 6 hours and thereafter treated with 20 μg/mL of actinomycin D with or without 200 nmol/L TSA or 2 mmol/L butyrate (Bu). RNAs were extracted from the cells at the times shown. RNAs were also extracted from the deprived cells as a control (C) and from the cells stimulated with IL-2 as a positive control (P). The 28S ribosomal RNAs are shown for the comparison of relative amounts of total RNA loaded.
Effect of butyrate and TSA on IL-2-mediated gene expression in ILT-Mat cells.
(A) Butyrate and TSA abrogated the IL-2-mediated induction of c-myc, LC-PTP, and bag-1 genes. ILT-Mat cells were treated with 2 mmol/L butyrate (Bu) or 200 nmol/L TSA 0.5 hours prior to stimulation with IL-2. Following IL-2 stimulation for 1 or 6 hours, RNAs were extracted from the cells. Dose dependency for suppression of LC-PTP mRNA induction was analyzed (right panel). Cells were treated with 50 to 200 nmol/L TSA 0.5 hours prior to stimulation with IL-2 and incubated for 6 hours. (B) Butyrate and TSA did not affect stability of c-myc and LC-PTP (right panel) mRNAs. ILT-Mat cells were stimulated with IL-2 for 1 or 6 hours and thereafter treated with 20 μg/mL of actinomycin D with or without 200 nmol/L TSA or 2 mmol/L butyrate (Bu). RNAs were extracted from the cells at the times shown. RNAs were also extracted from the deprived cells as a control (C) and from the cells stimulated with IL-2 as a positive control (P). The 28S ribosomal RNAs are shown for the comparison of relative amounts of total RNA loaded.
We then investigated the effects of HDAC inhibitors on the A region-mediated signal and the S region-mediated antiapoptotic signal, both of which are different from the signal responsible for c-myc gene induction. We previously demonstrated that IL-2 stimulation increases the mRNA levels of the cytoplasmic tyrosine phosphatase LC-PTP and the antiapoptotic proteinbag-1 genes. These increases in mRNAs became detectable within 3 hours and peaked at 6 to 9 hours after IL-2 stimulation.28 29 We also showed that the expression of these 2 genes was induced differently through 2 separate signaling pathways. Specifically, the mRNA expressions of LC-PTP andbag-1 required the presence of the A and the S regions within the IL-2Rβc chain, respectively. ILT-Mat cells were deprived of IL-2 for 24 hours, exposed to 2 mmol/L butyrate or 200 nmol/L TSA for 30 minutes and then stimulated with IL-2 for 6 hours. Although LC-PTP and bag-1 mRNAs were clearly induced by IL-2 stimulation in control cells, their induction was almost completely abrogated by treatment with butyrate or TSA (Figure 4A). In addition, TSA abrogated the IL-2–mediated LC-PTP mRNA expression in a dose-dependent manner, and a concentration as low as 200 nmol/L TSA was sufficient to suppress induction. These findings imply that the HDAC inhibitors abrogated the IL-2–mediated c-myc,LC-PTP, and bag-1 gene expression, although they use their own signaling pathway for their induction.
HDAC inhibitors did not affect c-myc and LC-PTP mRNA stability
To preliminary explore the mechanisms for suppression of IL-2–mediated gene expression, we investigated effects of HDAC inhibitors on c-myc and LC-PTP mRNA stability. To inhibit new transcription, actinomycin D was added to ILT-Mat cells that had been stimulated with IL-2 for 1 hour. Then, decay of c-mycmRNA levels was monitored either in the presence or absence of the HDAC inhibitors. Exposure of TSA or sodium butyrate to ILT-Mat cells did not virtually affect decay of the c-myc transcripts (Figure 4B). Similarly, there was no difference in decay of LC-PTP transcripts between TSA-treated cells and control cells, indicating that HDAC inhibitors did not affect the stability of these mRNAs.
HDAC inhibitors abrogated the IL-2–mediated gene expression prior to phosphatidylserine externalization
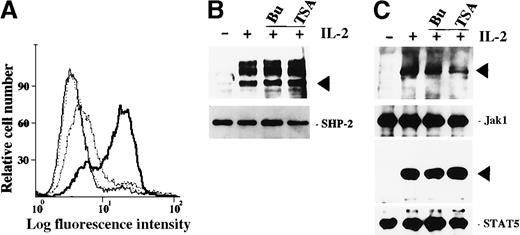
To exclude the possibility that abrogation of the IL-2–mediated gene expression was caused by HDAC inhibitors-induced cell damage, we investigated phosphatidylserine externalization (PE) in ILT-Mat cells treated with TSA. PE occurs in apoptotic cells at the early stage and thus is useful as an early detection signal of apoptosis. When ILT-Mat cells were exposed to 200 nmol/L TSA for 27 hours, PE began to increase and reached a point whereby at 36 hours after the exposure, more than 65% of the treated cells demonstrated PE (Figure5A). In contrast, ILT-Mat cells exposed to 200 nmol/L TSA for 18 hours showed no distinct PE, indicating that the HDAC inhibitors abrogated the IL-2–mediated gene expression prior to induction of apoptosis.
Roles of butyrate and TSA on IL-2-mediated signalings in ILT-Mat cells.
(A) Annexin V-FITC staining of ILT-Mat cells. ILT-Mat cells were incubated with 200 nmol/L TSA for 18 (stippled line), 27 (dashed line), or 36 hours (solid line) and stained with annexin V-fluorescein isothiocyanate (FITC). Untreated cells were also stained (thin line). Shown is a representative experiment. (B) HDAC inhibitors did not affect IL-2–mediated elevation of tyrosine phosphorylation levels in SHP-2. ILT-Mat cells were starved for 20 hours and treated with IL-2 (+) for 10 minutes with or without preincubation of 2 mmol/L butyrate (Bu) or 500 nmol/L TSA (TSA) for 0.5 hours. (C) HDAC inhibitors did not affect IL-2-mediated tyrosine phosphorylation of Jak1 and STAT5. ILT-Mat cells deprived of IL-2 for 20 hours were treated with IL-2 (+) for 15 minutes with or without preincubation of 2 mmol/L butyrate (Bu) or 500 nmol/L TSA (TSA) for 0.5 hours. SHP-2, Jak1, or STAT5 was immunoprecipitated from the lysates of ILT-Mat cells (lower panels) and their tyrosine phosphorylation levels were analyzed by immunoblotting using 4G10 (upper panels). The positions of SHP-2, Jak1, or STAT5 are indicated by arrows.
Roles of butyrate and TSA on IL-2-mediated signalings in ILT-Mat cells.
(A) Annexin V-FITC staining of ILT-Mat cells. ILT-Mat cells were incubated with 200 nmol/L TSA for 18 (stippled line), 27 (dashed line), or 36 hours (solid line) and stained with annexin V-fluorescein isothiocyanate (FITC). Untreated cells were also stained (thin line). Shown is a representative experiment. (B) HDAC inhibitors did not affect IL-2–mediated elevation of tyrosine phosphorylation levels in SHP-2. ILT-Mat cells were starved for 20 hours and treated with IL-2 (+) for 10 minutes with or without preincubation of 2 mmol/L butyrate (Bu) or 500 nmol/L TSA (TSA) for 0.5 hours. (C) HDAC inhibitors did not affect IL-2-mediated tyrosine phosphorylation of Jak1 and STAT5. ILT-Mat cells deprived of IL-2 for 20 hours were treated with IL-2 (+) for 15 minutes with or without preincubation of 2 mmol/L butyrate (Bu) or 500 nmol/L TSA (TSA) for 0.5 hours. SHP-2, Jak1, or STAT5 was immunoprecipitated from the lysates of ILT-Mat cells (lower panels) and their tyrosine phosphorylation levels were analyzed by immunoblotting using 4G10 (upper panels). The positions of SHP-2, Jak1, or STAT5 are indicated by arrows.
HDAC inhibitors did not affect IL-2–mediated tyrosine phosphorylation of SHP-2, Jak1, and STAT5
To explore further how HDAC inhibitors abrogate IL-2–mediated gene expression, we then investigated whether HDAC inhibitors affected IL-2–mediated tyrosine phosphorylation. In a previous study, we showed that the cytoplasmic tyrosine phosphatase, SHP-2, is phosphorylated in response to IL-2 on tyrosine residues by Src family kinases through the A region of the IL-2Rβc chain.30 When ILT-Mat cells were deprived of IL-2 for 24 hours and thereafter stimulated with IL-2, SHP-2 and its associated p90 and p130 proteins were immediately tyrosine phosphorylated (Figure 5B). Neither the addition of butyrate nor TSA before IL-2 stimulation affected the elevation of tyrosine phosphorylation levels in SHP-2 and its associated proteins. We also investigated whether HDAC inhibitors affected tyrosine phosphorylation of Jak1 and STAT5A in response to IL-2. Tyrosine phosphorylation of these proteins requires the S region within the IL-2Rβc chain.26 27 The addition of butyrate or TSA did not inhibit the elevation of tyrosine phosphorylation levels in Jak1 and STAT5 (Figure 5C). These findings indicated that the HDAC inhibitors did not affect the activation of Src and Jak family tyrosine kinases nor subsequent tyrosine phosphorylation of SHP-2 and STAT5. In turn, this implies that the HDAC inhibitors affected the IL-2-mediated signals at a level further downstream of Src/Jak family tyrosine kinases.
Sodium butyrate and TSA block IL-2–mediated gene expression in BAF cells that express IL-2Rβc chain
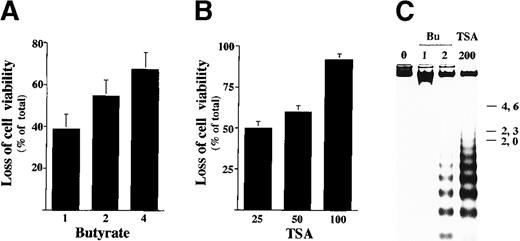
To investigate whether the HDAC inhibitors generally affected IL-2–mediated gene expression, we analyzed the effect of TSA in another IL-2-dependent cell BAF-B03 subclone (F7) that expresses the IL-2Rβc chain. Exposure of less than 2 mmol/L butyrate or 100 nmol/L TSA for 48 hours markedly reduced the viability of F7 cells (Figure6A,B). Cells treated with HDAC inhibitors for 28 hours showed a substantial amount of DNA fragmentation (Figure6C). The above findings indicated that the IL-2–dependent F7 cells showed a high sensitivity to HDAC inhibitors as the ILT-Mat cells did. To investigate the effect of TSA on IL-2–mediated gene expression in F7 cells, the cells were first deprived of IL-2 for 18 hours and then restimulated with IL-2 for 1 to 6 hours. As in the ILT-Mat cells, the F7 cells treated with 200 nmol/L TSA for 30 minutes prior to IL-2 stimulation showed no induction of c-myc mRNA expression (Figure 7A). To examine whether HDAC inhibitors specifically affect IL-2-mediated gene expression, we analyzed the effect of a representative apoptotic inducer ara-C on induction of c-myc mRNA in F7 cells. More than 80% of cells treated with 100 μmol/L ara-C for 48 hours revealed cell death (data not shown); however, exposure of ara-C for 30 minutes prior to IL-2 stimulation did not affect induction of c-myc mRNA expression (Figure 7A). In addition, treatment with 50 to 200 nmol/L TSA almost completely abrogated the induction of LC-PTP mRNAs (Figure7B). The above results imply that abrogation of the IL-2–mediated gene expression may be a unique event in HDAC inhibitor-induced apoptotic cells and may be commonly observed in IL-2–dependent cells.
Sodium butyrate and TSA induce apoptosis in F7 cells.
(A) Dose response of cell death in F7 cells by butyrate. F7 cells were treated with 1 to 4 mmol/L sodium butyrate for 48 hours and investigated by trypan blue dye exclusion assay. The results, 38.7 ± 7.0(1), 54.7 ± 7.6(2), and 67.3 ± 8.1%(4) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) F7 cells were treated with 25 to 100 nmol/L TSA for 48 hours and investigated by trypan blue dye exclusion assay. The results, 50.0 ± 4.0(25), 60.1 ± 3.9(50), and 91.7 ± 3.8%(100) of total cells, are averages of triplicate samples from 3 experiments ± SD. (C) DNA fragmentation in F7 cells. Proliferating F7 cells were incubated with sodium butyrate (Bu) or TSA at the indicated concentrations (mmol/L or nmol/L, respectively) for 48 hours. Genomic DNAs were extracted from the treated and untreated cells, and loaded onto 1.2% agarose gel with the size marker (λ phage/Hind III-digested DNA).
Sodium butyrate and TSA induce apoptosis in F7 cells.
(A) Dose response of cell death in F7 cells by butyrate. F7 cells were treated with 1 to 4 mmol/L sodium butyrate for 48 hours and investigated by trypan blue dye exclusion assay. The results, 38.7 ± 7.0(1), 54.7 ± 7.6(2), and 67.3 ± 8.1%(4) of total cells, are averages of triplicate samples from 3 experiments ± SD. (B) F7 cells were treated with 25 to 100 nmol/L TSA for 48 hours and investigated by trypan blue dye exclusion assay. The results, 50.0 ± 4.0(25), 60.1 ± 3.9(50), and 91.7 ± 3.8%(100) of total cells, are averages of triplicate samples from 3 experiments ± SD. (C) DNA fragmentation in F7 cells. Proliferating F7 cells were incubated with sodium butyrate (Bu) or TSA at the indicated concentrations (mmol/L or nmol/L, respectively) for 48 hours. Genomic DNAs were extracted from the treated and untreated cells, and loaded onto 1.2% agarose gel with the size marker (λ phage/Hind III-digested DNA).
Effect of TSA on IL-2–mediated gene expression in F7 cells.
(A) TSA abrogated the IL-2–mediated induction of c-myc gene expression. F7 cells were treated with (+) or without (−) 200 nmol/L TSA or 100 μmol/L ara-C 0.5 hour prior to stimulation with IL-2 (+) for 1 hour. (B) TSA abrogated the IL-2–mediated LC-PTP gene expression. F7 cells were treated with 50 or 200 nmol/L TSA 0.5 hour prior to stimulation with IL-2 for 6 hours. RNAs were extracted from the treated cells and also from the cells which received no treatment as a negative control (C) and from the cells stimulated with IL-2 alone as a positive control (P). The 28S ribosomal RNAs are shown for the comparison of relative amounts of total RNA loaded.
Effect of TSA on IL-2–mediated gene expression in F7 cells.
(A) TSA abrogated the IL-2–mediated induction of c-myc gene expression. F7 cells were treated with (+) or without (−) 200 nmol/L TSA or 100 μmol/L ara-C 0.5 hour prior to stimulation with IL-2 (+) for 1 hour. (B) TSA abrogated the IL-2–mediated LC-PTP gene expression. F7 cells were treated with 50 or 200 nmol/L TSA 0.5 hour prior to stimulation with IL-2 for 6 hours. RNAs were extracted from the treated cells and also from the cells which received no treatment as a negative control (C) and from the cells stimulated with IL-2 alone as a positive control (P). The 28S ribosomal RNAs are shown for the comparison of relative amounts of total RNA loaded.
Discussion
The present findings demonstrated that HDAC inhibitors strongly induced apoptosis in IL-2–dependent hematopoietic cells, whereas they induced apoptosis much less strongly in cytokine-independent cells. Inhibition of HDAC generally leads to histone hyperacetylation and a conformational change and ultimately to a relaxation of nuclear DNA.8 23 The relaxed form of nuclear DNA appears to be easily catalyzed by endonucleases, and thus these cells readily undergo apoptosis. The different apoptosis sensitivity to HDAC inhibitors may be caused by different acetylation levels of histones. However, histone hyperacetylation alone could not have accounted for the HDAC inhibitor-induced apoptosis, given that the butyrate-treated K562 cells demonstrated a substantial increase in H4 acetylation at a similar level as the same-treated ILT-Mat cells, whereas the K562 cells did not undergo apoptosis markedly. This leads us to conclude that HDAC inhibitor-induced apoptosis is not always linked to histone hyperacetylation and that other molecular events may be involved in the process.
To date, the understanding of how HDAC inhibitors induce apoptosis has been poor. In the present study, 2 HDAC inhibitors strongly suppressed the IL-2-mediated induction of gene expression. In contrast, a strong apoptotic inducer ara-C did not affect the gene expression. Furthermore, the inhibitory effect of TSA correlated well with its proapoptotic effect, given that 50 nmol/L TSA substantially induced apoptosis and suppressed the IL-2-mediated LC-PTP gene expression in F7 cells, whereas the same treatment did not markedly affect either cell viability or its gene expression in ILT-Mat cells. In contrast, treatment with 200 nmol/L TSA abrogated the IL-2-mediatedLC-PTP gene expression and induced a substantial level of cell death (87 ± 5.6%) in ILT-Mat cells. Given that IL-2–mediated gene expression is required for survival of IL-2–dependent cells, the above findings suggest that its abrogation may contribute to HDAC inhibitor-induced apoptosis. However, we are nevertheless not able to exclude the possibility that other molecular events may also be involved in the apoptotic process induced by HDAC inhibitors.
Recent findings have indicated that transcription is generally up-regulated as the levels of acetylated histones increase as mentioned above.1,2 In particular, TSA has been reported to increase the expression of gelsolin, histone H1, cytokeratin A, c-fos, c-myc, hsp70, and HDAC mRNAs.30-35 However, several studies have demonstrated that TSA down-regulates the expression of cytokine IL-2, IL-8, and the cyclin-dependent kinase inhibitor p57Kip2 mRNAs.35-37Thus, HDAC inhibitors are believed to modulate gene expression either positively or negatively in a gene-specific manner. In addition to these findings, the present study has demonstrated the negative effects of HDAC inhibitors on IL-2–mediated signalings. Although HDAC inhibitors broadly suppressed IL-2–mediated gene expression, for example, c-myc, LC-PTP, and bag-1 mRNAs, they did not affect the stability of these transcripts. Moreover, they did not suppress an increase in tyrosine phosphorylation levels of SHP-2, Jak1, and STAT5 in response to IL-2 stimulation. In this regard, HDAC inhibitors may selectively inhibit transcriptional machinery in the IL-2–mediated signalings.
Although the precise mechanism by which HDAC inhibitors abrogate IL-2–mediated gene expression remains to be elucidated, the results of the present study and those of past studies suggest the possibility of several mechanisms. First, IL-2–mediated gene expression may be sensitive to histone hyperacetylation, and thus preferentially suppressed by HDAC inhibitors. Secondly, the observed abrogation may be caused by hyperacetylation of transcription factors or molecules that participate in IL-2–mediated signaling. In particular, CBP (CREB-binding protein) has been reported to acetylate the transcription factor, TCF, resulting in repression of TCF function.38Finally, the abrogation may be caused by an inhibition of intracellular signaling processes.
The present findings suggest that HDAC inhibitors elicit apoptosis in IL-2–dependent cells via an inhibition of cell survival signals. Because TSA inhibits the IL-2 mRNA expression36 and IL-2–mediated gene expression, as the present study observed, TSA appears to be a strong inducer of apoptosis in T cells, and thus may be a good candidate as a therapeutic agent for T-cell malignancy. We propose that HDAC inhibitors should be included in trial studies to investigate whether they are useful for the treatment of hematopoietic malignancies, given that hematopoietic cells require cytokine-mediated signals for their survival and proliferation.
Acknowledgments
We thank Drs J. C. Reed and S. Takayama (The Burnham Institute, La Jolla, CA) for providing the BAG-1 cDNA probe. We also thank Drs K. Sugamura (Tohoku University, Sendai, Japan) and T. Taniguchi (Tokyo University, Tokyo, Japan) for providing the ILT-Mat cell line and BAF-B03 F7 transfectant, respectively.
Supported by a Research Grant of the Princess Takamatsu Cancer Research Fund (M.A.) and a Grant-in-Aid for Scientific Research on Priority Areas-Cancer of Ministry of Education, Science, Sports and Culture (K.I., M.A.), Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Masaaki Adachi, The First Department of Internal Medicine, Sapporo Medical University School of Medicine, S1, W16, Chuo-ku, Sapporo,060-8543 Japan; e-mail:adachi@sapmed.ac.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal