Abstract
Pluripotent hematopoietic stem cells have been studied extensively, but the events that occur during their differentiation remain largely uncharted. To develop a system that allows the differentiation of cultured multipotent progenitors by time-lapse fluorescence microscopy, myelomonocytic cells were labeled with green fluorescent protein (GFP) in vivo. This was achieved by knocking the enhanced GFP (EGFP) gene into the murine lysozyme M (lys) locus and using a targeting vector, which contains a neomycin resistant (neo) gene flanked by LoxP sites and “splinked” ends, to increase the frequency of homologous recombination. Analysis of the blood and bone marrow of thelys-EGFP mice revealed that most myelomonocytic cells, especially mature neutrophil granulocytes, were fluorescence-positive, while cells from other lineages were not. Removal of the neogene through breeding of the mice with the Cre-deleter strain led to an increased fluorescence intensity. Mice with an inactivation of both copies of the lys gene developed normally and were fertile.
During blood cell formation hematopoietic progenitors proliferate and differentiate, ultimately generating 8 well-defined lineages. While some bipotent intermediates in adult bone marrow have been defined by in vitro colony assays, the existence of other bi-, tri-, and quatri-potent progenitors remains contentious. The exact relationships between several hematopoietic lineages remain unclear because it has not yet been possible to directly observe individual steps of lineage specification and cell maturation during the division of mammalian hematopoietic cells. Such studies with developing nematodes have given invaluable information about lineage relationships, intermediate progenitors, and programmed cell death during embryogenesis.1 Time-lapse studies with colony-forming hematopoietic cells are difficult to perform because lineage assignments of live cells can only be performed during the late stages of maturation, when their morphologic features are clearly recognizable. And, although earlier stages can be recognized by immunostaining of lineage-specific cell surface markers, this procedure interrupts colony development. For this reason we have decided to generate mouse lines in which blood cells from a specific lineage are labeled in vivo, by expression of different fluorescent proteins. When placed in culture, multipotent progenitors from these mice should become fluorescence-positive as they differentiate into specific cell types, a process that can be followed by video microscopy.
We generated a mouse line in which EGFP was expressed specifically in the myelomonocytic lineage. This was accomplished by using homologous recombination to insert the EGFP gene into the lysozyme M ( lys) locus. This latter marker was used because it is expressed specifically in myelomonoytic cells (macrophages and neutrophil granulocytes)2 3 and is likely to encode a gene product that is not essential for viability of the animals. Indeed, thelys knock-in mouse line and the cultures derived from it showed specific fluorescence in macrophages and neutrophil granulocytes. These mice should become useful for the analysis of lineage relationships and for functional studies of myelomonocytic cells in transplantation experiments employing normal and leukemic mouse models.
Materials and methods
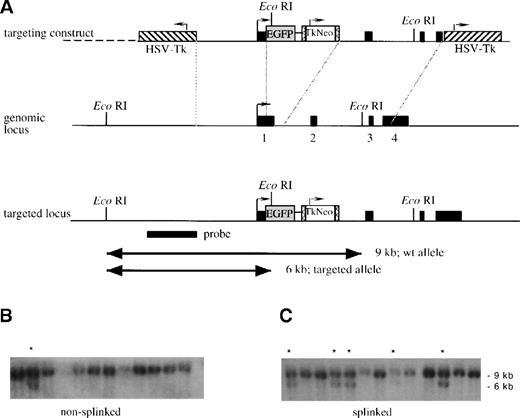
A genomic clone of the lys gene,2 covering the region from −6900 to +9150 relative to the transcriptional start site,2,4 was isolated from a 129/Sv mouse genomic library (lPS5) (gift from T. Boehm, Max Planck Institute, Freiburg, Germany). To generate the targeting construct, a 6.7-kb BamHI/HindIII fragment (position −2400 to +4300) was subcloned, simultaneously introducing a unique XbaI site at position +9 (20-bp [base pair] 5′ of the translation start) and deleting a 350-bp PvuII/NcoI fragment containing the coding part of exon 1 (including the start codon) and parts of intron 1. The NotI site of pEGFP-1 (Clontech Laboratories, Heidelberg, Germany) was inactivated, and a 1.3-kb fragment containing the thymidine kinase–neomycin resistant (Tk-neo) cassette from pPNT,6flanked by LoxP sites, was inserted into the DraIII site of this plasmid. The resulting EGFP-Lox-Tk-neo Lox cassette was cloned into the XbaI site of the modifiedlys subclone. To flank the targeting construct by Tk genes, it was inserted into the SalI site of the pKO vector.5 The targeting construct was linearized by NotI, and while half of the preparation was left untreated as a control, the other half was ligated to a NotI compatible splinker.7 The splinker was generated by self-annealing of the following oligonucleotide: 5′-GGCCGGGTACCGCTTTTGCGGTACCC-3′. The gel-purified fragments were then electroporated into R1 ES cells,8 and the clones were isolated after positive-negative selection with G418 and ganciclovir.9 The gene targeting event was identified by Southern blot analysis (Figure 1).
Strategy for insertion of the EGFP gene into thelys locus.
(A) A schematic representation of the linearized targeting construct and the genomic locus before and after recombination. The 4 exons of the lys gene and the probe used are shown as solid black boxes. The transcriptional direction of lysozyme is indicated by an arrow, theEGFP gene as a shaded box, and the LoxP-sites as boxes containing “less than” signs. The Tk-neo cassette, with its transcriptional direction (arrow), is indicated with an open box, and the HSV-Tk genes, with their transcriptional directions (arrows), are indicated with hatched boxes. After linearization, the targeting construct was either left untreated, nonsplinked (B), or splinked (C) by ligating to oligonucleotides forming hairpin “splinkers” in order to protect the fragment ends from exonucleolytic degradation. The fragments were then electroporated into R1 ES cells, the cells were cultured in G418 and ganciclovir, and double-resistant clones were isolated. Finally genomic DNA was prepared, digested with EcoRI, and analyzed by Southern blot analysis with the indicated probe. The 9-kb band in panels B and C corresponds to the fragment obtained from wild type clones; the 6-kb band corresponds to the fragment generated by homologous recombination in some clones. (These are indicated with a star).
Strategy for insertion of the EGFP gene into thelys locus.
(A) A schematic representation of the linearized targeting construct and the genomic locus before and after recombination. The 4 exons of the lys gene and the probe used are shown as solid black boxes. The transcriptional direction of lysozyme is indicated by an arrow, theEGFP gene as a shaded box, and the LoxP-sites as boxes containing “less than” signs. The Tk-neo cassette, with its transcriptional direction (arrow), is indicated with an open box, and the HSV-Tk genes, with their transcriptional directions (arrows), are indicated with hatched boxes. After linearization, the targeting construct was either left untreated, nonsplinked (B), or splinked (C) by ligating to oligonucleotides forming hairpin “splinkers” in order to protect the fragment ends from exonucleolytic degradation. The fragments were then electroporated into R1 ES cells, the cells were cultured in G418 and ganciclovir, and double-resistant clones were isolated. Finally genomic DNA was prepared, digested with EcoRI, and analyzed by Southern blot analysis with the indicated probe. The 9-kb band in panels B and C corresponds to the fragment obtained from wild type clones; the 6-kb band corresponds to the fragment generated by homologous recombination in some clones. (These are indicated with a star).
Genotypes of lys-EGFP-ki mice were determined by polymerase chain reaction (PCR) using the following primers:MLYSUP, 5′-AAGCTGTTGGGAAAGGAGGG-3′;EGFPDWN, 5′-GTCGCCGATGGGGGTGTTCT-3′; andMLP1, 5′-TCGGCCAGGCTGACT CCATA-3′. The reactions were performed with 100 ng tail DNA and all 3 primers (0.2 μmol/LMLYSUP, 0.1 μmol/L EGFPDWN, and 0.1μmol/LMLP1) for 30 cycles each at 30 seconds denaturation at 94°C, 30-second annealing at 60°C, and 60-second elongation at 72°C. The products were analyzed on a 1.5% agarose gel. PrimersMLYSUP and MLP1 amplify a 220-bp product from the wt allele, whereas MLYSUP and EGFPDWN together amplify a 680-bp product from the knock-in allele.
Blood was collected from tail veins using heparinized capillaries that were subsequently flushed with phosphate-buffered saline (PBS). The cells were then treated with lysis buffer (155 mmol/L NH4Cl, 10 mmol/L KHCO3, and 0.1 mmol/L EDTA [ethylenediamine tetraacetic acid]) to lyse mature erythroid cells. Cells for cytometric analyses were stained with ethidium bromide, and dead cells were gated out. To prepare the microscopic images, an inverted microscope with an oil immersion phase 3 objective (× 60; Olympus, Melville, NY) and an air-cooled CCD camera (Roper Scientifics, Tucson, AZ) was used. The fluorescent image was deconvoluted using Hazebuster software (Vaytek, Fairfield, IA) and Photoshop 3.0 software (Adobe Systems, San Jose, CA). For bone marrow preparations, mice were killed by cervical dislocation, and the bone marrow cells were flushed from femur cavities with PBS.
For antibody staining, 105 nucleated cells were pelleted by centrifugation, washed once in PBS containing 1% bovine serum albumin (BSA), and then resuspended in 10 μL appropriately diluted primary antibody. After 15 minutes of incubation on ice, 150 μL PBS/BSA was added, and the cells were pelleted, washed once with PBS/BSA, and resuspended in 10 μL secondary antibody (phycoerythrin-conjugated [PE-conjugated] goat antirat antibody) (Dianova, Hamburg, Germany). After 10 minutes of incubation on ice, 150 μL PBS/BSA was added, the cells were centrifuged again, washed once with PBS/BSA, and resuspended in 150 μL PBS/BSA. Subsequently, the cells were analyzed by 2-color flow cytometry using a fluorescence activated cell sorter (FACS) (FACScan, Becton Dickinson, San Jose, CA). Primary antibodies were directed against the following antigens: Mac-1,10 ER-MP12,11,12 ER-MP20,11Ly-6G,13 CD3, Ter119, B220, and Sca-1.14 Bone marrow cells used for sorting were pretreated with lysis buffer. Sorting was performed with a FACStar Plus (Becton Dickinson), excluding nonviable cells after propidium iodide staining.
For colony assays in plasma clots, 4 × 104 freshly prepared bone marrow cells were resuspended in 1 mL IMDM (Iscove's modified Dulbecco's medium) supplemented with 10% fetal calf serum (FCS), 0.3 mmol/L monothioglycerol, 50 μg/mL Vitamin C, 200 μg/mL transferrin, and 5% conditioned medium from L929 cells as a source of macrophage–colony-stimulating factor (M-CSF). We then added 60 μL citrated bovine plasma (Sigma Chemical Co, Poole, England) and 0.5 units of thrombin, and the mixture was quickly transferred to a 24-well plate, where it was allowed to clot. After incubation for 6 days at 37°C and 5% carbon dioxide (CO2) in a humidified incubator, the clots were transferred to slides and dried. Fluorescence micrographs were taken from some of the colonies before the clots were completely dried, and the colonies were marked. The cells were then fixed in methanol, stained with a May-Grünwald Giemsa stain (Diff-Quik; DADE Behring, Dudingen, Germany), and the marked colonies were photographed under brightfield.
Results
To generate mice expressing EGFP, specifically in cells of the myelomonocytic lineage, the gene targeting vector shown in Figure 1was constructed. The EGFP gene was inserted immediately downstream of the transcriptional start site of the lysgene.2 The targeting vector contained 2 copies of the herpes simplex virus (HSV) Tk gene at the 5′ and 3′ ends as a negative selectable marker,9 and the neogene was used a positive selection marker. The neo gene was flanked by LoxP sites, which are recognized by the bacteriophage P1 Cre recombinase. Homologous recombination will result in loss of theTk sequences. Therefore, stable transfectants that have recombined randomly and retain the Tk gene(s) should be susceptible to killing by ganciclovir.
In an attempt to enhance the efficiency of homologous recombination, we tested the effect of the “splinker ligation” technique. This method is based on the use of hairpin structure-forming oligonucleotides that protect the transfected construct from exonuclease attack. It has been shown to improve the efficiency of negative selection by ganciclovir in murine erythroleukemia cells transfected with targeting constructs containing the HSV Tk gene.7 To determine whether the technique can also be used to increase the relative frequency of homologous recombinants among G418/ganciclovir double-resistant ES cell colonies transfected with the targeting construct, 2 parallel experiments were performed. In the first, the NotI-linearized lysozyme targeting construct wastransfected without further treatment (“nonsplinked” construct). In the second, the construct was ligated to NotI-compatible splinkers, which are designed to protect the (open) DNA ends by hairpin structures (“splinked” construct). After transfection and selection with G418 or G418 plus ganciclovir, the colonies were counted. Although the number of G418-resistant clones was slightly lower with the splinked targeting construct, negative selection by ganciclovir was about 3 times more efficient in colonies transfected with this construct than with the nonsplinked construct (Table1).
The splinker ligation technique increases the yield of homologous recombinant ES cell clones
| . | No. of colonies/plate . | Enrichment factor . | No. of homologous recombinant clones/ no. of clones analyzed (%) . | |
|---|---|---|---|---|
| G418 . | G418 plus ganciclovir . | |||
| Nonsplinked | 820 | 360 | 2.3 | 2/25 (8) |
| Splinked | 560 | 80 | 7 | 22/42 (28) |
| . | No. of colonies/plate . | Enrichment factor . | No. of homologous recombinant clones/ no. of clones analyzed (%) . | |
|---|---|---|---|---|
| G418 . | G418 plus ganciclovir . | |||
| Nonsplinked | 820 | 360 | 2.3 | 2/25 (8) |
| Splinked | 560 | 80 | 7 | 22/42 (28) |
The targeting construct was linearized with NotI and either transfected without further treatment (nonsplinked) or after ligation to NotI-compatible splinkers (splinked). The cells were then transfected, and colonies were selected in the presence of G418 with or without negative selection by ganciclovir. The enrichment factor was calculated by dividing the number of colonies obtained by G418 selection over the number of colonies obtained by selection in G418 plus ganciclovir. Finally, surviving clones were isolated and analyzed by Southern blot analysis for homologous recombination.
To determine whether the proportion of clones with homologous recombination is enriched in the double-resistant ES cell colonies transfected with the splinked construct, we made a Southern blot analysis with an external probe. The targeting frequency of the nonsplinked construct was 8%, while the frequency for the splinked construct was 28% (Figure 1 and Table 1), which exactly reflects the factor of enrichment observed after negative selection. Five clones that had scored positively in the first round were subjected to additional Southern blot analyses with different restriction enzymes and a neo probe. This was done to rule out additional random integrations and to confirm the correct structure of the targeted locus (data not shown). After blastocyst injection, 2 of these ES cell clones resulted in germ line transmission, and the corresponding mice were designated lys-EGFP-ki.
To analyze EGFP expression in lys-EGFP-ki–positive orlys-EGFP-ki–negative mice, peripheral blood was collected from the tail veins of 1 litter of mice at weaning. Peripheral blood leukocytes were analyzed by flow cytometry, and the mice were genotyped by PCR. Between 14% and 44% fluorescent cells could be detected in the peripheral blood leukocytes of heterozygous lys-EGFP-kimice, whereas no fluorescent-positive cells were seen in their wild type littermates (less than 0.1%) (Figure2A,B). The PCR bands identified the genotypes of knock-in mice (Figure 2E). Most fluorescence-positive cells were larger than erythrocytes. They could be identified by their morphology under epifluorescent illumination as polymorphonuclear granulocytes because they exhibited nuclear structures characteristic for this cell type (Figure 2F). In addition, about 1 in 50 of the green cells showed band-shaped nuclei characteristic of monocytes. Cells isolated from the peritoneum contained 5%-15% EGFP+cells. This percentage increased to about 60% when the cells were seeded in tissue culture plates for 1 day. Nonadherent cells were removed, thereby revealing cells with the morphology typical of peritoneal macrophages. Approximately 10% of positive cells were extremely bright (Figure 3A). The proportion of EGFP+ cells did not further increase after a 48-hour incubation with 500 ng/mL lipopolisaccharide (F.V. and T.G., unpublished data, July, 1999). Cryosections revealed the presence of EGFP+ cells in the lung and liver and in cell suspensions from uteri of mice in estrus (F.V., T.G., and Jian Li, unpublished data, July, 1999), thereby reflecting the presence of resident macrophages in these organs.
Analysis of blood leukocytes from lys-EGFP-kimice.
(A-D) FACS analysis of blood samples collected from the tail veins. Red cells were lysed to obtain the leukocyte fraction. After suspension in PBS, the samples were analyzed for GFP fluorescence, and the data were plotted as relative fluorescence intensity (Y axis) versus the relative cell number (X axis). The numbers in each panel represent the percentage of fluorescent-positive cells in the respective sample. The following animals were used: (A) control mice; (B) heterozygous lys-EGFP mice, not neo-deleted; (C) heterozygous lys-EGFP mice, neo-deleted; (D) homozygous lys-EGFP mice, neo-deleted. The following average fluorescence intensities plus or minus SD were calculated from 4-5 animals in each group: (A) less than 2, (B) 92.6 ± 51.5, (C) 194.6 ± 50.1, and (D) 625 ± 396.3. (E) Genotypic analysis. Genomic DNA was prepared from mouse tails and analyzed by PCR using primers that amplify a 220-bp fragment from the wild type allele and a 680-bp fragment from the targeted allele. (F) Micrographs of a fresh blood sample from a lys-EGFP+/kimouse were made using phase contrast (left) and fluorescence microscopy (middle). The picture shown on the right was obtained by overlaying the 2 images. (Scale bar, 10 μm.)
Analysis of blood leukocytes from lys-EGFP-kimice.
(A-D) FACS analysis of blood samples collected from the tail veins. Red cells were lysed to obtain the leukocyte fraction. After suspension in PBS, the samples were analyzed for GFP fluorescence, and the data were plotted as relative fluorescence intensity (Y axis) versus the relative cell number (X axis). The numbers in each panel represent the percentage of fluorescent-positive cells in the respective sample. The following animals were used: (A) control mice; (B) heterozygous lys-EGFP mice, not neo-deleted; (C) heterozygous lys-EGFP mice, neo-deleted; (D) homozygous lys-EGFP mice, neo-deleted. The following average fluorescence intensities plus or minus SD were calculated from 4-5 animals in each group: (A) less than 2, (B) 92.6 ± 51.5, (C) 194.6 ± 50.1, and (D) 625 ± 396.3. (E) Genotypic analysis. Genomic DNA was prepared from mouse tails and analyzed by PCR using primers that amplify a 220-bp fragment from the wild type allele and a 680-bp fragment from the targeted allele. (F) Micrographs of a fresh blood sample from a lys-EGFP+/kimouse were made using phase contrast (left) and fluorescence microscopy (middle). The picture shown on the right was obtained by overlaying the 2 images. (Scale bar, 10 μm.)
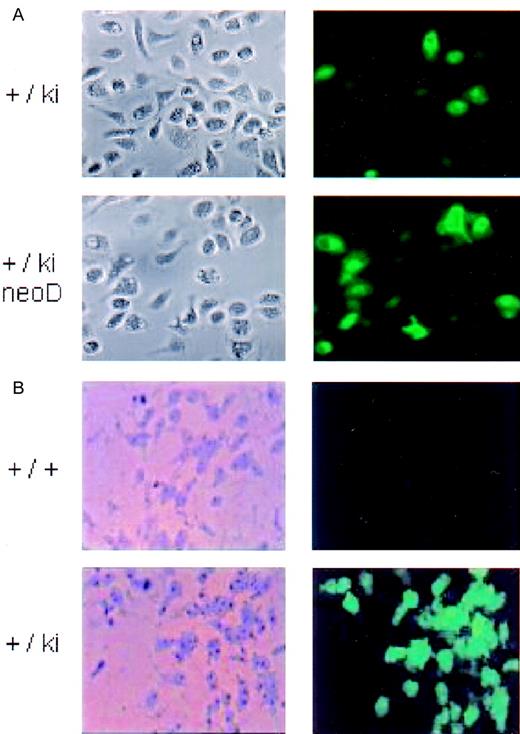
Micrographs of cultured peritoneal macrophages and bone marrow–derived colonies.
Left image, bright field; right image, fluorescence. (A) Peritoneal blood macrophages from heterozygous lys-EGFP not neo-deleted (+/ki) and neo-deleed (+/ki=neo D) mice 1 day after seeding in culture. (B) Macrophage colonies. Bone marrow cells prepared from a wild type (+/+) and from a lys-EGFP+/ki mouse were seeded in plasma clot cultures containing M-CSF. After 7 days, the plasma clots were partially dehydrated, and fluorescence pictures were taken of the colonies (right panels). Subsequently, the clots were fixed in methanol and stained with Diff-Quik, and the identical colonies were photographed under bright field (left panels).
Micrographs of cultured peritoneal macrophages and bone marrow–derived colonies.
Left image, bright field; right image, fluorescence. (A) Peritoneal blood macrophages from heterozygous lys-EGFP not neo-deleted (+/ki) and neo-deleed (+/ki=neo D) mice 1 day after seeding in culture. (B) Macrophage colonies. Bone marrow cells prepared from a wild type (+/+) and from a lys-EGFP+/ki mouse were seeded in plasma clot cultures containing M-CSF. After 7 days, the plasma clots were partially dehydrated, and fluorescence pictures were taken of the colonies (right panels). Subsequently, the clots were fixed in methanol and stained with Diff-Quik, and the identical colonies were photographed under bright field (left panels).
To study the EGFP expression in colonies developed in culture, bone marrow cells of lys-EGFP-ki mice were seeded in plasma clot cultures containing M-CSF. The resulting macrophage colonies contained bright green fluorescing cells that were not seen in similar colonies obtained from a control wild type mouse (Figure 3B). However, only about half of the adherent macrophage colonies from the knock-in mice were fluorescence-positive, with some variegation in fluorescence intensity.
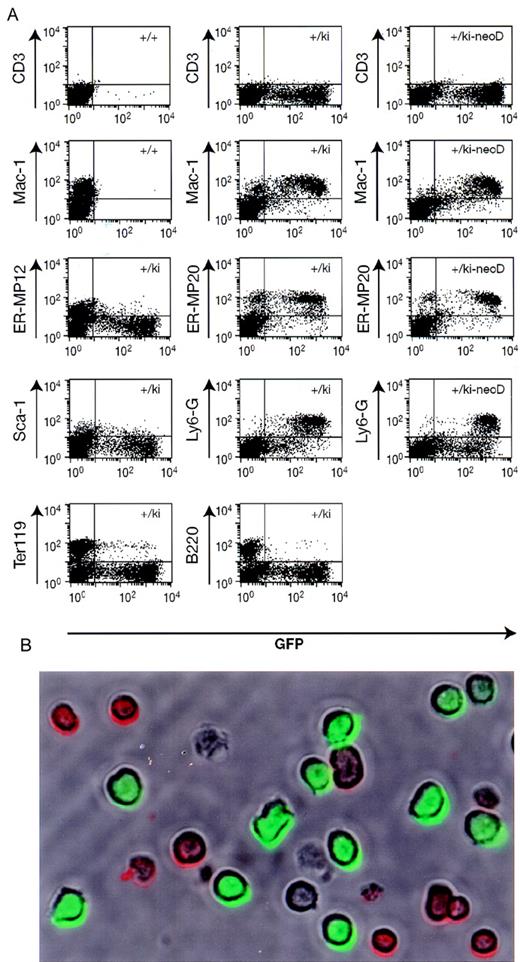
To confirm that the fluorescence is confined to cells of the myelomonocytic lineage, bone marrow of heterozygous lys-EGFP-kimice was observed by phase contrast and fluorescence microscopy. Approximately 20% of the cells were fluorescence-positive, of which about 20% were brightly fluorescent and resembled myelocytes and neutrophil granulocytes, with occasional monocyte-like cells. The remaining 80% of the cells were weakly positive, showed mostly cytoplasmic fluorescence, and had a blast-like morphology with larger nuclei. In addition, we analyzed bone marrow cells of the same mice by staining them with lineage-specific antibodies (detected by PE-coupled secondary antibodies), and flow cytometry demonstrated that the EGFP+ cells expressed the myelomonocytic-specific antigen Mac-1, the granulocyte-specific Ly6-G (or Gr-1) antigen, as well as the ER-MP20 antigen, which is specific for macrophage precursors from the monoblast stage onwards (Figure4).11 In contrast, the cells did not significantly express the ER-MP12 antigen, which is present on more immature progenitors but is no longer found on monoblasts.11 Likewise, the GFP+ cells were largely negative for Sca-1, a marker for early hematopoietic progenitors. Finally, they neither expressed the B-cell specific marker B220 nor the erythroid-specific marker Ter119, although in both cases a small proportion of the population scored double-positive.
Immunofluorescence analysis of bone marrow cells.
(A) Bone marrow was prepared from wild type mice (+/+), lys-EGFP+/ki mice, and mice in which the neo gene had been deleted (+/ki-neoD). The cells were stained with various primary antibodies (indicated on the left), and a PE-labeled secondary antibody was followed by flow-cytometry analysis. The profiles show GFP fluorescence (green) on the horizontal axis and antibody-mediated fluorescence (red) on the vertical axis. The percentage of cells in each of the 3 quadrants containing fluorescence-positive cells (antibody only, antibody/GFP, and GFP only) is indicated either within the respective quadrant or immediately adjacent to it. Specificity of the antibodies: CD3, T cells; Mac-1, macrophages and myelomonocytic cells; ER-MP12, immature monocytic cells; ER-MP20, monocytic (and some granulocytic) cells; Ly6-G (Gr-1), neutrophil granulocytes; Sca-1, early multilineage progenitors; Ter119, erythroid cells; and B220, cells of the B-cell lineage. (B) Micrograph of live cells from alys-EGFP-ki/ki-neoD mouse. Cells were stained with anti-B220 antibody coupled to PE and photographed under a brightfield to reveal antibody-positive cells (red surface staining) as well as under epifluorescence illumination to reveal EGFP+ cells (green cells). The field contains 9 B220+ cells, 11 EGFP+ cells, and 2 double-negative cells (as well as 2 cell ghosts).
Immunofluorescence analysis of bone marrow cells.
(A) Bone marrow was prepared from wild type mice (+/+), lys-EGFP+/ki mice, and mice in which the neo gene had been deleted (+/ki-neoD). The cells were stained with various primary antibodies (indicated on the left), and a PE-labeled secondary antibody was followed by flow-cytometry analysis. The profiles show GFP fluorescence (green) on the horizontal axis and antibody-mediated fluorescence (red) on the vertical axis. The percentage of cells in each of the 3 quadrants containing fluorescence-positive cells (antibody only, antibody/GFP, and GFP only) is indicated either within the respective quadrant or immediately adjacent to it. Specificity of the antibodies: CD3, T cells; Mac-1, macrophages and myelomonocytic cells; ER-MP12, immature monocytic cells; ER-MP20, monocytic (and some granulocytic) cells; Ly6-G (Gr-1), neutrophil granulocytes; Sca-1, early multilineage progenitors; Ter119, erythroid cells; and B220, cells of the B-cell lineage. (B) Micrograph of live cells from alys-EGFP-ki/ki-neoD mouse. Cells were stained with anti-B220 antibody coupled to PE and photographed under a brightfield to reveal antibody-positive cells (red surface staining) as well as under epifluorescence illumination to reveal EGFP+ cells (green cells). The field contains 9 B220+ cells, 11 EGFP+ cells, and 2 double-negative cells (as well as 2 cell ghosts).
Figure 4 also shows that a small percentage of ER-MP20+cells scored negative for EGFP fluorescence. This could be due to the fact that EGFP is only expressed by the most mature myelomonocytic cells or because levels of EGFP were below detection. Alternatively, it might be due to an interference of the neo selection marker with expression of the targeted gene, as had been demonstrated for a similar knock-in of the β globin locus.15 We therefore deleted the floxed neo gene by crossing lys-EGFP-ki mice with a Cre-deleter strain.16 Heterozygous animals of both strains were crossed, and the offspring was analyzed by Southern blot analysis. Out of 18 animals, 10 had inherited the lys-EGFP-ki allele. Of these, 6 animals showed 100% deletion of the neo cassette in their tail DNA, whereas 4 animals showed no detectable deletion (data not shown). It is thus highly likely that the neo cassette was deleted in all mice that had inherited the lys-EGFP-ki allele as well as the Cre transgene. Animals that showed deletion of theneo gene in their tail DNA transmitted only theneo-deleted form to their progeny, indicating that the deletion had also occurred with 100% efficiency in germ cells.
There was a slight increase in the number of EGFP+ cells in the blood leukocytes of heterozygous Lys EGFP mice (Figure 2C). However, there was a statistically significant (2-fold) increase in fluorescence intensity, which increased another 3-fold in theneo-deleted homozygous lys EGFP knock-in animals (Figure 2, legend). The proportion of EGFP+ peritoneal macrophages also increased in the cells derived fromneo-deleted animals (from about 60% to 95%) (Figure 3B), although the very bright cells seen before deletion were no longer apparent. Also, the proportion of EGFP+myeloid type colonies obtained from bone marrow increased significantly. The overall expression level of EGFP in the bone marrow increased somewhat again, with approximately a 2-fold increase in the EGFP fluorescence intensity of ER-MP20+ and Ly6-G+ cells in neo-deleted mice (Figure 4, right column). We therefore conclude that expression of the knocked-inEGFP gene was impaired by the neo cassette.
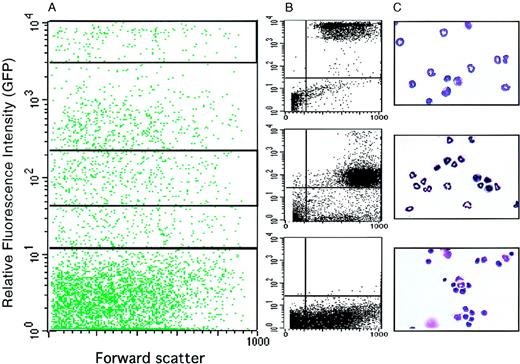
To further characterize the identity of the EGFP-expressing cells, bone marrow cells were sorted by FACS into GFP-high, GFP-low, and GFP− populations (Figure5A,B). They were then cytocentrifuged onto slides, stained with benzidine and Diff-Quik, and evaluated by microscopic inspection (Figure 5C). The highly GFP+fraction contained 52% mature neutrophil granulocytes, 40% myelocytes (promyelocytes and metamyelocytes), 4% monocytes, and 3% nonmyeloid cells. The weakly GFP+ cells contained 13% granulocytes, 80% myelocytes, and 4% other cells. Finally, the GFP− cells comprised 85% erythroid and lymphoid cells and only 4.5% myelomonocytic cells.
Morphology of EGFP+ cells sorted from the bone marrow of lys-EGFP+/ki mice.
Bone marrow cells were prepared and sorted by FACS. (A) FACS profile showing relative fluorescence intensity of GFP+ cells versus forward scatter, a parameter that is proportional to the cell's volume. The profile was subdivided by gating into a highly positive fraction (2 × 103 to 1 × 104, top quadrant), a moderately positive fraction (4.5 × 101 to 2.3 × 102, middle quadrant), and a negative fraction (100 to 101, bottom quadrant). (B) Cell profiles are shown from these 3 gated areas after sorting. (C) Micrographs of the sorted cells stained with Diff-Quik (a May-Grünwald Giemsa–like stain).
Morphology of EGFP+ cells sorted from the bone marrow of lys-EGFP+/ki mice.
Bone marrow cells were prepared and sorted by FACS. (A) FACS profile showing relative fluorescence intensity of GFP+ cells versus forward scatter, a parameter that is proportional to the cell's volume. The profile was subdivided by gating into a highly positive fraction (2 × 103 to 1 × 104, top quadrant), a moderately positive fraction (4.5 × 101 to 2.3 × 102, middle quadrant), and a negative fraction (100 to 101, bottom quadrant). (B) Cell profiles are shown from these 3 gated areas after sorting. (C) Micrographs of the sorted cells stained with Diff-Quik (a May-Grünwald Giemsa–like stain).
Discussion
Hematopoietic cells from the myelomonocytic lineage can be labeled in vivo by inserting the EGFP gene into the lys locus. Perhaps surprisingly, the most brightly positive cells were found to be mature neutrophil granulocytes, followed by their precursors and by monocytic cells. This mouse line should enable us to visualize the transition of multipotent progenitors to myelomonocytic cells in culture by observing the onset of EGFP expression using time-lapse fluorescence microscopy.
The observed 3-fold increase in the proportion of homologous recombinant ES cell clones, through the use of a splinker-protected targeting construct, indicates that this technique will be useful for introducing other targeting vectors that contain the Tk gene as a negative selection marker. This should also make it easier to generate mice expressing different GFP forms in various hematopoietic lineages because the number of clones to be analyzed can be significantly reduced, and thus several transfections can be performed in parallel. It has been suggested that the splinkers act by protecting the transfected DNA fragment from exonucleolytic degradation before integration, thus preventing destruction of the Tk gene. If the gene is destroyed, ganciclovir-resistant colonies will exhibit random integrations.7 A positive effect of the splinkers on the general transfection efficiency would be expected if the neogene, which is located more centrally in the construct, would be affected by exonuclease attack. However, this was not the case in our experiments; the total number of neo-resistant colonies was not elevated in the cultures transfected with the splinked construct relative to the nonsplinked control.
Following the cross of lys-EGFP-ki mice with aCre-deleter strain, deletion of the neo cassette from the integrated targeting construct resulted in a significant increase in fluorescence intensity without altering the specificity. This has also been observed for the β globin locus flanked by a floxedneo cassette.15 However, the effect seems to be more complex because cell populations from the original mice contain a fraction of extremely bright fluorescent cells that are no longer seen in the neo-deleted populations (compare Figure2B,C). The molecular mechanism by which the neo cassette modulates EGFP expression remains to be determined.
As expected from the expression of the lysgene,2,17 the inserted EGFP gene was specifically activated in myelomonocytic cells. These data are also in agreement with a recently described knock-in of the Cre gene into thelys locus, which led to Cre activity in macrophages and granulocytes.18 For reasons that are unclear, the proportion of EGFP+ cells in the peripheral blood of the lys-EGFP knock-in mice showed great variability between animals, while there was less variability between bone marrow preparations. One possibility is that these variations reflect differences in exposure to bacterial pathogens.2 19Indeed, the 2 animals with the highest proportion of myeloid cells observed (the last 2 animals shown in Figure 2D) were males that exhibited wounds incurred through fighting.
A small proportion of erythroid cells (defined by expression of the Ter119 marker) and B cells (expressing the B220 marker) were alsoEGFP+. Reconstruction experiments with lysates from EGFP-expressing cells incubated with normal erythrocytes (F.V. and T.G., unpublished data, May, 1999) ruled out the possibility that the double-positives represent erythroid cells that had nonspecifically bound EGFP. Another possibility is that they correspond to myelomonocytic cells that have ingested erythroid cells.20 However, the most likely explanation is that they represent artifacts of aggregation because the proportion of double-positives was dramatically reduced when the blood samples were diluted before FACS analysis.
The targeting vector was constructed in such a way that the lysgene of the targeted locus is no longer transcribed or translated. Therefore, mice homozygous for the knock-in vector should not make a functional lysozyme protein. Nine homozygous animals (4 containing theneo gene and 5 without the gene) grew to normal sizes, they could be bred as homozygous strains, and all contained macrophages. However, a more detailed examination of specific macrophage and neutrophil functions (such as phagocytic capacity and bactericidal action) of the lys-defective animals remains to be performed.
The lys-EGFP-ki mice will be useful in reconstitution/transplantation experiments of myelomonocytic cells because small numbers of donor-derived fluorescence-positive cells can be easily identified in recipient animals. If the mice are crossed with appropriate myeloid leukemia models, theEGFP+ cells might help to identify leukemic cells and to monitor their ablation during treatments aimed at curing disease. They might also be useful in monitoring granulocyte infiltration as a response to bacterial infections. Most importantly, however, the lys-EGFP-ki mouse model will enable us to perform time-lapse fluorescence microscopy of developing colonies in vitro, under conditions that avoid interfering with the spatial arrangements of cells within the colony. This technique could be combined with the staining of hemoglobin-expressing cells in the colonies, thereby revealing cells of erythroid origin (A. Sivunen, F.V., and T.G., unpublished data, July, 1999). Then it would be possible to reconstruct the path by which these cells are formed from the colony founder, to clarify whether hematopoiesis proceeds in a hierarchical manner, and to determine the influence of cytokines on lineage commitment. The answers to some of these questions should not only improve our general understanding of the differentiation and function of myelomonocytic cells, but they might ultimately lead to practical applications such as gene therapy approaches.
Acknowledgments
We thank Kelly McNagny for help with flow cytometry and cytospins, Jonathon Homeister for the Ly-6G antibody, Karen Brennan for blastocyst injections, Ruediger Klein and Richard Stanley's group for reagents and discussions, and Michael Cammer, Analytical Imaging Facility of AECOM, for help with the imaging.
F.V. is a recipient of the Beca de Perfeccionamiento de Doctores of the Ministerio de Educación y Cultura de España, Spain. S.H. was sponsored by a postdoctoral fellowship of the DAAD, Bonn, Germany.
Reprints: Thomas Graf, Albert Einstein College of Medicine, 1300 Morris Park Ave, Chanin 302, Bronx, NY 10461; e-mail:graf@aecom.yu.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal