Abstract
In protein S Heerlen, an S-to-P (single-letter amino acid codes) mutation at position 460 results in the loss of glycosylation of N458. This polymorphism has been found to be slightly more prevalent in thrombophilic populations than in normal controls, particularly in cohorts of patients having free protein S deficiency. This suggests that carriers of the Heerlen allele may have an increased risk of thrombosis. We have now characterized the expression in cell cultures of recombinant protein S Heerlen and investigated the anticoagulant functions of the purified recombinant protein in vitro. Protein S Heerlen was synthesized and secreted equally well as wild-type protein S by transiently transfected COS-1 cells. The recombinant protein S Heerlen interacted with conformation-dependent monoclonal antibodies and bound C4b-binding protein to the same extent as wild-type protein S. Protein S Heerlen displayed reduced anticoagulant activity as cofactor to activated protein C (APC) in plasma-based assays, as well as in a factor VIIIa–degradation system. In contrast, protein S Heerlen functioned equally well as an APC cofactor in the degradation of factor Va as wild-type protein S did. However, when recombinant activated factor V Leiden (FVa:Q506) was used as APC substrate, protein S Heerlen was found to be a poor APC cofactor as compared with wild-type protein S. These in vitro results suggest a possible mechanism of synergy between protein S Heerlen and factor V Leiden that might be involved in the pathogenesis of thrombosis in individuals carrying both genetic traits.
In hereditary thrombophilia, different genetic factors predispose to the development of thromboembolic events.1Resistance to activated protein C (APC) is a highly prevalent inherited hypercoagulable state caused by a single-point mutation in the factor V (FV) gene (FV R506Q,[single-letter amino acid codes] FV:Q506, or FV Leiden). This mutation is found in 2% to 15% of different healthy white populations.2 The high prevalence of FV Leiden mutation facilitates studies of multiple gene-gene and gene-environment interactions among thrombosis patients.3-6 Although not as prevalent as APC resistance, many mutations in the genes of antithrombin, protein C, and protein S have been described as pathogenic risk factors of thrombosis. In the heterozygous form, these gene modifications have a mild clinical expression by themselves, but the likelihood of thrombosis increases when they are associated with a second prothrombotic genetic defect.7 Recently, the concept that thrombophilia is a multigenic disorder has been reinforced by several studies of genetic risk factors in thrombotic individuals.8-12
Congenital protein S deficiency is found in 1% to 5% of patients with thrombosis.13,14 The important anticoagulant role of protein S is dramatically illustrated by the severe thrombotic tendency in homozygous or compound heterozygous cases. Heterozygous carriers have a fivefold to tenfold increased frequency of thrombosis, as compared with their healthy relatives.15,16 Protein S is a plasma glycoprotein that exerts its major role via the APC anticoagulant pathway.17 It works as a nonenzymatic cofactor to APC, enhancing the degradations of activated factor V (FVa) and activated factor VIII (FVIIIa), thereby limiting thrombin generation. Full anticoagulant activity of APC in the degradation of FVIIIa requires not only protein S but also FV, with the 2 proteins functioning as synergistic APC cofactors.18 Thus, FV has the potential of working as both procoagulant and anticoagulant protein. Protein S has also been reported to regulate hemostasis by APC-independent inhibition of tenase and prothrombinase.19-22
Human protein S is a single-chain multimodular protein, composed of a γ-carboxyglutamic acid–containing domain (Gla domain), a thrombin-sensitive region (TSR), 4 epidermal growth factor (EGF)–like domains, and a region homologous to sex-hormone binding globulin (SHBG).23 The Gla domain is involved in membrane interaction, whereas TSR and EGF1 have been demonstrated to interact with APC.24-27 The SHBG-like domain interacts with the β-chain of C4b-binding protein (C4BP), a regulator of the classical pathway of complement.28 In normal plasma, approximately 70% of the protein S molecules are bound to C4BP.17 Only the free form of protein S is active as cofactor to APC.29The concentrations of β-chain–containing C4BP and protein S determine the concentration of free protein S in plasma; the free form of protein S is the molar surplus of protein S over C4BP.30Specific tests for the free and the total (free plus bound-to-C4BP) forms of protein S have been developed. On the basis of these measurements, protein S deficiency is classified according to 3 subtypes. Type I deficiency is characterized by a decrease in the total protein S antigen and, concomitantly, of free protein S. Type II or qualitative deficiency is characterized by normal antigen levels and reduced protein S activity due to a dysfunctional protein S in plasma. Type III deficiency is characterized by low free protein S levels, while the total plasma concentration of protein S is normal. The distinction between type I and type III could be of clinical importance in assessing the risk of thrombosis in a given individual, but its biological basis and significance have been controversial. On the one hand, type I and III deficiencies are commonly found in the same groups of kindred and have in some cases been demonstrated to be associated with the same single mutation in the protein S gene (PROS1).31-34 On the other hand, several studies have identified kindred groups in which protein S deficiency appears only as type III. A relatively frequent protein S variant called protein S Heerlen35 is often found associated with type III deficiency.36
Protein S Heerlen contains an S460P mutation that results in the loss of the N-linked glycosylation site at N458,35 which is probably the same variant protein S reported by Schwarz et al.37 Protein S Heerlen was reported as an immunologic polymorphism detected during the screening of thrombophilic patients with a monoclonal antibody–based enzyme–linked immunosorbent assay (ELISA). In that study, no association was observed between the presence of protein S Heerlen and an increased risk of thrombosis,35 and loss of this glycosylation has not been found to affect the anticoagulant function of protein S or the C4BP binding.38 These facts, together with the prevalence of the S460P mutation in healthy populations, which has been found to range between 0.5% and 0.8%, have led to the mutation's being considered a neutral polymorphism or protein variant. However, subsequent family studies have shown that the Heerlen allele is significantly more frequent in thrombophilic families diagnosed with protein S deficiency than in the normal population.36,39,40 Nevertheless, in families having the Heerlen variant and type III protein S deficiency, the 2 traits do not always cosegregate,37 40 calling into question the mutation-phenotype relationship between the Heerlen polymorphism and protein S deficiency.
The frequent association between protein S Heerlen and the FV Leiden mutation among thrombophilic patients suggests a cooperative effect between these 2 traits.39,41 However, even though this association has been found in particular cases and FV Leiden is known to increase the risk of thrombosis in protein S deficiency,4 5 a direct biochemical basis for a synergism between the 2 traits is lacking. The aim of the present study was to understand the possible basis of thrombosis in symptomatic carriers of the protein S Heerlen allele. We expressed recombinant protein S Heerlen in mammalian cell lines and analyzed its secretion profile. The purified recombinant protein S Heerlen was characterized for functional anticoagulant activity. Furthermore, we studied the in vitro effect of the combination between protein S Heerlen and FV Leiden and demonstrate a possible synergistic prothrombotic effect of the 2 mutations.
Materials and methods
Protein S assays
Plasma samples collected from carriers of the protein S Heerlen variant and their relatives were stored at −70°C before analysis. Determination of the Heerlen variant was made as described previously.40 Plasma samples from 15 protein S Heerlen carriers (14 heterozygous and 1 homozygous) belonging to 5 thrombophilic families previously described34,40,41 were analyzed for the concentration of free protein S. As a control, the plasma concentration of protein S was measured in 12 of their relatives not carrying the Heerlen allele. Free protein S was measured by means of 3 previously described assays: a polyethyleneglycol precipitation followed by radioimmunoassay (RIA),42 an enzyme-linked ligand sorbent assay (ELSA) using immobilized C4BP as catcher,43 and an ELISA based on a monoclonal antibody specific for the free form of protein S (Asserachrome free protein S, Stago, Asnières, France).44
Site-directed mutagenesis
The Quikchange site-directed mutagenesis kit from Stratagene (La Jolla, CA) was used to substitute S460 by P (S460P), essentially as described before for other protein S mutants.45 The sense primer, 5′-GAT TAT AAT AAT GTA CCC AGT GCT GAG GGT TG -3′, was used together with its antisense primer, 5′-CAA CCC TCA GCA CTG GGT ACA TTA TTA TAA TC -3′, for mutagenesis. Polymerase chain reaction was performed according to the instruction manual. Human protein S cDNA subcloned in the pcDNA3 vector (Invitrogen, Carlsbad, CA) was used as template for mutagenesis. The mutation was confirmed by DNA sequencing by means of the ABIprism Taq polymerase–based sequencing kit with fluorescent dye terminators (Perkin Elmer–Applied Biosystems, Foster, CA).
Cell culture and transient expression
The protein S Heerlen variant and wild-type protein S were transiently expressed in monkey kidney COS-1 cells, and the expression levels were determined with an ELSA as described.43Pulse-chase experiments of recombinant protein S including radioactive labeling with [35S] methionine and [35S] cysteine, immunoprecipitation, and electrophoresis were performed essentially as previously described,46 except that a transient expression system was used instead of a stable expression system.45 In brief, the expression vectors were transiently transfected into COS-1 cells, and the cells were divided into several dishes on the following day. On the third day after transfection, the cells were pulse-labeled with [35S] methionine and [35S] cysteine and then chased for the indicated times. Radiolabeled protein S in the culture media and cell lysates were immunoprecipitated with anti–protein S polyclonal antibodies (Dako, Glostrup, Denmark) and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Gels were dried and a Phospho Imager (Molecular Dynamics, Sunnyvalle, CA) system was used to quantify the radioactivity of the bands on the gels. Wild-type FV and R506Q mutant FV were also transiently expressed, and the expression levels were determined by an ELISA as described.47
Stable expression of protein S, purification and characterization of recombinant proteins
The protein S Heerlen variant and wild-type protein S were purified from conditioned media of stably transformed human kidney cell line 293, with the use of methods described previously.25,45 The expression levels were approximately 1.5 mg/L. The concentration of the purified protein S was determined by amino acid composition analysis, with the use of methods as previously described.45Determination of the content of Gla residues was performed as described.45 The purified recombinant proteins were subjected to 10% SDS-PAGE and stained with Coomassie Brilliant Blue R-250 (BDH, Poole, England) to check their purity. The recognition of the protein S Heerlen variant and wild-type protein S by monoclonal antibodies24 directed against Gla domain, TSR, and EGF1 and the ability of the protein S Heerlen variant to interact with C4BP were analyzed with the ELSA technique.43 The stability at 55°C of the wild-type protein S and the Heerlen variant was evaluated by following the decay of protein S activity.48 In brief, 100 μL aliquots of wild-type or Heerlen protein S (10 nmol/L) diluted in 50 mmol/L Tris-HCl, 150 mmol/L NaCl, pH 7.5 (TBS), containing 0.1% bovine serum albumin (BSA), were incubated at 55°C in a water bath, removed at 30-minute intervals, and kept on ice. After 2.5 hours, all aliquots were analyzed for their APC-cofactor activity in an APC-dependent FVIIIa-degradation assay, as described below.
Coagulation assays
APC-cofactor activity of protein S in an activated partial thromboplastin time (APTT)–based analysis.
A previously described procedure was followed with minor modifications.45 In this assay, increasing concentrations of the protein S variants were added to protein S–deficient plasma. The plasma aliquots (50 μL) were incubated with an APTT reagent (50 μL of the APTT reagent present in the APC resistant kit, Chromogenix, Mölndal, Sweden) for 180 seconds at 37°C before the clotting was initiated by the addition of the APC/CaCl2 mixture (50 μL) from the APC resistance kit. The clotting times were measured by means of an Amelung-Coagulometer KC 10, Lemgo, Germany.
APC-cofactor activity of protein S in a prothrombin time–based analysis.
Protein S–depleted plasma (50 μL) was incubated at 37°C for 120 seconds in the Amelung-Coagulometer KC-10 sample cups. Mixtures containing 10 μL wild-type or protein S Heerlen (with final concentrations of 0 to 100 nmol/L), 50 μL APC/CaCl2 (from the APC resistance kit), and 50 μL Simplastin Excel (Organon Teknika, RM Boxtel, Netherlands) were prepared at room temperature and added to the wells, and the clotting time was recorded. The Simplastin was diluted 1:10 or 1:100 with TBS containing 0.1% BSA and 25 mmol/L CaCl2.
Effects of protein S variants in APC-catalyzed inactivation of wild-type and mutant FVa.
A previously described procedure was followed with minor modifications.45 As APC substrate, either plasma-derived or recombinant FVa was used. In the assay, 30 nmol/L wild-type FVa or FVa Leiden, phospholipid (20 μmol/L), wild-type protein S or protein S Heerlen (final concentration from 0 to 80 nmol/L), and human APC (0.3 nmol/L, final concentration) were incubated at 37°C. Aliquots of this mixtures were removed at intervals and diluted 1:10 in ice-cold TBS, and the remaining FVa activity was determined in a standard clotting-based FV assay using FV deficient plasma. The remaining FVa in the incubation mixture with only APC was considered as being 100%. The transiently expressed wild-type FV and FV Leiden were activated prior to each experiment by incubating the FV (100 μL at 45 nmol/L) with 5 nmol/L of α thrombin (Haematologic Technologies, Essex Junction, VT) in the presence of 5 mmol/L CaCl2 and 10 μm phospholipid vesicles at 37°C. After 20 minutes, the reaction was stopped by the addition of 5 nmol/L PPACK (Calbiochem, La Jolla, CA). The phosholipid vesicles, composed of 25% (wt/wt) phosphatidylserine, 37.5% (wt/wt) phosphatidylcholine, and 37.5% (wt/wt) phosphatidylethanolamine, were prepared as described.49
Effects of wild-type or mutant protein S in APC-catalyzed inactivation of FVIIIa.
The FVIIIa-degradation assay was performed as described previously,50 with minor modifications.45 Human FVIII, activated factor IX, and phospholipids were incubated with α thrombin for 3 minutes to activate the FVIII. The thrombin activation was stopped by the addition of hirudin. The FVIIIa degradation was induced by the addition of APC (3 nmol/L final concentration) together with wild-type protein S or protein S Heerlen (final concentration from 0 to 10 nmol/L) and with or without FV (5 nmol/L). After 2.5 minutes' incubation at 37°C, the remaining FVIIIa activity was determined by the addition of bovine factor X (FX). After 8 minutes' incubation, the substrate S-2222 was added and the color resulting from its hydrolysis measured after 10 minutes. Under these conditions, degradation of S-2222 correlated linearly with the FVIIIa activity. The FVIIIa activity remaining after incubation with only APC (3 nmol/L) was considered as being 100%.
Results
Free protein S concentrations in carriers of the protein S Heerlen allele as measured by different free protein S methods
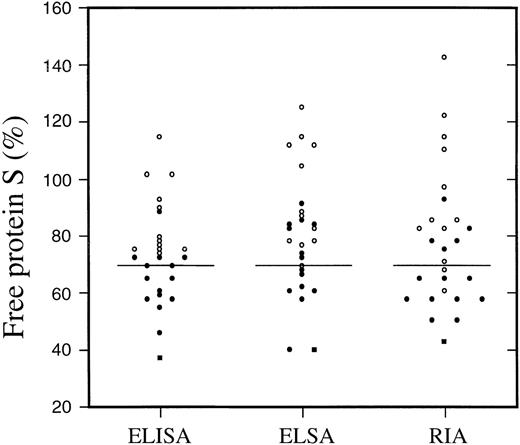
There is an established association between the presence of the protein S Heerlen allele and an increased prevalence of type III protein S deficiency. To elucidate the possibility that the protein S Heerlen is less efficiently detected than wild-type protein S by different assays, plasma samples of individuals carrying the Heerlen mutation in heterozygous state (n = 14) or in homozygous form (n = 1) were analyzed together with 12 control samples by 3 different protein S assays. The 3 assays were based on different principles of detection of the free form of protein S. In the RIA assay, C4BP-protein S complexes were precipitated by polyethylene glycol, and the free protein S present in the supernatant was measured by a standard RIA, with the use of polyclonal antibodies.42 The free protein S ELISA relied on a monoclonal antibody specific for free protein S as catcher.44 The more recently developed free protein S ELSA used immobilized C4BP as catcher of free protein S and subsequent detection of the bound protein S by a monoclonal antibody.43 The 3 methods were found to give similar results both for protein S Heerlen carriers and for normal controls, suggesting that protein S Heerlen can be detected equally well by the 3 methods (Figure 1). In a pairwise comparison, the correlation coefficients between the 3 methods were high (r2 ≥ 0.95). In all 3 methods, the mean concentrations of free protein S among the heterozygous carriers of protein S Heerlen were significantly lower than those estimated for normal controls. The single homozygous case of protein S Heerlen yielded the lowest free protein S values in all 3 methods (around 40%). From these results, it can be concluded that the slightly low free protein S levels found among carriers of the protein S Heerlen allele are real and not the result of systematic laboratory errors.
Free protein S analysis in patients with protein S Heerlen and in their relatives.
Plots of free protein S levels as measured by 3 different methods (ELISA, ELSA, and RIA) in plasma samples from a protein S Heerlen homozygous carrier (filled square) and from 14 heterozygous carriers (filled circles) belonging to 5 thrombophilic families.40 41 In addition, the plasma concentration of protein S was measured in 12 of the relatives (open circles) not carrying the Heerlen allele. The vertical lines at 70% represent the lower normal limit.
Free protein S analysis in patients with protein S Heerlen and in their relatives.
Plots of free protein S levels as measured by 3 different methods (ELISA, ELSA, and RIA) in plasma samples from a protein S Heerlen homozygous carrier (filled square) and from 14 heterozygous carriers (filled circles) belonging to 5 thrombophilic families.40 41 In addition, the plasma concentration of protein S was measured in 12 of the relatives (open circles) not carrying the Heerlen allele. The vertical lines at 70% represent the lower normal limit.
Transient expression of recombinant protein S in COS-1 cells
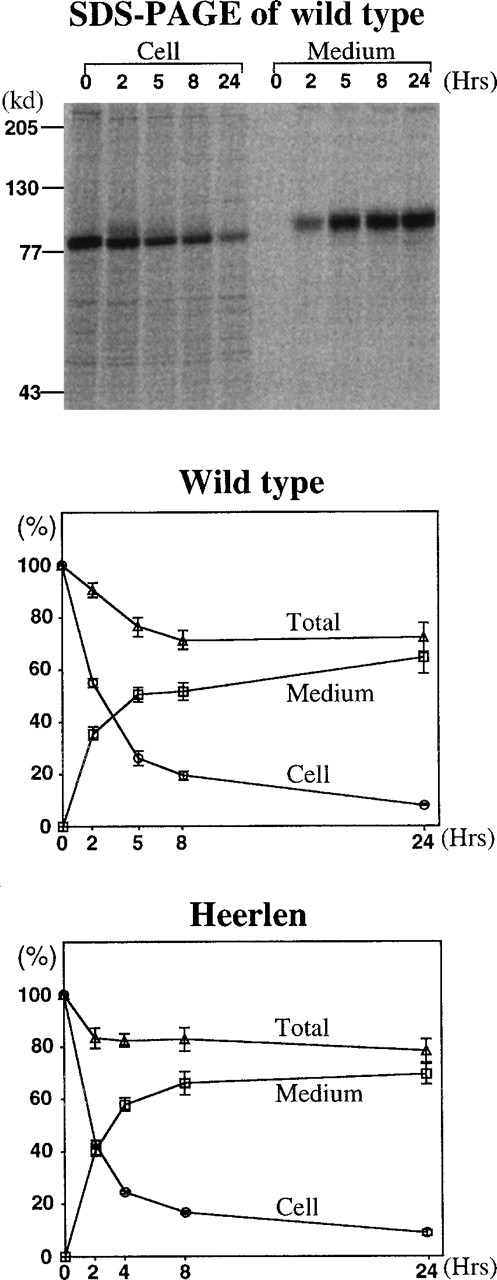
Expression vectors (pcDNA3) containing either wild-type or protein S Heerlen cDNA were used for transient expression in COS-1 cells. The protein S concentration in the conditioned media was analyzed by the ELSA method. The concentrations of wild-type and protein S Heerlen were found to be approximately 110 to 120 ng/mL, and there was no statistically significant difference between the concentrations. When the transfected cells were pulse-chased for protein S, the patterns of expression were very similar for the 2 protein S constructs, and the recovery of protein S in the media after 24 hours was approximately 70% for both (Figure 2). In conclusion, the pulse-chase experiments did not indicate any reduced efficiency in secretion of protein S Heerlen; if such a reduction had been found, it could have provided an explanation for the slightly low protein S levels in protein S Heerlen carriers (Figure 1).
Expression of wild-type protein S and protein S Heerlen.
Quantitative analysis of pulse-chase experiments using transient expression in COS-1 cells. Radiolabeled protein S in the media and cell lysates were immunoprecipitated and electrophoresed on 7.5% SDS-PAGE. The radioactivity of the protein S bands in the dried gels was analyzed with a Phospho Imager. The amount of radioactive protein S present in the cell lysates at the beginning of each individual experiment (0 hours) was assigned a value of 100%. Cell indicates the amount of radioactive protein S in cell lysates (○); medium, amount of radioactive protein S in media (□); and total, the sum of protein S in cell and medium (▵). The means ± SEM of 4 independent experiments are shown.
Expression of wild-type protein S and protein S Heerlen.
Quantitative analysis of pulse-chase experiments using transient expression in COS-1 cells. Radiolabeled protein S in the media and cell lysates were immunoprecipitated and electrophoresed on 7.5% SDS-PAGE. The radioactivity of the protein S bands in the dried gels was analyzed with a Phospho Imager. The amount of radioactive protein S present in the cell lysates at the beginning of each individual experiment (0 hours) was assigned a value of 100%. Cell indicates the amount of radioactive protein S in cell lysates (○); medium, amount of radioactive protein S in media (□); and total, the sum of protein S in cell and medium (▵). The means ± SEM of 4 independent experiments are shown.
Characterization of the purified recombinant protein S variants
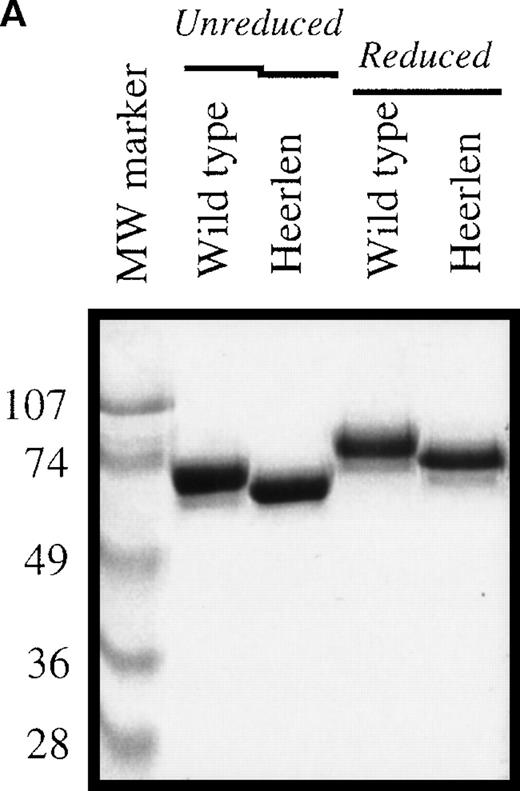
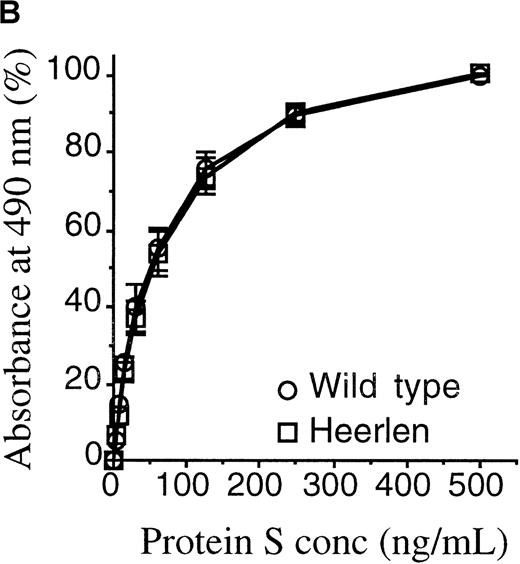
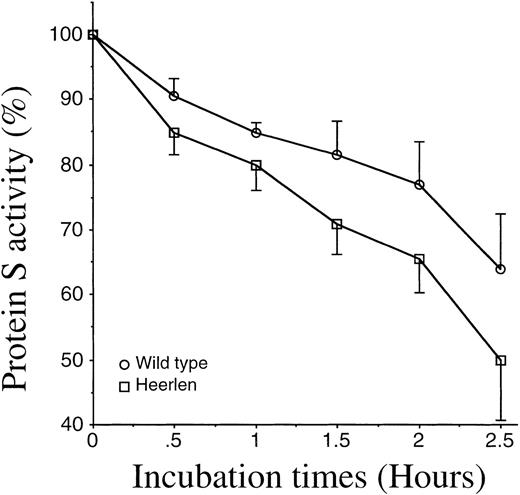
Protein S Heerlen and wild-type protein S were expressed in stably transfected eucaryotic cell cultures; the proteins were purified to homogeneity; and the final products were visualized on SDS-PAGE stained with Coomassie Brilliant Blue (Figure 3A). The migration of protein S Heerlen in the gel was slightly faster than that of wild-type protein S owing to the lack of N-glycosylation at residue 458. Gla analysis revealed that the γ-carboxylation of glutamic acid residues was similar for protein S Heerlen and wild-type protein S (approximately 10 Gla/molecule). Recognition of the recombinant protein S Heerlen by 3 different monoclonal antibodies, against the Gla-domain (HPS 21), the TSR (HPS 67), or the EGF1 (HPS 54), was analyzed with the use of purified protein S bound to immobilized C4BP-coated microtiter wells. The 3 monoclonal antibodies recognized conformation-dependent epitopes, and none of them interfered with the binding of C4BP to protein S.24 All monoclonal antibodies were found to recognize protein S Heerlen and wild-type protein S equally well; the result obtained with one (HPS 54) is shown (Figure 3B). This experiment also demonstrated that recombinant protein S Heerlen and wild-type protein S bound C4BP equally well, suggesting that the S460P mutation did not influence the ability of protein S to bind C4BP. To test whether the stability of protein S in vitro was affected by the S460P mutation, wild-type protein S and protein S Heerlen were incubated at 55°C and their activities recorded at different time intervals (Figure 4). A small but consistent difference was observed: the Heerlen variant was denatured slightly faster than the wild-type protein S. Approximately 35% of the APC-cofactor activity of wild-type protein S was lost after 2.5 hours' incubation, while the Heerlen variant had lost 50% of its activity.
Electrophoretic characterization of protein S and binding to C4BP.
(A) The purified wild-type protein S and protein S Heerlen (5 μg/well) were run on 10% SDS-PAGE gels under unreduced and reduced conditions and were visualized with the use of Coomassie Brilliant Blue stain. (B) Binding of wild-type protein S and protein S Heerlen to immobilized C4BP. Purified C4BP was immobilized in microtiter plates. Following the binding of the protein S to the C4BP, the monoclonal antibody (HPS 54) was used to detect the bound protein S. A value of 100% is assigned for the maximum absorbance of wild-type protein S. Means ± SEM of 3 independent experiments performed in duplicate are shown.
Electrophoretic characterization of protein S and binding to C4BP.
(A) The purified wild-type protein S and protein S Heerlen (5 μg/well) were run on 10% SDS-PAGE gels under unreduced and reduced conditions and were visualized with the use of Coomassie Brilliant Blue stain. (B) Binding of wild-type protein S and protein S Heerlen to immobilized C4BP. Purified C4BP was immobilized in microtiter plates. Following the binding of the protein S to the C4BP, the monoclonal antibody (HPS 54) was used to detect the bound protein S. A value of 100% is assigned for the maximum absorbance of wild-type protein S. Means ± SEM of 3 independent experiments performed in duplicate are shown.
Heat stability of protein S Heerlen and wild-type protein S.
The APC-cofactor activity of protein S Heerlen and wild-type protein S was analyzed by means of a FVIIIa-degradation assay (described in “Materials and methods”) after exposure of the protein S to 55°C for various amounts of time. The 100% value represents the activity of protein S before the heat exposure. Means ± SEM of 3 independent experiments performed in duplicate are shown.
Heat stability of protein S Heerlen and wild-type protein S.
The APC-cofactor activity of protein S Heerlen and wild-type protein S was analyzed by means of a FVIIIa-degradation assay (described in “Materials and methods”) after exposure of the protein S to 55°C for various amounts of time. The 100% value represents the activity of protein S before the heat exposure. Means ± SEM of 3 independent experiments performed in duplicate are shown.
Recombinant protein S Heerlen demonstrated decreased APC-cofactor activity in plasma-based coagulation assays
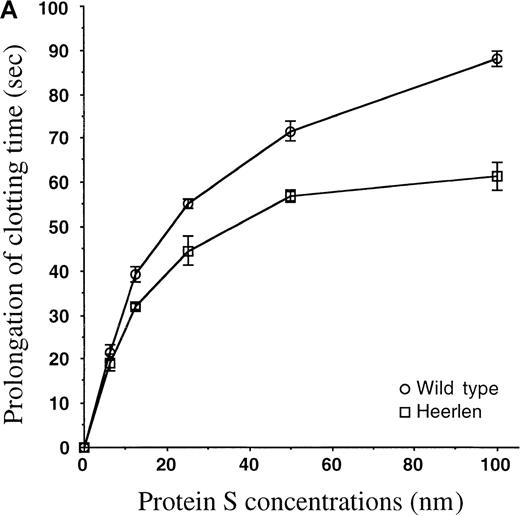
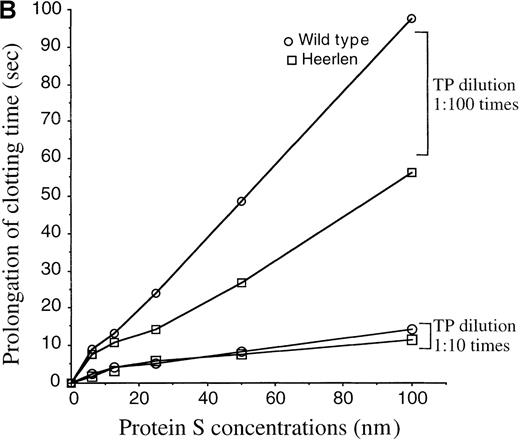
To investigate whether the Heerlen mutation had any effect on the APC-cofactor function of protein S, the activities of wild-type protein S and protein S Heerlen were tested in plasma-based assays with the use of APC and protein S–deficient plasma supplemented with the purified recombinant protein S preparations. In an APTT-based assay, where clotting was triggered via the intrinsic pathway, protein S prolonged the clotting time in a dose-dependent manner in the presence of APC (Figure 5A). In the absence of protein S, the addition of APC yielded a clotting time of 77 ± 1 seconds, as compared with 35.4 ± 0.2 seconds in the absence of added APC. In the presence of both APC and 100 nm wild-type protein S, the clotting time was 165.3 ± 3.5 seconds. Under similar conditions but with protein S Heerlen, the mean clotting time was 138.6 ± 3.2 seconds. The assay did not allow an accurate estimation of the exact activity loss, because the 2 dose-response curves were not parallel. However, it was apparent that protein S Heerlen demonstrated reduced APC-cofactor activity as compared with wild-type protein S, a difference that was statistically significant in an unpaired Student t test (P = .011). In the absence of APC, the presence or absence of protein S (100 nm) did not affect the clotting times (36.5 ± 0.2 seconds versus 35.4 ± 0.2 seconds). Next, we tested the APC-cofactor activities of the expressed protein S variant in a clotting assay based on the tissue-factor pathway. APC was unable to prolong the clotting time after induction of coagulation by undiluted tissue-factor reagent (Simplastin). To be able to record the anticoagulant activity of APC, we therefore diluted the Simplastin reagent 1:10 and 1:100. In the absence of APC, the presence or absence of protein S (100 nmol/L) did not affect the clotting time (19.4 ± 0.2 seconds versus 20 ± 0.5 seconds) when the 1:10 dilution was used. In the presence of APC, the clotting time was 26.5 ± 1 seconds in the absence of protein S and 40.5 ± 1.5 seconds in the presence of 100 nmol/L wild-type protein S. The protein S Heerlen was essentially equally efficient as wild-type protein S (Figure 5B). However, when the Simplastin reagent was diluted 1:100, the Heerlen variant demonstrated approximately 50% of the activity of wild-type protein S (Figure 5B). Under these conditions, neither of the recombinant proteins demonstrated APC-independent anticoagulant activity. Thus, the clotting time was prolonged by 34 seconds, and protein S (100 nmol/L) alone did not have any independent effect. Addition of APC alone yielded a clotting time of approximately 51 seconds.
APC-cofactor activity of protein S Heerlen in plasma-based assays.
(A) Protein S–dependent prolongation of the clotting time in an APTT-based assay. Wild-type protein S and protein S Heerlen were added at increasing concentrations to protein S–depleted plasma in the presence of APC, and the clotting times were recorded by the APTT-based assay. The concentrations of protein S refer to the final concentrations in the assay. The means ± SEM of 3 independent experiments performed in duplicate are shown. (B) Protein S–dependent prolongation of clotting time in the tissue-factor pathway. Wild-type protein S or protein S Heerlen were added to protein S–depleted plasma (final concentrations in the assay of between 0 and 100 nmol/L), and the APC-cofactor activities were measured after initiation of clotting by thromboplastin (TP) reagent diluted 1:10 and 1:100. The means of 2 independent experiments performed in duplicate are shown.
APC-cofactor activity of protein S Heerlen in plasma-based assays.
(A) Protein S–dependent prolongation of the clotting time in an APTT-based assay. Wild-type protein S and protein S Heerlen were added at increasing concentrations to protein S–depleted plasma in the presence of APC, and the clotting times were recorded by the APTT-based assay. The concentrations of protein S refer to the final concentrations in the assay. The means ± SEM of 3 independent experiments performed in duplicate are shown. (B) Protein S–dependent prolongation of clotting time in the tissue-factor pathway. Wild-type protein S or protein S Heerlen were added to protein S–depleted plasma (final concentrations in the assay of between 0 and 100 nmol/L), and the APC-cofactor activities were measured after initiation of clotting by thromboplastin (TP) reagent diluted 1:10 and 1:100. The means of 2 independent experiments performed in duplicate are shown.
APC-cofactor activities of recombinant protein S in degradation of wild-type FVa or FVa Leiden
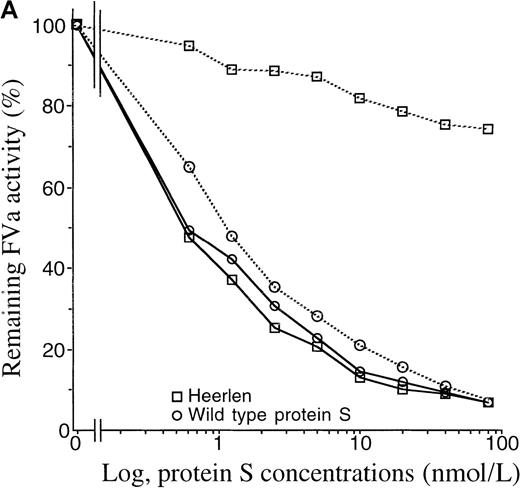
Protein S Heerlen was found to be equally efficient as wild-type protein S in supporting APC-mediated inactivation of plasma-purified FVa (data not shown). Approximately 80% of the initial FVa activity was lost in the presence of 0.3 nmol/L APC and 10 nmol/L wild-type protein S after 5 minutes' incubation. Subsequently, we tested the influence of protein S on the APC-mediated degradation of FVa Leiden, which carries the R506Q mutation. In this experiment, recombinant wild-type FV and FV Leiden were produced in parallel in a transient expression system. The conditioned media were collected and concentrated and the FV levels were then determined with an ELISA for FV. Wild-type FV and FV Leiden were activated by thrombin and thereafter incubated with APC and the protein S variants in the presence of phospholipid vesicles. Wild-type protein S and protein S Heerlen were equally active as APC cofactors in the inactivation of wild-type FVa (Figure 6A), which was in accordance with the experiment described above using plasma-purified FVa. However, when FVa Leiden was used as the APC substrate, protein S Heerlen displayed poor APC-cofactor activity. In the presence of wild-type protein S, approximately 90% of the initial FVa Leiden activity was lost, which was similar to the results obtained with the wild-type FVa. However, under the same conditions, protein S Heerlen failed to support the APC-mediated degradation of FVa Leiden, whereas the degradation of wild-type FVa was efficiently stimulated by protein S Heerlen (Figure 6A).
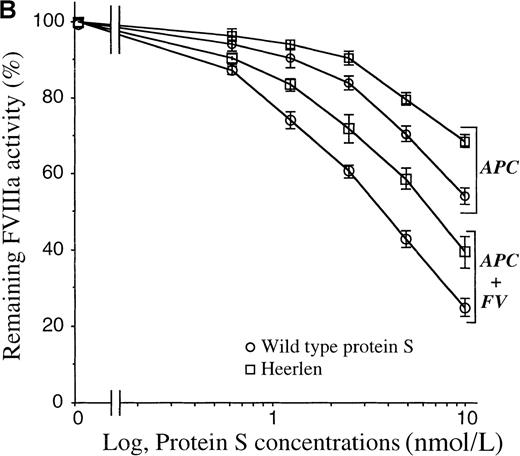
APC-cofactor activity of protein S Heerlen in FVa- and FVIIIa-degradation assays.
(A) FVa-degradation assay. APC-mediated inactivation of wild-type FVa (solid lines) or FVa Leiden (dotted lines) was measured in the presence of wild-type protein S or protein S Heerlen. The mean of 2 independent experiments performed in duplicate are shown. (B) FVIIIa-degradation assay. Inactivation of FVIIIa was performed in either the absence or presence of FV, together with increasing concentrations of wild-type protein S or protein S Heerlen. The means ± SEM of 3 independent experiments performed in duplicate are shown.
APC-cofactor activity of protein S Heerlen in FVa- and FVIIIa-degradation assays.
(A) FVa-degradation assay. APC-mediated inactivation of wild-type FVa (solid lines) or FVa Leiden (dotted lines) was measured in the presence of wild-type protein S or protein S Heerlen. The mean of 2 independent experiments performed in duplicate are shown. (B) FVIIIa-degradation assay. Inactivation of FVIIIa was performed in either the absence or presence of FV, together with increasing concentrations of wild-type protein S or protein S Heerlen. The means ± SEM of 3 independent experiments performed in duplicate are shown.
APC-cofactor activities of protein S variants in APC-mediated inactivation of factor VIIIa
Efficient inhibition of FVIIIa by APC requires the synergistic cofactor activities of protein S and FV. To test the effect of the protein S Heerlen mutation in this reaction, increasing concentrations of either wild-type protein S or protein S Heerlen were added in a FVIIIa-inactivation assay. In both the presence and the absence of FV, protein S Heerlen demonstrated approximately 50% APC-cofactor activity as compared with wild-type protein S (Figure 6B). In this assay system, APC alone yielded approximately 10% reduction in FVIIIa activity after 2.5 minutes' incubation, which is consistent with results on record showing that APC alone is an inefficient inhibitor of FVIIIa.50 51 Protein S (10 nmol/L) alone had essentially no APC-independent inhibitory activity in the FVIIIa assay system. In contrast, after 2.5 minutes' incubation in the presence of APC, 10 nmol/L wild-type protein S, and 5 nmol/L FV, approximately 75% of the FVIIIa activity was lost. Under these assay conditions, protein S Heerlen demonstrated reduced APC-cofactor activity, and at 10 nmol/L protein S Heerlen, approximately 50% FVIIIa activity remained (Figure6B).
Discussion
Protein S Heerlen (S460P) has been circumstantially linked to thrombosis,36 despite not being significantly more prevalent in populations with venous thrombosis than in the general population.35 Protein S Heerlen, and other mutations in the region near residue 460, have been reported to be associated with type III protein S deficiency, ie, low levels of free protein S despite normal levels of total protein S.36,39,40 It has been suggested that the mechanism underlying type III deficiency in these mutants involves increased affinity for C4BP with a resulting decrease in the level of free protein S, or that each C4BP molecule is able to bind more than 1 protein S Heerlen molecule.36 However, in this study, which used recombinant wild-type protein S and protein S Heerlen produced and purified in parallel, no difference in binding to C4BP could be detected (Figure 3B). This is in accordance with results obtained with other recombinant protein S mutants, eg, N458Q or S460G in the SHBG-like domain of protein S, in which the absence of carbohydrate at position 460 does not affect binding to C4BP.38 The SHBG-like region of protein S is known to fully contain the binding site for C4BP, but a detailed molecular understanding of the binding site is missing.28,52Recently, we found evidence for the involvement of both laminin-G–type domains of the SHBG-like region in the formation of the complex.52 Thus, the binding site in protein S for C4BP is complex, involving more than 1 domain, and the loss of a carbohydrate side chain does not appear to affect the binding affinity to any significant degree. Furthermore, the affinity of protein S for C4BP is very high,28 and the free protein S concentration is the molar surplus of protein S over the β-chain containing C4BP in plasma.30 Thus, a change in affinity of protein S for C4BP appears to be a less likely mechanism for the slightly decreased levels of free protein S associated with the Heerlen allele.
We found heterozygous carriers of the Heerlen mutation to have lower values of free protein S than their normal relatives, displaying values slightly below the lower normal limit or in the lower part of the normal range of protein S (Figure 1), which is in agreement with data on record.36,39 It is noteworthy that in its homozygous state, the Heerlen mutation produced a clear type I deficiency.41 Three different assays for free protein S yielded similar results, making it less likely that the lower values were due to a procedural bias (Figure 1), eg, poor recognition of protein S Heerlen by a monoclonal antibody. Other possible explanations for the lower free protein S levels associated with protein S Heerlen are decreased synthesis of protein S or decreased half-life of protein S Heerlen in the circulation. It has been reported that the mRNA expression of the Heerlen allele is almost equal to that of wild-type allele in vivo.53 Protein S Heerlen was found to be synthesized and secreted as efficiently as wild-type protein S in vitro (Figure 2). Even though we cannot excluded the existence of a synthesis or secretion defect in vivo, these results argued against a defective synthesis or secretion as the cause of the mild quantitative deficiency. This stands in contrast to other protein S deficiency causative missense mutations in the functional protein S genePROS1, resulting in poor secretion.45,54,55The other possible mechanism for the low free protein S values, ie, a decreased half-life in circulation of the mutant protein, could not be tested for. However, it is noteworthy that the Heerlen mutant demonstrated slightly lower heat stability as compared with wild-type protein S (Figure 4). It is known that N-glycosylation increases the stability of proteins.56 In conclusion, the Heerlen allele seems to produce a mild quantitative deficiency, whose the underlying mechanism is not fully understood. It has been shown that missense mutations resulting in mild protein S deficiency are manifested more frequently as type III.55 This is probably the reason the Heerlen allele is more often diagnosed as type III than as type I protein S deficiency.
We found that protein S Heerlen decreased anticoagulant activity, as compared with wild-type protein S, in plasma-based clotting assays as well as in a system looking specifically at FVIIIa inhibition. However, in a system investigating the inhibition of normal FVa, no significant difference between wild-type protein S and protein S Heerlen was observed. In the APTT-based plasma assay, the activity of protein S Heerlen was significantly lower than the activity of wild-type protein S (Figure 5A), but the nonparallel dose-response curves did not allow an accurate quantification of the activity loss. In the tissue-factor–based plasma system, a difference between wild-type protein S and protein S Heerlen was observed only when highly diluted tissue-factor regent was used (Figure 5B). The APTT assay and the diluted tissue-factor system are both sensitive to degradation of both FVIIIa and FVa, whereas there is no or minimal influence of FVIIIa degradation in the system using high concentrations of tissue factor.57 Thus, results from both the purified systems and the plasma-based assays suggested that the mutation in the Heerlen variant affects the APC-mediated degradation of FVIIIa more than the FVa degradation. The difference in FVIIIa degradation in the presence of wild-type protein S and protein S Heerlen was observed in both the presence and the absence of FV (Figure 6B), which is known to synergistically stimulate the APC-cofactor activity of protein S in the FVIIIa degradation.18,50 It has been shown that the SHBG-like domain of protein S is involved in the degradation of FVIIIa.58,59 It is possible that a FVIII interaction site is present in protein S near the 460 region and that this site is affected by the Heerlen mutation. A FVa-binding site has recently been demonstrated to be present in the C-terminal part of the SHBG-like domain of protein S.60
It is noteworthy that the degradation of FVa Leiden was severely impaired in the presence of protein S Heerlen as compared with what was seen in the presence of wild-type protein S (Figure 6A). This difference suggests that the protein S Heerlen is less efficient than wild-type protein S in stimulating the APC-mediated cleavage at R306 in FVa Leiden. The degradation of normal FVa was less affected by the Heerlen mutation (Figure 6A). Normal FVa is initially cleaved by APC at R506 before APC cleaves at the R306 site, and the latter cleavage site is specifically stimulated by protein S.61 It is interesting to note that influence of the Heerlen mutation was seen only in FVa Leiden, which cannot be cleaved at R506. The molecular mechanism for the different rates of cleavages at R306 in normal FVa and FVa Leiden is not understood.
The now-observed deficient APC-cofactor activity of protein S Heerlen in the degradation of FVa Leiden suggests a possible synergistic pathogenic mechanism between these 2 genetic traits resulting in increased risk of thrombosis. It is well known that the pathogenesis of thrombosis often involves more than 1 genetic factor.3,5-12 Two genetic risk factors act in synergy when their combination confers a risk of thrombosis that is higher than the sum of the risk conferred by each isolated trait. The now-reported in vitro synergism between the protein S Heerlen and FV Leiden suggests that there may be a synergistic pathogenic effect between these 2 mutations in vivo. This would provide a reasonable explanation for the relatively high prevalence of the Heerlen mutation in the general population, as the risk conferred by the mutation alone could be low or absent. On the other hand, the coexistence of FV Leiden and the protein S Heerlen would confer a higher risk of thrombosis as compared with FV Leiden alone. Actually, several thrombosis patients have been reported to carry the 2 genetic traits.39 41 In summary, carriers of the protein S Heerlen mutation display mild protein S deficiency with a mixed phenotype. The inability of protein S Heerlen to support APC-mediated degradation of FVa Leiden offers a possible mechanism of synergism between the 2 genetic factors, resulting in an increased risk of thrombophilia.
Acknowledgments
We thank Prof R. J. Kaufman for the kind gift of human FV wild-type and Leiden cDNA; and Dr M. Borrell, from Hospital de la Sanat Creu i Sant Pau, and Dr G. Navarro, from Laboratori de Referència de Catalunya, Barcelona, Spain, for plasma samples from carriers of the protein S Heerlen allele and their relatives.
Supported by the Swedish Medical Research Council (Grants 07143, 12561, and 13000), a Senior Investigators Award from the Swedish Foundation for Strategic Research, research funds from the University Hospital in Malmö, the Fondation Louis-Jeantet de Médecine, the Alfred Österlund Trust, and the Albert Påhlsson Trust.
Reprints:Björn Dahlbäck, Department of Laboratory Medicine, Division of Clinical Chemistry, Lund University, University Hospital, S-20502 Malmö, Sweden; e-mail:bjorn.dahlback@klkemi.mas.lu.se.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal