Abstract
To study human immunodeficiency virus (HIV)–specific cellular immunity in vivo, we transferred syngeneic lymphocytes after ex vivo expansion and transduction with a chimeric receptor gene (CD4/CD3-ζ) between identical twins discordant for HIV infection. Single and multiple infusions of 1010 genetically modified CD8+ T cells resulted in peak fractions in the circulation of approximately 104 to 105modified cells/106 mononuclear cells at 24 to 48 hours, followed by 2- to 3-log declines by 8 weeks. In an effort to provide longer high-level persistence of the transferred cells and possibly enhance anti-HIV activity, we administered a second series of infusions in which both CD4+ and CD8+ T cells were engineered to express the chimeric receptor and were costimulated ex vivo with beads coated with anti-CD3 and anti-CD28. Sustained fractions of approximately 103 to 104 modified cells/106 total CD4+ or CD8+cells persisted for at least 1 year. Assessment of in vivo trafficking of the transferred cells by lymphoid tissue biopsies revealed the presence of modified cells in proportions equivalent to or below those in the circulation. The cell infusions were well tolerated and were not associated with substantive immunologic or virologic changes. Thus, adoptive transfer of genetically modified HIV-antigen–specific T cells was safe. Sustained survival in the circulation was achieved when modified CD4+ and CD8+ T cells were infused together after ex vivo costimulation, indicating the important role played by antigen-specific CD4+ T cells in providing “help” to cytotoxic effectors.
Antiretroviral therapies have radically altered the course of patients with human immunodeficiency virus (HIV) infection.1 However, these treatment regimens, commonly referred to as highly active antiretroviral therapy (HAART), may be limited by toxic effects, cost, inconvenience, and incomplete viral suppression leading to drug resistance. Despite often dramatic virologic and CD4+ T-cell responses after instituting HAART, development of effective HIV-specific immune responses and replacement of elements of the immunologic repertoire depleted by HIV infection do not occur consistently.2 3 For these reasons, adjunctive approaches to treatment, including immunomodulatory therapies, are under investigation.
Virus-specific T cells play an important role in host defense against certain viral infections. HIV-specific T-cell activity is likely a major component of the host immune response associated with control of virus replication after acute infection.4,5 Furthermore, HIV-specific cytotoxic T lymphocytes (CTL) isolated from infected patients showed specific cytotoxic activity against HIV-infected target cells, suggesting a possible role for HIV-specific CTL in more advanced infection.6 In experiments conducted in simian immunodeficiency virus (SIV)–macaque models of acute and chronic lentiviral infection, SIV-specific CD8+ T-cell depletion with an anti-CD8 antibody was associated with acceleration of death during acute infection and increased plasma viremia in chronically infected animals; moreover, control of viral replication in the plasma was associated with the return of SIV-specific CTL.7 8These observations suggest that enhanced HIV-specific CTL activity may be of potential therapeutic benefit in reversing or preventing further immunologic decline in patients with HIV infection.
How best to augment CTL activity in vivo is unknown. In tissue culture, in response to stimulation with viral peptides, virus-specific CTL express cytokines such as interferon γ and show cytolytic activity toward virally infected targets. Evidence of in vivo Epstein-Barr virus (EBV)–specific9 and cytomegalovirus-specific10antiviral activity and of EBV-specific anti-tumor activity11 has been found in patients who have undergone allogeneic bone marrow transplantation and subsequently received donor-derived virus-specific CTL. These observations suggest that CTL can be fully functional on their own. Other studies, however, indicated that CD8+ CTL rely on “help” in the form of antigen-specific CD4+ T cells and interleukin 2 (IL-2) to clear chronic viral infections.10 12
T cells bearing chimeric antigen-receptor proteins have been developed in the laboratory in an effort to redirect T-cell antigen specificity and circumvent major histocompatibility complex (MHC) restriction. An example of this technology involves the CD4/CD3-ζ chimeric receptor, which contains the extracellular targeting domain, human CD4 (accounting for HIV specificity), linked to the cytoplasmic signaling domain, CD3-ζ, enabling T-cell activation on binding of the receptor regardless of MHC haplotype.13 On binding to the HIV envelope, CD8+ T cells genetically engineered to express the CD4/CD3-ζ receptor proliferate and initiate effector functions such as cytokine secretion and HIV-specific cytolytic activity.13 14
To explore the role of HIV-specific T-cell immunity in vivo, we genetically engineered HIV-specific, CD4/CD3-ζ–bearing, syngeneic T cells obtained from healthy adult donor twins and administered these cells to their HIV-infected identical twins. We sought to assess the safety and feasibility of these cell transfers, as well as the in vivo survival, distribution, and activity of the ex vivo–modified cells in the setting of HIV infection.
Patients, materials, and methods
Patients
Sets of twins were determined to be identical on the basis of HLA and erythrocyte marker phenotypes. HIV infection in the recipient twin was confirmed by enzyme-linked immunosorbent assay (ELISA) and Western blotting. Donor twins were confirmed to be negative for HIV by ELISA, Western blotting, and polymerase chain reaction (PCR) using 3 sets of primer pairs amplifying regions for gag, env, and long terminal repeats (LTRs). Other criteria for enrollment included seronegativity of the donor for EBV, cytomegalovirus, and hepatitis B and C viruses, unless the recipient was seropositive for the organism. Written informed consent was obtained from all participants after the nature and risks of the study were explained. The study protocol and procedures were reviewed and approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board, the National Institutes of Health (NIH) Institutional Biosafety Committee, and the NIH Recombinant DNA Advisory Committee.
The first 3 twin recipients enrolled received a single infusion of 107 HIV-specific, genetically modified CD8+CTL. Thereafter, twin recipients were randomly assigned in a 1:1:1:1 ratio to receive ex vivo–stimulated and –expanded, but unmodified, CD8+ T cells or either 108, 109, or 1010 HIV-specific, genetically modified CD8+CTL. After completion of the single-infusion phase of the study, patients randomly assigned to the unmodified-cell group received infusions of 1010 unmodified cells every 8 weeks for 1 to 6 infusions (mean, 5.4 infusions; median, 6 infusions) and patients randomly assigned to the modified-cell group received 2 to 6 infusions (mean, 5.3 infusions; median, 6 infusions) of 1010HIV-specific, modified cells according to the same schedule. A subset of 8 patients participated in a second randomization to receive either modified cells alone or modified cells plus IL-2 infusions. IL-2 was given by continuous intravenous administration as a starting dose of 9 million IU/day for 5 days beginning the day of each cell infusion, every 8 weeks.
After completing 1 year of treatment, patients were offered an additional 3 infusions of 1010 HIV-specific, genetically modified CD4+ and CD8+ T cells administered biweekly on an open-label basis. Patients who received IL-2 with previous cell infusions received IL-2 with the third of the modified CD4+ and CD8+ infusions.
CD8+ CTL isolation, enrichment, and growth
Lymphocytapheresis in the donor twins was performed with an automated cell separator (CS-3000; Baxter Healthcare, Deerfield, IL). Approximately 10 L of whole blood was processed for each procedure to collect a minimum of 5 × 109 peripheral blood mononuclear cells (PBMC). CD8+ T cells were enriched with 2-staged immunoselection using immunoaffinity columns (CellPro, Bothell, WA) containing anti-CD8 monoclonal antibodies (anti-Leu2a; Becton Dickinson, San Jose, CA) and then CD4 depletion of the effluent on columns containing anti-CD4 monoclonal antibodies (anti-Leu3a; Becton Dickinson). A minimum of 5 × 107CD8-enriched cells were recovered and cultured ex vivo in AR medium (1:1 AIM V [Gibco, Grand Island, NY]:RPMI [Gibco]–8.8% human AB serum [Gibco]) supplemented with recombinant IL-2 (700 IU/mL; Chiron, Emeryville, CA). Cells were stimulated with syngeneic irradiated PBMC and anti-CD3 antibody (OKT3, 10 ng/mL; Ortho Biotech, Raritan, NJ) and cultured at 37°C. Cells from twins randomly assigned to the control group were then expanded to approximately 2 × 1010total cells.
Growth and transduction of HIV-specific CTL
Cells from the donors of patients randomly assigned to receive HIV-specific, genetically modified cells were processed as follows. Supernatant containing the rkat4SVGF3e− retroviral vector15 was added to the cells 5 times during 2 days, for exposure times of 3, 3, 18, 3, and 3 hours, respectively, on those days. The rkat4SVGF3e− vector contains the following sequences 5′ to 3′: pBR322 plasmid backbone; Moloney murine leukemia virus (MMLV) U3, Moloney murine sarcoma virus LTR, and 5′ untranslated region; MMLV env splice acceptor; CD4/CD3-ζ chimeric-receptor coding sequence; MMLV U3 region; MMLV LTR; and pBR322 plasmid backbone. The vector titer was 105to 106/mL, and the multiplicity of infection (MOI) was 0.7 to 1.4 vector particles/cell (minimum cell number, 5 × 107); the mean transduction efficiency was 18% (range, 7%-38%). After the final exposure to retroviral vector, fresh AR medium was added to the cells, which then expanded to a target dose of 108 cells. A final column purification was performed with selection for CD4+ expression, thereby capturing a population of CD8 cells enriched for transduction and expressing both CD8 and CD4 surface markers. A minimum of 106 transduced cells were cultured in AR medium containing IL-2 and anti-CD3 to yield a final cell number of 2 × 1010, with a mean time in culture of 50 days (range, 35-93 days).
Secondary expansion of purified CD8+ cells
Culture-expanded control cells and HIV-specific transduced cells were divided into 10 aliquots of approximately 1.5 × 109 each and cryopreserved in liquid nitrogen in AIM V media with 10% dimethyl sulfoxide (DMSO) (Sigma) and 4.5% human serum albumin (HSA) (Alpha Therapeutics, Los Angeles, CA). Before freezing, representative samples were obtained for routine testing for viability, sterility, and mycoplasma contamination; CTL assays; and flow cytometry. Testing for replication-competent retrovirus (RCR) was also performed on samples of transduced cells.
Approximately 1 to 2 weeks before infusion, 1 aliquot of cells was thawed and cultured in AR medium with IL-2 and anti-CD3 to reach the target cell dose (mean number of days in culture, 12; range, 4-28 days). On the day of infusion, the cells were harvested, filtered through a 170-μm filter, suspended in 400 mL of normal saline containing 2.5% HSA, and stored at 2°C to 8°C until infusion. A cell sample was again tested for viability, percentage of CD4 and CD8 coexpression, cytolytic activity, sterility, mycoplasma, RCR (transduced cells), and endotoxin contamination. When predefined release criteria were met (including percentage of total cells expressing both CD4 and CD8 surface markers ≥ 70% and cytolytic activity against gp120–expressing 293 target cells ≥ 100 lytic units/107 cells), the cells were infused by vein into the recipient over 1 hour, usually within 4 to 6 hours after they were thawed.
Preparation of CD4/CD3-ζ–modified CD4+ and CD8+ T cells
Donor twins underwent a second lymphocytapheresis as described above. PBMC were separated with Ficoll gradient separation, and monocytes were removed by adherence to plastic. The enriched T-cell population was stimulated to proliferate in serum-free AIM V medium containing recombinant IL-2 (200 IU/mL) and immunomagnetic beads (Dynal, Oslo, Norway) loaded by tosyl conjugation with equal amounts of anti-CD3 OKT3 and anti-CD28 (provided by C. June, Jackson Foundation) at a ratio of 3 beads/cell.16 On day 3, the stimulated cells were removed by using a magnetic bead separator (MaxSep, Baxter Healthcare). On days 5 to 7 of stimulation, the cells were exposed torkat4SVGF3e− retroviral vector supernatant for 3 days at a MOI of 2, then expanded in serum-free medium containing recombinant IL-2 until a total cell number of at least 2 × 1010was achieved (mean days in culture, 13; range, 12-17 days). Cells were then suspended in Plasmalyte A (Baxter) containing 10% DMSO, 1% dextran (Baxter), and 5% HSA and divided into aliquots of 5 × 109 (50 mL) for cryopreservation. At the time of cryopreservation, a sample of cells was tested for viability, sterility, mycoplasma contamination, transduction efficiency, and RCR and subjected to CTL assays and flow cytometry. For infusion, 2 aliquots (total of 1010 cells) were thawed in a 37°C water bath and immediately administered to the recipient over 10 to 20 minutes.
Preparation and analysis of radiolabeled cells
An aliquot containing 2.5 × 109 expanded gene-modified or unmodified CD8+ T cells was removed from the harvested product. The cells were pelleted, washed, resuspended in 20 mL Hanks balanced salt solution, and incubated for 15 minutes at room temperature with 0.74 MBq of indium 111–oxine/108 cells (Amersham, Arlington Heights, IL) to a maximum of 0.37 MBq/kg of body weight. Labeled cells were then pelleted, resuspended in 100 mL normal saline with 2% HSA, and filtered through a 140-μm filter (Monoject; Sherwood Medical, St Louis, MO). The radiolabeled cell suspension was then infused intravenously over 5 to 10 minutes, and this was followed by infusion of the remaining unlabeled cells. Approximately 2 to 4, 24, and 48 hours after administration of radiolabeled cells, images were obtained by using either a BIAD (Trionix, Twinsburg, OH) or Genesis (ADAC Labs, Milpitas, CA) dual-headed γ camera equipped with medium-energy collimators with 20% windows centered at the 174- and 247-keV γ-ray energies of indium 111. Whole-body and spot images were obtained.
For analysis, geometric mean images were generated from paired anterior and posterior whole-body scans. Liver, left and right lung, spleen, and whole-body regions of interest were drawn on these geometric mean images, and ratios of organ to whole-body findings were obtained at each time point.
Analysis of the fraction of CD4/CD3-ζ–positive cells
CD4/CD3-ζ DNA copy numbers in peripheral blood and lymphoid tissue were measured by quantitative competitive PCR using DNA from the e+ cell line as the competitor. The e+ cell line contains 10 copies of the CD4/CD3-ζ gene per cell with a 102-base-pair (bp) DNA insert. DNA lysates from varying numbers of e+ cells were added to equal aliquots of DNA from patient samples (unfractionated PBMC or lymphoid cells, and immunoselected CD4+ and CD8+ cells) and amplified for 30 cycles by using Taq polymerase and primer pairs located in the U3 region of the 3′ LTR and the CD4/CD3-ζ genes. The primer sequences used were 5′-GGTTCACTCTTCTCAGCCACTGAAG-3′ and 5′-TAGCTTGCCAAACCTACAGGTGGG-3′. These yielded amplified products of 325 bp from the e+ cells and of 223 bp from PBMC or lymphoid cells. PCR products were labeled with phosphorus 32–deoxycytidine triphosphate during the amplification and were electrophoresed on a 10% polyacrylamide gel in 90 mmol/L Tris-borate and 2 mmol/L EDTA (pH 8.4) buffer. Radioactivity in the resulting bands was converted to photo-stimulated luminescence units (PSL) by using a phosphor imager (BAS 1000; Fuji Medical Systems, Stamford, CT). The CD4/CD3-ζ gene copy number in an individual sample was derived from a linear regression curve relating the ratio of PSL for the patient sample to the e+ control on the y-axis to PSL of the e+ control on the x-axis; the value for the sample was the corresponding x-axis value to a PSL ratio equal to 1.
Immunoselection for CD8+ T cells from whole blood was performed by first depleting monocytes from the samples with use of using anti-CD14–coated beads (Dynal) and selecting CD8+ T cells with anti-CD8–coated beads. Immunoselection for CD4+T cells from whole blood was performed by first depleting monocytes and CD8+ T cells from the samples by using anti-CD14–coated beads and anti-CD8–coated beads and selecting CD4+ T cells with anti-CD4–coated beads.
Immunologic and virologic monitoring
Enumeration of fractions of CD3+, CD4+, and CD8+ subpopulations in processed cells and peripheral blood samples from the patients was performed by standard 3-color flow cytometry using the following combination of monoclonal antibodies: anti-CD3–fluorescein isothiocyanate, anti-CD8–phycoerythrin (Becton Dickinson), and anti-CD4–Cychrome (Pharmingen, San Diego, CA). HIV RNA in plasma was quantified with a commercially available reverse transcriptase-PCR assay (Amplicor HIV Monitor; Roche Diagnostics, Branchburg, NJ) by using previously described methods with a sensitivity of 50 copies/mL.17
Statistical analysis
The observed data were derived from sequences of measurements done over time in the patients in the study. The statistical approaches used to analyze the data account for the correlation of values within a patient. Statistical analyses focused on data from the series of repeated infusions of activated CD8+ T cells and the second series of infusions of activated CD4+ and CD8+T cells. Measures of CD4/CD3-ζ gene signal, plasma viral load, and total CD4+ T-cell count were analyzed.
Values below the detection limit for CD4/CD3-ζ gene signal and viral load were replaced with random numbers from an exponential distribution ranging from 0 to the detection limit.18 This method helps preserve the variability of the measure. Values of 0 were replaced with one tenth of the minimum of the nonzero values when converting to the log10 scale. Missing values were imputed by the models described below or excluded, as appropriate. Wilcoxon rank-sum tests were used to compare study group populations at baseline.
Persistence of signal over time was modeled with a linear mixed model18-20 of the log10-transformed measure of CD4/CD3-ζ gene signal. This approach constructs models that recognize observations belonging to individual patients but do not make assumptions about the order and length of time between study points. A simple variance structure with few assumptions was employed. The model-based least-squares means at each of the study points were plotted over time to provide a visual representation of the long- and short-term effects of repeated infusions of cells. Similar methods were used to model log10 viral load and total CD4+T-cell counts over time.
Area-under-the-curve (AUC) analyses were performed for CD4/CD3-ζ gene signal and viral load. The area for each patient was calculated by using the trapezoidal rule with the observed values in the original scale for each time point. The AUC represented the total level of signal or viral load integrated over time. Two-sample t tests were used to compare the mean AUC for the different groups. The AUC analyses included patients who had more than 2 infusions in a series.
Another set of linear mixed models, accounting for order and interval between points, were fit to log10 CD4/CD3-ζ gene signal, log10 HIV RNA, and total CD4+ T-cell count. These models recognize the set of observations belonging to a patient as a unit, and include terms for study group, time point, and the interaction of group and time point. Slopes of change over time were calculated by using patients' fitted points from these interval-adjusted linear mixed models. The model t test for the interaction term tested the difference in slopes between the groups. All statistical analyses were conducted with SAS, version 6.12.19
Results
Dose escalation of CD4/CD3-ζ–modified CD8+ T cells
Twenty-seven subjects participated in the initial dose-escalation phase of the study: 3 subjects received 107 modified cells in a nonrandomized pilot phase, and 24 subjects were randomly assigned to 1 of 4 groups and received single infusions of either gene-modified cells (10,8 10,9 or 1010 total cells) or unmodified cells (1010). When a quantitative competitive PCR assay was used to measure CD4/CD3-ζ DNA copy number in PBMC (with a limit of quantitation of 30 copies/million cells), CD4/CD3-ζ DNA was detected at the limit of quantitation in 1 of 3 recipients of 107 cells and in 4 of 6 recipients of 108 cells. In 3 of these patients, CD4/CD3-ζ DNA was no longer detected after the first 1 to 3 days, whereas in 2 patients, CD4/CD3-ζ DNA persisted for 2 and 24 weeks, respectively.
All 12 subjects who received either 109 or 1010modified cells had CD4/CD3-ζ DNA detected in their PBMC. Peak copy number usually occurred 24 to 48 hours after infusion and ranged from 1460 to 41 179 copies/million PBMC. A dose-response relation was evident in that median copy numbers for recipients of 109and 1010 cells were 2973 and 28 245, respectively. CD4/CD3-ζ DNA persisted in 11 of 12 evaluable subjects, albeit at much lower copy numbers (median, 54 copies/million PBMC; range, ≤ 30-6587) for at least 15 to 40 weeks, when they received additional infusions of modified cells.
Repeated infusions of activated CD8+ T cells
Thirty-three patients enrolled in the multiple CD8+T-cell–infusions phase of the study: 25 had previously participated in the dose-escalation phase, and 8 were newly enrolled and randomly assigned. Three patients did not receive cell infusions during this study period: 2 died from HIV-related complications before the first scheduled infusion and 1 voluntarily withdrew from the study. Table1 shows baseline characteristics of the 30 patients who received at least 1 cell infusion (1010cells/infusion).
Baseline characteristics of patients with human immunodeficiency virus (HIV) infection who received modified or unmodified CD8+ T cells
| Characteristic . | Modified cells (n = 21) . | Unmodified cells (n = 9) . | P . |
|---|---|---|---|
| Median CD4 count, (range) | 241 (52-746) | 193 (14-674) | .28* |
| HIV RNA† | |||
| Median log10 copies/mL (range) | 3.64 (1.50-5.23) | 3.31 (1.70-6.01) | .87* |
| No. (%) of patients with <50 copies/mL | 3 (14) | 0 | |
| Antiretroviral therapy, no. of patients | |||
| None | 1 | 1 | |
| NRTIs only | 6 | 2 | |
| NRTIs and PI | 14 | 6 |
| Characteristic . | Modified cells (n = 21) . | Unmodified cells (n = 9) . | P . |
|---|---|---|---|
| Median CD4 count, (range) | 241 (52-746) | 193 (14-674) | .28* |
| HIV RNA† | |||
| Median log10 copies/mL (range) | 3.64 (1.50-5.23) | 3.31 (1.70-6.01) | .87* |
| No. (%) of patients with <50 copies/mL | 3 (14) | 0 | |
| Antiretroviral therapy, no. of patients | |||
| None | 1 | 1 | |
| NRTIs only | 6 | 2 | |
| NRTIs and PI | 14 | 6 |
NRTIs indicates nucleoside reverse transcriptase inhibitors; and PI, protease inhibitor.
By Wilcoxon rank-sum test.
HIV RNA values are based on 20 patients in the modified-cell group and 9 in the unmodified-cell group. One recipient of modified cells who did not have a baseline sample available was excluded from the analysis.
Figure 1A shows the persistence of CD4/CD3-ζ DNA in the circulation of recipients of either gene-modified cells or gene-modified cells plus IL-2, expressed as log10 mean copy number estimated from mixed models. A reproducible pattern was observed in which copy number peaked 24 hours after infusion at approximately 105/million CD8+ cells and then fell steadily to approximately 103copies before the next infusion 56 days later. By the fourth and fifth infusions, the nadir copy number in the group of patients who received cells without IL-2 declined even further. Although mean copy number for the 4 subjects who received modified cells plus IL-2 also showed 2-log declines from peak values in the days after infusion, this group had a numerically higher mean nadir DNA copy number. Analysis of the model-based least-squares means, excluding the day 1 peaks for both groups, yielded slopes of 0.0009 ± 0.0013 and −0.0088 ± 0.0013 log10 copies/day for the IL-2 recipients and nonrecipients, respectively (P = .0012 for the interaction of study time and IL-2 use). An AUC analysis showed no difference between the 2 groups (P = .48 by 2-sample t test).
Results of repeated infusions of receptor-modified syngeneic T cells.
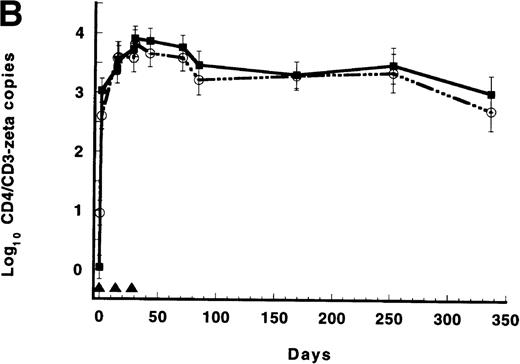
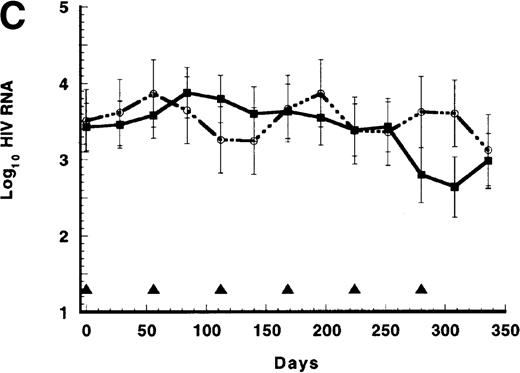
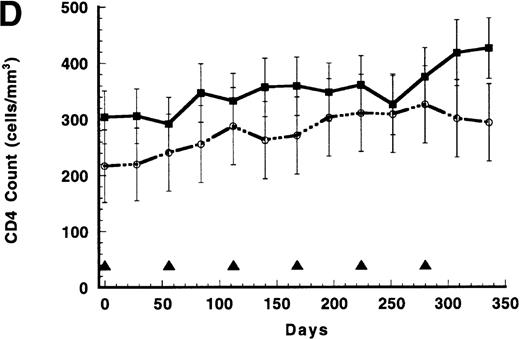
(A) Persistence of CD4/CD3-ζ–modified CD8+ T cells in the 17 patients who received gene-modified CD8+T cells without interleukin 2 (IL-2) (closed squares and solid line) and in the 4 patients who received gene-modified CD8+ T cells with IL-2 (open circles and broken line). Quantitative competitive DNA polymerase chain reaction (PCR) for the CD4/CD3-ζ gene was performed on peripheral blood mononuclear cells (PBMC), and results were expressed as log10 DNA copies per million CD8+T cells. The closed triangles spaced at 56-day intervals represent the timing of each cell infusion with and without IL-2. Data were modeled for individual patients, and a mixed-model analysis was used to derive the mean of each time point for the 2 groups. SEs are between 0.40 and 0.52 at each point. (B) Persistence of CD4/CD3-ζ–modified CD4+ T cells (closed squares and solid line) and CD8+ T cells (open circles and broken line) in the 14 patients who received modified CD4+ and CD8+ cells without IL-2. Data were modeled as described for Figure 1A and results expressed as log10 DNA copies per million CD4+ or CD8+ cells. The closed triangles spaced at 14-day intervals represent the timing of the 3 cell infusions. SEs are between 0.02 and 0.03 at each point. (C) Mean plasma human immunodeficiency virus (HIV) RNA levels in recipients of modified CD8+ T-cell infusions (closed squares and solid line, n = 17) and in recipients of unmodified CD8+ T cells (open circles and broken line, n = 9). Data were modeled as described for Figure 1A and results expressed as log10 HIV RNA copies per milliliter. Closed triangles represent the timing of cell infusions. P = .29 for the differences between the slopes of the 2 curves estimated with an interval-adjusted linear mixed model. (D) Mean CD4+ T lymphocyte counts in recipients of modified CD8+ T-cell infusions (closed squares and solid line, n = 17) and in recipients of unmodified CD8+ T cells (open circles and broken line, n = 9). Data were modeled as described for Figure 1A and results expressed as cells per milliliter. Closed triangles represent the timing of cell infusions. P = .89 for the difference between the slopes of the 2 curves estimated with an interval-adjusted linear mixed model as described for Figure 1C.
Results of repeated infusions of receptor-modified syngeneic T cells.
(A) Persistence of CD4/CD3-ζ–modified CD8+ T cells in the 17 patients who received gene-modified CD8+T cells without interleukin 2 (IL-2) (closed squares and solid line) and in the 4 patients who received gene-modified CD8+ T cells with IL-2 (open circles and broken line). Quantitative competitive DNA polymerase chain reaction (PCR) for the CD4/CD3-ζ gene was performed on peripheral blood mononuclear cells (PBMC), and results were expressed as log10 DNA copies per million CD8+T cells. The closed triangles spaced at 56-day intervals represent the timing of each cell infusion with and without IL-2. Data were modeled for individual patients, and a mixed-model analysis was used to derive the mean of each time point for the 2 groups. SEs are between 0.40 and 0.52 at each point. (B) Persistence of CD4/CD3-ζ–modified CD4+ T cells (closed squares and solid line) and CD8+ T cells (open circles and broken line) in the 14 patients who received modified CD4+ and CD8+ cells without IL-2. Data were modeled as described for Figure 1A and results expressed as log10 DNA copies per million CD4+ or CD8+ cells. The closed triangles spaced at 14-day intervals represent the timing of the 3 cell infusions. SEs are between 0.02 and 0.03 at each point. (C) Mean plasma human immunodeficiency virus (HIV) RNA levels in recipients of modified CD8+ T-cell infusions (closed squares and solid line, n = 17) and in recipients of unmodified CD8+ T cells (open circles and broken line, n = 9). Data were modeled as described for Figure 1A and results expressed as log10 HIV RNA copies per milliliter. Closed triangles represent the timing of cell infusions. P = .29 for the differences between the slopes of the 2 curves estimated with an interval-adjusted linear mixed model. (D) Mean CD4+ T lymphocyte counts in recipients of modified CD8+ T-cell infusions (closed squares and solid line, n = 17) and in recipients of unmodified CD8+ T cells (open circles and broken line, n = 9). Data were modeled as described for Figure 1A and results expressed as cells per milliliter. Closed triangles represent the timing of cell infusions. P = .89 for the difference between the slopes of the 2 curves estimated with an interval-adjusted linear mixed model as described for Figure 1C.
We considered that if IL-2 was promoting cell survival, it was likely doing so by substituting for HIV-specific CD4+ T-cell help. Therefore, a second series of cell infusions was done, in which CD4/CD3-ζ–modified CD4+ and CD8+ T cells were given together. Seventeen of the 30 subjects who were given repeated infusions of CD8+ T cells participated in this nonrandomized and uncontrolled study; Table2 shows their baseline characteristics before they received the modified CD4+ and CD8+cell infusions. Four of these patients had previously been given CD8+ cells plus IL-2, and in this phase of the study, 3 received IL-2 with the third of 3 CD4+ and CD8+cell infusions.
Baseline characteristics of the 17 patients with HIV infection who received gene-modified CD4+ and CD8+ T cells
| Characteristic . | Value . |
|---|---|
| Previous therapy, no. (%) of patients | |
| Gene-modified CD8+ T cells | 7 (41) |
| Interleukin 2 plus gene-modified CD8+ T cells | 4 (24) |
| Unmodified CD8+ T cells | 6 (35) |
| Median CD4 count, (range) | 415 (169-1867) |
| HIV RNA | |
| Median log10 copies/mL (range) | 2.37 (1.37-5.25) |
| No. (%) of patients with <50 copies/mL | 6 (35) |
| Antiretroviral therapy, no. of patients | |
| NRTIs only | 3 |
| NRTIs and PI | 14 |
| Characteristic . | Value . |
|---|---|
| Previous therapy, no. (%) of patients | |
| Gene-modified CD8+ T cells | 7 (41) |
| Interleukin 2 plus gene-modified CD8+ T cells | 4 (24) |
| Unmodified CD8+ T cells | 6 (35) |
| Median CD4 count, (range) | 415 (169-1867) |
| HIV RNA | |
| Median log10 copies/mL (range) | 2.37 (1.37-5.25) |
| No. (%) of patients with <50 copies/mL | 6 (35) |
| Antiretroviral therapy, no. of patients | |
| NRTIs only | 3 |
| NRTIs and PI | 14 |
NRTIs indicates nucleoside reverse transcriptase inhibitors; and PI, protease inhibitor.
When CD4/CD3-ζ–modified CD4+ and CD8+ T cells (1010 cells/infusion) were administered at the same time, a persistence pattern different from that resulting from modified CD8+ T-cell infusions was observed (Figure 1B). Of note, these cells were stimulated ex vivo by using costimulation through CD3 and CD28 receptors16 rather than by anti-CD3 and IL-2 stimulation. CD4/CD3-ζ DNA copy number in circulating CD4+ and, to a lesser extent, in CD8+ T cells increased slightly in the interval between cell infusions (Table3). The persistence curves for the 3 patients who received exogenous IL-2 and cells were indistinguishable from the curves of patients who received cells alone (data not shown). Gene-containing peripheral blood CD4+ and CD8+T cells were detected for at least 1 year after the cell infusions, in fractions ranging from 45 to 44 208 DNA copies/millioncells.
Survival of CD4/CD3-ζ–transduced peripheral blood mononuclear cells in 14 recipients of gene-modified CD4+and CD8+ T cells
| Day of study . | CD4+ cells . | CD8+ cells . |
|---|---|---|
| 03-150 | 0 ± 0.2 | 1.0 ± 0.2 |
| 1 | 3.0 ± 0.2 | 2.6 ± 0.2 |
| 143-150 | 3.4 ± 0.2 | 3.6 ± 0.3 |
| 15 | 3.5 ± 0.2 | 3.6 ± 0.3 |
| 283-150 | 3.7 ± 0.2 | 3.6 ± 0.2 |
| 29 | 3.9 ± 0.2 | 3.8 ± 0.2 |
| 42 | 3.9 ± 0.2 | 3.7 ± 0.2 |
| 70 | 3.8 ± 0.2 | 3.6 ± 0.2 |
| 84 | 3.5 ± 0.2 | 3.2 ± 0.3 |
| 168 | 3.3 ± 0.2 | 3.3 ± 0.2 |
| 252 | 3.5 ± 0.3 | 3.4 ± 0.3 |
| 336 | 3.0 ± 0.3 | 2.7 ± 0.3 |
| Day of study . | CD4+ cells . | CD8+ cells . |
|---|---|---|
| 03-150 | 0 ± 0.2 | 1.0 ± 0.2 |
| 1 | 3.0 ± 0.2 | 2.6 ± 0.2 |
| 143-150 | 3.4 ± 0.2 | 3.6 ± 0.3 |
| 15 | 3.5 ± 0.2 | 3.6 ± 0.3 |
| 283-150 | 3.7 ± 0.2 | 3.6 ± 0.2 |
| 29 | 3.9 ± 0.2 | 3.8 ± 0.2 |
| 42 | 3.9 ± 0.2 | 3.7 ± 0.2 |
| 70 | 3.8 ± 0.2 | 3.6 ± 0.2 |
| 84 | 3.5 ± 0.2 | 3.2 ± 0.3 |
| 168 | 3.3 ± 0.2 | 3.3 ± 0.2 |
| 252 | 3.5 ± 0.3 | 3.4 ± 0.3 |
| 336 | 3.0 ± 0.3 | 2.7 ± 0.3 |
Values are model-based least-squares means ± SE log10 DNA copy number per 106 cells analyzed.
Indicates day on which cells were infused.
In vivo trafficking of cells
To study homing patterns of the infused cells, an aliquot of ex vivo–expanded CD8+ T cells was labeled with indium 111–oxine before infusion in 4 subjects, and γ-camera imaging was performed approximately 2 to 4, 24, and 48 hours after the cells were given. Figure 2 shows results from 1 representative series of scans. Radiolabeled cells initially distributed to reticuloendothelial organs (liver and spleen) and the lungs. By 20 and 43 hours, the lungs had substantial clearing, whereas the liver and spleen continued to show preferential uptake. At these later time points, the labeled cells also distributed to the bone marrow (spine and iliac bones). Peripheral lymph nodes showed no evidence of preferential uptake of the labeled cells at any of the time points studied. The distribution pattern of the CD8+ T cells in this study was similar to that described after autologous PBMC infusions in healthy volunteers,21 in whom cells were neither activated nor expanded ex vivo, and in HIV-infected patients given ex vivo–activated, autologous CD8+cells.22
Whole-body γ-camera images from 1 representative patient who received 2.5 × 109 indium 111–oxine–labeled CD4/CD3-ζ–modified CD8+ T cells in a total of 1010 gene-modified cells at time 0.
The images shown are anterior projection geometric means from 4, 20, and 43 hours after infusion of cells.
Whole-body γ-camera images from 1 representative patient who received 2.5 × 109 indium 111–oxine–labeled CD4/CD3-ζ–modified CD8+ T cells in a total of 1010 gene-modified cells at time 0.
The images shown are anterior projection geometric means from 4, 20, and 43 hours after infusion of cells.
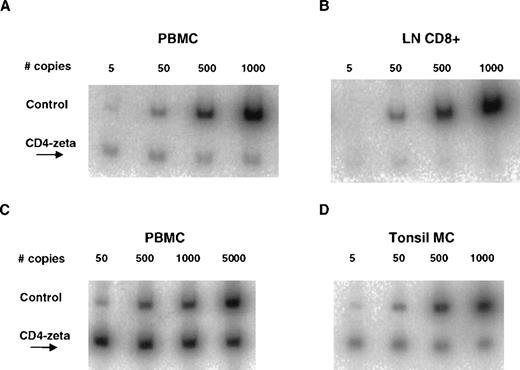
Lymph node or tonsillar biopsies were performed in 5 patients between 3 and 21 weeks after infusion of CD4/CD3-ζ–transduced CD8+T cells to detect the presence or absence or preferential lymphoid distribution of the transduced cells. In all patients studied, CD4/CD3-ζ DNA copy numbers in tissue were either equivalent to or below simultaneously determined values in PBMC and were usually below the limit of quantitation (< 30 copies/million cells) for the PCR assay (Figure 3A-B). We also assessed in vivo trafficking in 2 patients who received gene-modified CD4+ and CD8+ T cells. A lymph node biopsy was performed in 1 patient 2 weeks after cell infusion, and a tonsillar biopsy was performed 1 day after infusion in the other patient. As was observed after infusion of CD8+ T cells alone, CD4/CD3-ζ–containing T cells were present in these lymphoid tissue samples at fractions equal to or below that in simultaneously collected PBMC (Figure 3C-D).
Comparison of CD4/CD3-ζ DNA copy number obtained from PBMC and lymphoid tissue by quantitative competitive PCR.
(A) PBMC sample (142 copies/million cells) and (B) lymph node purified CD8+ cell fraction (below the limits of quantitation) obtained from 1 patient 10 weeks after an infusion of 109CD4/CD3-ζ–modified CD8+ T cells. Similar data are shown in panel C for PBMC (> 5000 copies/million cells) and in panel D for unfractionated mononuclear cells (MC) from tonsillar tissue (567 copies/million cells) for a different patient 2 weeks after an infusion of 1010 CD4/CD3-ζ–modified CD4+ and CD8+ T cells. The e+ cell line, which contains 10 copies of the CD4/CD3-ζ gene with a 102-base-pair DNA insert per cell, was used for standardization.
Comparison of CD4/CD3-ζ DNA copy number obtained from PBMC and lymphoid tissue by quantitative competitive PCR.
(A) PBMC sample (142 copies/million cells) and (B) lymph node purified CD8+ cell fraction (below the limits of quantitation) obtained from 1 patient 10 weeks after an infusion of 109CD4/CD3-ζ–modified CD8+ T cells. Similar data are shown in panel C for PBMC (> 5000 copies/million cells) and in panel D for unfractionated mononuclear cells (MC) from tonsillar tissue (567 copies/million cells) for a different patient 2 weeks after an infusion of 1010 CD4/CD3-ζ–modified CD4+ and CD8+ T cells. The e+ cell line, which contains 10 copies of the CD4/CD3-ζ gene with a 102-base-pair DNA insert per cell, was used for standardization.
Virologic and immunologic effects of the cells
As shown in Figure 1C, a small decline in mean HIV RNA levels occurred in patients who received repeated infusions of either modified CD8+ cells or unmodified CD8+ cells (slope, −0.0022 log10 HIV RNA copies/day for the modified-cell group versus −0.0006 copies/day for the unmodified-cell group; P = .29 by t test for interaction in the interval-adjusted linear mixed model). Although (despite randomization) the groups differed in their baseline mean total CD4+ T-cell counts, the slopes of the 2 CD4+ T-cell curves were nearly the same (slope, 0.28 cells/day for the modified group versus 0.30 cells/day for the unmodified group; P = .89 by t test for the interaction in the interval-adjusted linear mixed model; Figure 1D). The 4 patients who received modified CD8+ cells and IL-2 infusions during this period had similar changes in mean plasma HIV RNA levels (slope, −0.0007 log10 copies/day) while showing the expected IL-2–induced rise in mean CD4+ T-cell count over the year (slope, 1.44 cells/day).23
In the patients who received modified CD4+ and modified CD8+ cells without IL-2, HIV RNA levels were essentially unchanged from baseline values at 6 and 10 weeks after the start of cell infusions (baseline log10 copies/mL, 2.87 ± 0.38; week 6 log10 copies/mL, 3.03 ± 0.41; and week 10 log10 copies/mL, 2.81 ± 0.39), whereas CD4 counts increased from a mean of 390 ± 71 at week 0 to 503 ± 77 at week 6 and to 471 ± 74 at week 10. Three patients received an infusion of exogenous IL-2 at week 4 with the third infusion of modified CD4+ and modified CD8+ cells. By week 10, CD4 counts had increased in all 3 patients by 166 to 509 cells/μL; in the 2 patients with detectable plasma viremia (baseline HIV RNA, 2.97 and 3.40 log10 copies/mL, respectively), HIV RNA levels fell 0.46 and 1.64 log10, respectively, by week 10.
Clinical effects and tolerance of the cell infusions
A total of 212 T-cell infusions were given (161 of CD8+ cells and 51 of CD4+ plus CD8+ cells). No serious adverse event related to the cell infusions occurred. Minor side effects, such as fever, chills, and other events listed in Table 4, accompanied 28% of the infusions (occurrence rate, 30% for CD8+ T cells and 22% for CD4+ plus CD8+ T cells). No cell infusion was interrupted because of side effects. Premedication with acetaminophen, ibuprofen, or meperidine was offered to patients who previously had fever, chills, or other influenza-like symptoms with infusion.
Tolerance of the cell infusions: adverse clinical events
| Clinical event . | CD8+ cells . | CD4+plus CD8+ cells . |
|---|---|---|
| Chills | 15 | 1 |
| Fever | 10 | 2 |
| Headache | 6 | 0 |
| Nausea | 5 | 0 |
| Rigors | 4 | 1 |
| Bronchospasm | 3 | 2 |
| Myalgias | 4 | 0 |
| Pruritus | 0 | 2 |
| Rash | 0 | 2 |
| Blurred vision | 1 | 0 |
| Hypertension | 0 | 1 |
| Total episodes | 48 | 11 |
| Total episodes/total infusions (%) | 48/161 (30) | 11/51 (22) |
| Clinical event . | CD8+ cells . | CD4+plus CD8+ cells . |
|---|---|---|
| Chills | 15 | 1 |
| Fever | 10 | 2 |
| Headache | 6 | 0 |
| Nausea | 5 | 0 |
| Rigors | 4 | 1 |
| Bronchospasm | 3 | 2 |
| Myalgias | 4 | 0 |
| Pruritus | 0 | 2 |
| Rash | 0 | 2 |
| Blurred vision | 1 | 0 |
| Hypertension | 0 | 1 |
| Total episodes | 48 | 11 |
| Total episodes/total infusions (%) | 48/161 (30) | 11/51 (22) |
Values are number of episodes unless otherwise indicated.
Although this study did not have enough power to reliably detect differences in clinical end points between groups, HIV-related clinical events and deaths were recorded. No major differences between the modified-cell and unmodified-cell groups were observed. In the modified-cell group, 1 case each of non-Hodgkin lymphoma (NHL), progressive multifocal leukoencephalopathy (PML), and anorectal carcinoma occurred. In the unmodified group, 2 cases of presumptive PML (resulting in 1 death) and 1 case of NHL (also resulting in death) occurred.
Discussion
We demonstrated that genetically engineered, HIV-specific, ex vivo–expanded and –activated CD8+ and CD4+cells can be administered safely at doses of up to 1010cells without producing evidence of substantive acute or cumulative toxicity. We did not study cell numbers above 1010 in this trial, so the maximal tolerated dose was not determined. HIV-specific CD8+ T-cell therapy resulted in long-term persistence of gene-modified cells in the bloodstream, but the fraction of gene-modified cells declined with time, even during the course of repeated cell administrations. This observation raises the possibility that immune mechanisms leading to shortened cell survival with reexposure may be a limiting factor.24,25 Providing CD4+ T-cell help in the form of exogenous IL-2 infusions or HIV-specific, CD4/CD3-ζ–modified CD4+ T cells appeared to alter the in vivo survival of the modified cells. This finding was particularly striking when HIV-specific CD4+ T cells were coadministered with CD8+ T cells after ex vivo costimulation with anti-CD3 and anti-CD28; in this setting, transduced CD8+ and CD4+ T-cell fractions increased transiently and persisted at relatively high levels (0.1%-0.01% of circulating cells) for many months. We also showed, using indium 111 radiolabeling studies, that ex vivo manipulation, including cell transduction, did not alter the typical tissue-distribution pattern observed with nonmanipulated PBMC in healthy volunteers21or ex vivo–expanded autologous CD8+ T-lymphocyte infusions in patients with HIV infection.26
We used the least-squares method with linear mixed-effect modeling to estimate the immunologic and virologic effects of the cell infusions and to characterize the persistence of gene-modified cells in the circulation. We also imputed values for CD4/CD3-ζ DNA and HIV RNA levels below detection limits by substituting random numbers from an exponential distribution ranging from 0 to the detection limit. These complicated approaches were important for this study because they account for interdependence of longitudinal observations within patients and the heavy-tailed distributions of the variables being analyzed while not underestimating variance.18 In the case of HIV RNA, when we repeated the analysis substituting 49 for each value below 50 copies/mL rather than using exponential replacements, the least-squares means were slightly higher and the SEs slightly smaller; the overall difference in the results was small, however, and did not change our conclusions.
The cells administered in this study did not appear to have an antiretroviral effect in vivo. Other investigators have reported similar findings when HIV p24 antigenemia or plasma HIV RNA levels were assayed after treatments with either polyclonal, ex vivo–expanded CD8+ cells,22,27 HIV-specific CTL lines,28 or HIV-specific CTL clones.29,30 One patient who received infusions of an autologous Nef-specific CTL clone and IL-2 had clinical deterioration.29 Several possibilities could account for the lack of correlation between in vitro and in vivo cytotoxicity. Suppression of transcription from the transgene resulting in gene silencing was one consideration.31 We analyzed a small number of PBMC samples from our patients and were consistently able to detect CD4/CD3-ζ messenger RNA, suggesting that this was not a major factor (data not shown). Apoptosis is another mechanism purported to account for lack of in vivo killing32-34; however, survival of transduced cells would be expected to shorten in this instance and this was not observed in our study, particularly when CD4+ and CD8+ T cells were coadministered. A survival pattern consistent with immune elimination24,25 was observed with the CD8+T-cell infusions alone, and this might have been responsible for the limited activity of those cells. However, this same survival pattern was not evident when CD4+ and CD8+ T cells were given together or when CD8+ T cells were given with IL-2, raising the possibility that IL-2–induced anergy to the CD4/CD3-ζ transgene accounted for increased cell survival.35
Optimal survival of genetically engineered HIV-specific CTL was achieved by culturing the cells with anti-CD3 and anti-CD28 costimulation16 and coadministering the gene-modified CD4+ and CD8+ T cells. Costimulation was shown previously to enhance ex vivo expansion of CD4+ T cells, to provide transient resistance of the cells to HIV infection, and to prevent antigen-induced apoptosis.16 36 Future studies investigating the potential therapeutic role of culture-expanded CTL should incorporate these modifications. The lack of measurable in vivo antiretroviral activity remains unexplained. Studies in progress will explore potential antiviral activity by evaluating HIV-specific CTL therapy in the setting of controlled viremia and measuring HIV load in compartments and cell populations believed to represent latent reservoirs of replication-competent HIV.
Acknowledgments
We thank the patients and their referring physicians for their willingness to participate in and support this study; the staff of the NIAID inpatient unit and the outpatient research clinic of the NIAID and Critical Care Medicine Department; and A. S. Fauci, R. A. Morgan, and C. S. Carter for their helpful discussions and support.
Funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract NO1-C0-56000.
Reprints:H. Clifford Lane, Building 10, Room 11S231, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892-1894; e-mail: clane@niaid.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal