Abstract
Although considerable effort has been devoted to characterizing alloantibodies specific for the Rhesus D (RhD) blood group antigen, virtually nothing is known about the helper response that drives their production. Therefore, the aim of this study was to map alloreactive T-cell epitopes on the RhD protein. Peripheral blood mononuclear cells (PBMCs) were obtained from 22 RhD-negative volunteers in whom anti-D alloantibodies had developed after deliberate immunization or RhD-incompatible pregnancy. The PBMCs were stimulated with a panel of up to 68 overlapping synthetic 15-mer peptides spanning the complete sequence of the RhD protein. One or more peptides elicited proliferative responses by PBMCs from all 22 of the alloimmune volunteers but from only 2 of 8 alloantibody-negative control donors. Proliferation of PBMCs from the alloimmune donors was mediated by major histocompatibility complex class II–restricted T cells expressing the CD45RO marker of previous activation or memory. The number of peptides that induced proliferative responses was unrelated to either the frequency of, or time since, exposure to RhD-positive red blood cells, but it correlated strongly (Rs = 0.75;P < .003) with the level of anti-D antibodies in deliberately immunized donors. The patterns of stimulatory peptides varied among alloimmune volunteers, but particular sequences were commonly recognized, with 4 peptides each eliciting a response in more than 50% of these donors. Identification of such peptides containing dominant alloreactive helper epitopes is the first step in the development of improved or new approaches to preventing hemolytic disease of the newborn that are based on modulating the T-cell response to the RhD protein.
Introduction
The most immunogenic and clinically important blood group borne by the Rhesus complex is the Rhesus D (RhD) antigen.1 Not only is this antigen routinely considered in transfusion medicine, but RhD incompatibility between mother and fetus is the classic cause of hemolytic disease of the newborn (HDN). Although infant mortality due to HDN has been dramatically reduced by passive administration of human anti-D immunoglobulin to women at risk of alloimmunization, this prophylactic program is threatened by growing ethical concerns over the deliberate immunization of RhD-negative volunteers to produce human blood products.2 In particular, fears about the transmission of viral disease and the spongiform encephalopathies have recently resulted in difficulties in recruiting volunteers for immunization programs, or cessation of such programs, with potential for shortages of anti-D immunoglobulin. Furthermore, despite the use of anti-D immunoglobulin, alloimmunization still occurs in 1% to 1.5% of women at risk and cannot be reversed once an immune response has been initiated. Although monoclonal antibodies (mAbs) specific for RhD have been developed, it remains unclear whether these preparations will be safe, efficacious, or affordable replacements for polyclonal anti-D immunoglobulin.1 3 There is therefore an urgent need to more fully understand the immune response to RhD in order to design better strategies to prevent HDN.
The major focus for research on the alloimmune response to RhD has been characterization of the red blood cell (RBC) membrane antigen that is recognized by anti-D alloantibodies. With the development of modern immunochemical techniques, it was demonstrated that anti-D antiserum or mAbs immunoprecipitate a nonglycosylated protein of approximately 30 kd that is exposed on the surface of RhD-positive, but not RhD-negative, RBCs.4 The amino acid sequence of this RhD protein was deduced from complementary DNA (cDNA) analysis,5,6 and transfection experiments confirmed that it does indeed encode the RhD antigen.7 Studies using Southern analysis revealed the genetic basis of RhD-positive and RhD-negative polymorphism by demonstrating that the corresponding RHD gene is deleted in RhD-negative individuals and that no alternative allelic form exists at the same locus in Caucasians.8 Characterization of anti-D mAbs binding to RBCs bearing rare RhD subtypes revealed that the RhD antigen includes several distinct epitopes,9 and testing of additional mAbs suggested that there may be up to 30 B-cell determinants.10
In striking contrast to the emphasis on elucidating the molecular structure of the epitopes recognized by anti-D antibodies, virtually nothing is known about the specificity of RhD protein–reactive helper T cells. However, it is clear that T-cell help must play a crucial role in the production of anti-D antibodies, since the generation of most IgG antibodies depends on T cells and responses to RBC antigens appear to be no exception. Thus, in mice, T-cell depletion blocks the antibody response to injected rat RBCs11 and anti-RBC autoantibody production can be abrogated by anti-CD4 mAb.12,13 Helper T cells recognize short processed peptides bound to the products of major histocompatibility complex (MHC) class II genes on antigen-presenting cells (APCs),14 and with the determination of the primary structure of the components of the polymorphic Rh complex,5,6,15,16 it should now be possible to identify alloreactive helper epitopes on the RhD protein. Indeed, a panel of synthetic peptides spanning the sequence of the RhD protein has already been used to map the epitopes that elicit in vitro proliferative responses by autoreactive T cells from patients with autoimmune hemolytic anemia (AIHA).17
Mapping the helper T-cell epitopes on the RhD protein would enable at least 2 novel strategies for HDN prevention to be explored. First, it may be possible to produce safe, synthetic peptide immunogens to boost the helper response and replace the RBCs given to volunteers in the current program of prophylactic anti-D antibody production. Second, the ability to induce immunologic tolerance in animals by administration of peptides corresponding to T-cell epitopes or substituted epitope analogues18-20 has raised the possibility that alloimmunization in RhD-negative women could be controlled in a similar way. However, neither of these novel approaches will be possible until the fine specificity of the T-helper response that drives anti-D alloantibody production has been determined. Therefore, the purpose of this project was to map the epitopes on the RhD protein that are recognized by helper T cells from a series of RhD-alloimmunized donors.
Materials and methods
Donors
Blood samples were obtained from 22 healthy, RhD-negative volunteers (cde phenotype) whose serum contained anti-D alloantibodies. Nineteen of these volunteers were plasma donors from the program to produce anti-D immunoglobulin for the Scottish National Blood Transfusion Service, and 13 of them had received boosters of deliberate immunizations with 1 mL packed RhD-positive RBCs. The remaining 6 plasma donors from the program and an additional 3 whole-blood donors had produced anti-D antibodies as a result of RhD-incompatible pregnancy and had not been deliberately alloimmunized. Information about the deliberately and naturally immunized donors is given in Table1 and Table2, respectively. Concentrations of anti-D antibodies in serum samples were determined by the Scottish National Blood Transfusion Service, and where more than one measurement was obtained during the study, the highest level was recorded. The presence or absence of anti-C and anti-E in addition to anti-D in response to boosting was also noted. Negative-control blood samples were collected from healthy, RhD-negative volunteers (cde phenotype) with no serologic evidence of anti-D alloantibodies. All blood was obtained by venipuncture and was drawn into tubes containing citrate or preservative-free heparin anticoagulant.
Rhesus D peptides that stimulate peripheral blood mononuclear cells from deliberately alloimmunized RhD-negative plasma donors to proliferate in vitro
| Donor . | Sex . | HLA-DR type . | Serum anti-D antibody (IU/mL*) + other antibody . | Deliberate immunizations with RhD+RBCs . | RhD peptides stimulating proliferation† . | |
|---|---|---|---|---|---|---|
| No. times/type RBCs . | Date of most recent immunization . | |||||
| 1 | M | DRB1*15;11 | 699 + C‡ + E | 5/R2 cells | 7/1996 | 2, 3, 4, 5, 6, 10, 13, 18, 21, 26, 28, 40, 42, 43, 44, 47, 52, 54, 68 |
| 2 | M | DRB1*1;11 | 462 + C + E | 7/NK | 9/1994 | 5,8, 10, 12, 17, 46, 63 |
| 3 | M | DRB1*15;7 | 37 + C | 5/NK | 7/1996 | 13, 28, 63 |
| 4 | M | DRB1*3;11 | 1190 + C‡ + E | 6/R2 cells | 7/1997 | 3, 5, 6, 7, 9, 12, 13, 17, 22, 25, 27, 29, 33, 40,46, 47, 49, 51, 52, 53, 55, 59, 63, 65, 66, 67, 68 |
| 5 | M | DRB1*3;7 | 3 + C + E | 3/R1R2 cells | 6/1997 | 16 |
| 6 | M | DRB1*15 | 196 + C‡ + E | 6/R2 cells | 7/1997 | 1, 25, 6, 7, 8, 9, 10, 13, 14, 17, 19, 22, 24, 25, 26, 27, 28, 37, 44, 46, 47, 51, 53, 59, 61, 63,64, 67, 68 |
| 7 | M | DRB1*7;13 | 304 + C | 5/R1 cells | 7/1997 | 4,17, 52 |
| 8 | M | DRB1*15;11 | 2724 + C‡ + E | 4/R2 cells | 8/1997 | 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 20, 22, 23, 24, 26, 27, 28, 32, 37, 39, 40, 44, 45,46, 48, 49, 52, 54, 55, 56, 58, 59, 61, 62, 63,66, 67 |
| 9 | F | DRB1*15;7 | 215 + C‡ + E | 5/R2 cells | 7/1994 | 53 |
| 10 | F | DRB1*15 | 590 + C | 9/R1 cells | 7/1997 | 5,6, 10, 11, 17, 27 |
| 11 | F | DRB1*7 | 128 + C | 4/R1 cells | 7/1997 | 9, 13, 16, 17, 26, 28, 46, 47, 63, 68 |
| 12 | F | DRB1*15;13 | 699 + C‡ | 3/R2 cells | 8/1993 | 2, 3, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 19, 20,21, 22, 24, 26, 27, 28, 29, 30, 32, 35, 36, 38, 40, 41, 45,46, 47, 48, 49, 51, 53, 55, 56, 58, 59, 60, 61, 62, 63,64 |
| 13 | F | DRB1*15 | 1528 + C | 2/R1 cells | 7/1994 | 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 20, 21, 22, 23, 26, 27, 28, 30, 33, 38, 39, 40, 41, 44, 45, 46, 48, 49, 54, 55, 56, 57, 58, 61, 62, 63, 67, 68 |
| Donor . | Sex . | HLA-DR type . | Serum anti-D antibody (IU/mL*) + other antibody . | Deliberate immunizations with RhD+RBCs . | RhD peptides stimulating proliferation† . | |
|---|---|---|---|---|---|---|
| No. times/type RBCs . | Date of most recent immunization . | |||||
| 1 | M | DRB1*15;11 | 699 + C‡ + E | 5/R2 cells | 7/1996 | 2, 3, 4, 5, 6, 10, 13, 18, 21, 26, 28, 40, 42, 43, 44, 47, 52, 54, 68 |
| 2 | M | DRB1*1;11 | 462 + C + E | 7/NK | 9/1994 | 5,8, 10, 12, 17, 46, 63 |
| 3 | M | DRB1*15;7 | 37 + C | 5/NK | 7/1996 | 13, 28, 63 |
| 4 | M | DRB1*3;11 | 1190 + C‡ + E | 6/R2 cells | 7/1997 | 3, 5, 6, 7, 9, 12, 13, 17, 22, 25, 27, 29, 33, 40,46, 47, 49, 51, 52, 53, 55, 59, 63, 65, 66, 67, 68 |
| 5 | M | DRB1*3;7 | 3 + C + E | 3/R1R2 cells | 6/1997 | 16 |
| 6 | M | DRB1*15 | 196 + C‡ + E | 6/R2 cells | 7/1997 | 1, 25, 6, 7, 8, 9, 10, 13, 14, 17, 19, 22, 24, 25, 26, 27, 28, 37, 44, 46, 47, 51, 53, 59, 61, 63,64, 67, 68 |
| 7 | M | DRB1*7;13 | 304 + C | 5/R1 cells | 7/1997 | 4,17, 52 |
| 8 | M | DRB1*15;11 | 2724 + C‡ + E | 4/R2 cells | 8/1997 | 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 20, 22, 23, 24, 26, 27, 28, 32, 37, 39, 40, 44, 45,46, 48, 49, 52, 54, 55, 56, 58, 59, 61, 62, 63,66, 67 |
| 9 | F | DRB1*15;7 | 215 + C‡ + E | 5/R2 cells | 7/1994 | 53 |
| 10 | F | DRB1*15 | 590 + C | 9/R1 cells | 7/1997 | 5,6, 10, 11, 17, 27 |
| 11 | F | DRB1*7 | 128 + C | 4/R1 cells | 7/1997 | 9, 13, 16, 17, 26, 28, 46, 47, 63, 68 |
| 12 | F | DRB1*15;13 | 699 + C‡ | 3/R2 cells | 8/1993 | 2, 3, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 19, 20,21, 22, 24, 26, 27, 28, 29, 30, 32, 35, 36, 38, 40, 41, 45,46, 47, 48, 49, 51, 53, 55, 56, 58, 59, 60, 61, 62, 63,64 |
| 13 | F | DRB1*15 | 1528 + C | 2/R1 cells | 7/1994 | 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 20, 21, 22, 23, 26, 27, 28, 30, 33, 38, 39, 40, 41, 44, 45, 46, 48, 49, 54, 55, 56, 57, 58, 61, 62, 63, 67, 68 |
Peptides containing Rhesus D–specific polymorphisms are in boldface. The results are combined if more than one sample was tested.
RhD indicates Rhesus D; RBCs, red blood cells; NK, not known.
Quantified against national reference standards; if tested more than once, highest level is shown.
Peptides 34, 35, and 36 containing Met218 were not tested in donors 1-5, 7, 9, or 13.
The presence of anti-C when given only R2 cells indicates anti-G.
Rhesus D peptides that stimulate peripheral blood mononuclear cells from naturally alloimmunized RhD-negative women to proliferate in vitro
| Donor . | HLA-DR type . | Serum anti-D antibody (IU/mL*) + other antibody . | No. of pregnancies/most recent pregnancy (y) with RhD+ infant . | RhD peptides stimulating proliferation† . |
|---|---|---|---|---|
| 14 | DRB1*1;4 | 86 + C | 2/1993 | 6 |
| 15 | DRB1*4 | 43 + C | 2/1999 | 2, 3, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 20, 21, 22, 23, 25, 26, 27, 28, 40, 45, 46, 48, 56, 58, 60, 61, 62, 63, 65, 68 |
| 16 | DRB1*3;11 | 6.1 + C | 2/1987 | 2, 3, 4, 5, 6, 7, 8, 11, 13, 16, 17, 18, 19, 21, 22, 23, 24, 28, 29, 32, 38, 40, 46, 47, 52, 53, 55, 56, 58, 59,61, 64 |
| 17 | DRB1*15;3 | 132 + C | 3/1996 | 2, 3,6, 12, 1619, 22, 26, 28, 32, 40, 53, 54, 56, 59, 60, 62 |
| 18 | DRB1*1;15 | 7 | 1/1984 | 17 |
| 19 | DRB1*3;4 | 5 | NK/NK | 6, 7, 9, 10, 13, 14, 16, 17, 21, 22, 28, 46, 53, 55, 60, 61, 62 |
| 20 | NK | 1/1 | 1/1994 | 6 |
| 21 | DRB1*4;13 | 1/2560 | 1/1997 | 9, 13, 16, 63, 68 |
| 22 | DRB1*7;8 | 1/32 | 3/1971 | 2, 17, 28 |
| Donor . | HLA-DR type . | Serum anti-D antibody (IU/mL*) + other antibody . | No. of pregnancies/most recent pregnancy (y) with RhD+ infant . | RhD peptides stimulating proliferation† . |
|---|---|---|---|---|
| 14 | DRB1*1;4 | 86 + C | 2/1993 | 6 |
| 15 | DRB1*4 | 43 + C | 2/1999 | 2, 3, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 20, 21, 22, 23, 25, 26, 27, 28, 40, 45, 46, 48, 56, 58, 60, 61, 62, 63, 65, 68 |
| 16 | DRB1*3;11 | 6.1 + C | 2/1987 | 2, 3, 4, 5, 6, 7, 8, 11, 13, 16, 17, 18, 19, 21, 22, 23, 24, 28, 29, 32, 38, 40, 46, 47, 52, 53, 55, 56, 58, 59,61, 64 |
| 17 | DRB1*15;3 | 132 + C | 3/1996 | 2, 3,6, 12, 1619, 22, 26, 28, 32, 40, 53, 54, 56, 59, 60, 62 |
| 18 | DRB1*1;15 | 7 | 1/1984 | 17 |
| 19 | DRB1*3;4 | 5 | NK/NK | 6, 7, 9, 10, 13, 14, 16, 17, 21, 22, 28, 46, 53, 55, 60, 61, 62 |
| 20 | NK | 1/1 | 1/1994 | 6 |
| 21 | DRB1*4;13 | 1/2560 | 1/1997 | 9, 13, 16, 63, 68 |
| 22 | DRB1*7;8 | 1/32 | 3/1971 | 2, 17, 28 |
Peptides containing Rhesus D–specific polymorphisms are in boldface. The results are combined if more than one sample was tested.
RhD indicates Rhesus D; NK, not known.
Quantified against national reference standards; if tested more than once, highest level is shown. For donors 20, 21, and 22 (whole-blood donors), the value shown is the antibody titer determined by indirect antiglobulin test.
Peptides 34, 35, and 36 containing Met218 were not tested.
Antigens and mitogens
A complete panel of 68 15-mer peptides with 5 to 10 amino acid overlaps was synthesized (Department of Biochemistry, University of Bristol, United Kingdom), spanning the sequence of the 30-kd Rh protein associated with expression of the D antigen (Table3). The amino acid sequence for the RhD protein was previously deduced independently by 2 laboratories on the basis of cDNA analyses5,6; the findings differ only at residue 218. The variant Ile2185 (GenBank accession number M34015) was initially chosen for the peptide panel, since it is widely cited21; however, after our study was completed, it became known that this sequence is probably the result of a sequencing error8 and that the alternative, Met21822(GenBank accession number L08429), may be correct. For this report, therefore, we omitted the original results obtained with peptides 34, 35, and 36 containing Ile218 and, where possible, replaced them with data from repeated assays using the corresponding Met218 substituted peptides.
Amino acid sequences of the peptide panel spanning the Rhesus D protein56
| Peptide . | Sequence . | Residues . |
|---|---|---|
| 1 | SSKYPRSVRRCLPLW | 2-16 |
| 2 | CLPLWALTLEAALIL | 12-26 |
| 3 | AALILLFYFFTHYDA | 22-36 |
| 4 | THYDASLEDQKGLVA | 32-46 |
| 5 | KGLVASYQVGQDLTV | 42-56 |
| 6 | QDLTVMAAIGLGFLT | 52-66 |
| 7 | MAAIGLGFLTSSFRR | 57-71 |
| 8 | LGFLTSSFRRHSWSS | 62-76 |
| 9 | SSFRRHSWSSVAFNL | 67-81 |
| 10 | HSWSSVAFNLFMLAL | 72-86 |
| 11 | FMLALGVQWAILLDG | 82-96 |
| 12 | ILLDGFLSQFPSGKV | 92-106 |
| 13 | FLSQFPSGKVVITLF | 97-111 |
| 14 | PSGKVVITLFSIRLA | 102-116 |
| 15 | VITLFSIRLATMSAL | 107-121 |
| 16 | SIRLATMSALSVLIS | 112-126 |
| 17 | TMSALSVLISVDAVL | 117-131 |
| 18 | SVLISVDAVLGKVNL | 122-136 |
| 19 | VDAVLGKVNLAQLVV | 127-141 |
| 20 | GKVNLAQLVVMVLVE | 132-146 |
| 21 | MVLVEVTALGNLRMV | 142-156 |
| 22 | VTALGNLRMVISNIF | 147-161 |
| 23 | NLRMVISNIFNTDYH | 152-166 |
| 24 | ISNIFNTDYHMNMMH | 157-171 |
| 25 | NTDYHMNMMHIYVFA | 162-176 |
| 26 | MNMMHIYVFAAYFGL | 167-181 |
| 27 | IYVFAAYFGLSVAWC | 172-186 |
| 28 | AYFGLSVAWCLPKPL | 177-191 |
| 29 | SVAWCLPKPLPEGTE | 182-196 |
| 30 | LPKPLPEGTEDKDQT | 187-201 |
| 31 | PEGTEDKDQTATIPS | 192-206 |
| 32 | DKDQTATIPSLSAML | 197-211 |
| 33 | ATIPSLSAMLGALFL | 202-216 |
| 343-150 | LSAMLGALFLWMFWP | 207-221 |
| 353-150 | GALFLWMFWPSFNSA | 212-226 |
| 363-150 | WMFWPSFNSALLRSP | 217-231 |
| 37 | SFNSALLRSPIERKN | 222-236 |
| 38 | LLRSPIERKNAVFNT | 227-241 |
| 39 | IERKNAVFNTYYAVA | 232-246 |
| 40 | AVFNTYYAVAVSVVT | 237-251 |
| 41 | YYAVAVSVVTAISGS | 242-256 |
| 42 | AISGSSLAHPQGKIS | 252-266 |
| 43 | SLAHPQGKISKTYVH | 257-271 |
| 44 | QGKISKTYVHSAVLA | 262-276 |
| 45 | KTYVHSAVLAGGVAV | 267-281 |
| 46 | SAVLAGGVAVGTSCH | 272-286 |
| 47 | GTSCHLIPSPWLAMV | 282-296 |
| 48 | WLAMVLGLVAGLISV | 292-306 |
| 49 | LGLVAGLISVGGAKY | 297-311 |
| 50 | GLISVGGAKYLPGCC | 302-316 |
| 51 | GGAKYLPGCCNRVLG | 307-321 |
| 52 | LPGCCNRVLGIPHSS | 312-326 |
| 53 | NRVLGIPHSSIMGYN | 317-331 |
| 54 | IPHSSIMGYNFSLLG | 322-336 |
| 55 | IMGYNFSLLGLLGEI | 327-341 |
| 56 | FSLLGLLGEIIYIVL | 332-346 |
| 57 | LLGEIIYIVLLVLDT | 337-351 |
| 58 | IYIVLLVLDTVGAGN | 342-356 |
| 59 | LVLDTVGAGNGMIGF | 347-361 |
| 60 | VGAGNGMIGFQVLLS | 352-366 |
| 61 | QVLLSIGELSLAIVI | 362-376 |
| 62 | LAIVIALTSGLLTGL | 372-386 |
| 63 | LLTGLLLNLKIWKAP | 382-396 |
| 64 | LLNLKIWKAPHEAKY | 387-401 |
| 65 | IWKAPHEAKYFDDQV | 392-406 |
| 66 | HEAKYFDDQVFWKFP | 397-411 |
| 67 | FDDQVFWKFPHLAVG | 402-416 |
| 68 | DDQVFWKFPHLAVGF | 403-417 |
| Peptide . | Sequence . | Residues . |
|---|---|---|
| 1 | SSKYPRSVRRCLPLW | 2-16 |
| 2 | CLPLWALTLEAALIL | 12-26 |
| 3 | AALILLFYFFTHYDA | 22-36 |
| 4 | THYDASLEDQKGLVA | 32-46 |
| 5 | KGLVASYQVGQDLTV | 42-56 |
| 6 | QDLTVMAAIGLGFLT | 52-66 |
| 7 | MAAIGLGFLTSSFRR | 57-71 |
| 8 | LGFLTSSFRRHSWSS | 62-76 |
| 9 | SSFRRHSWSSVAFNL | 67-81 |
| 10 | HSWSSVAFNLFMLAL | 72-86 |
| 11 | FMLALGVQWAILLDG | 82-96 |
| 12 | ILLDGFLSQFPSGKV | 92-106 |
| 13 | FLSQFPSGKVVITLF | 97-111 |
| 14 | PSGKVVITLFSIRLA | 102-116 |
| 15 | VITLFSIRLATMSAL | 107-121 |
| 16 | SIRLATMSALSVLIS | 112-126 |
| 17 | TMSALSVLISVDAVL | 117-131 |
| 18 | SVLISVDAVLGKVNL | 122-136 |
| 19 | VDAVLGKVNLAQLVV | 127-141 |
| 20 | GKVNLAQLVVMVLVE | 132-146 |
| 21 | MVLVEVTALGNLRMV | 142-156 |
| 22 | VTALGNLRMVISNIF | 147-161 |
| 23 | NLRMVISNIFNTDYH | 152-166 |
| 24 | ISNIFNTDYHMNMMH | 157-171 |
| 25 | NTDYHMNMMHIYVFA | 162-176 |
| 26 | MNMMHIYVFAAYFGL | 167-181 |
| 27 | IYVFAAYFGLSVAWC | 172-186 |
| 28 | AYFGLSVAWCLPKPL | 177-191 |
| 29 | SVAWCLPKPLPEGTE | 182-196 |
| 30 | LPKPLPEGTEDKDQT | 187-201 |
| 31 | PEGTEDKDQTATIPS | 192-206 |
| 32 | DKDQTATIPSLSAML | 197-211 |
| 33 | ATIPSLSAMLGALFL | 202-216 |
| 343-150 | LSAMLGALFLWMFWP | 207-221 |
| 353-150 | GALFLWMFWPSFNSA | 212-226 |
| 363-150 | WMFWPSFNSALLRSP | 217-231 |
| 37 | SFNSALLRSPIERKN | 222-236 |
| 38 | LLRSPIERKNAVFNT | 227-241 |
| 39 | IERKNAVFNTYYAVA | 232-246 |
| 40 | AVFNTYYAVAVSVVT | 237-251 |
| 41 | YYAVAVSVVTAISGS | 242-256 |
| 42 | AISGSSLAHPQGKIS | 252-266 |
| 43 | SLAHPQGKISKTYVH | 257-271 |
| 44 | QGKISKTYVHSAVLA | 262-276 |
| 45 | KTYVHSAVLAGGVAV | 267-281 |
| 46 | SAVLAGGVAVGTSCH | 272-286 |
| 47 | GTSCHLIPSPWLAMV | 282-296 |
| 48 | WLAMVLGLVAGLISV | 292-306 |
| 49 | LGLVAGLISVGGAKY | 297-311 |
| 50 | GLISVGGAKYLPGCC | 302-316 |
| 51 | GGAKYLPGCCNRVLG | 307-321 |
| 52 | LPGCCNRVLGIPHSS | 312-326 |
| 53 | NRVLGIPHSSIMGYN | 317-331 |
| 54 | IPHSSIMGYNFSLLG | 322-336 |
| 55 | IMGYNFSLLGLLGEI | 327-341 |
| 56 | FSLLGLLGEIIYIVL | 332-346 |
| 57 | LLGEIIYIVLLVLDT | 337-351 |
| 58 | IYIVLLVLDTVGAGN | 342-356 |
| 59 | LVLDTVGAGNGMIGF | 347-361 |
| 60 | VGAGNGMIGFQVLLS | 352-366 |
| 61 | QVLLSIGELSLAIVI | 362-376 |
| 62 | LAIVIALTSGLLTGL | 372-386 |
| 63 | LLTGLLLNLKIWKAP | 382-396 |
| 64 | LLNLKIWKAPHEAKY | 387-401 |
| 65 | IWKAPHEAKYFDDQV | 392-406 |
| 66 | HEAKYFDDQVFWKFP | 397-411 |
| 67 | FDDQVFWKFPHLAVG | 402-416 |
| 68 | DDQVFWKFPHLAVGF | 403-417 |
Rhesus D–specific polymorphisms are in boldface.
Two published sequences5,6 differ only at position 218. The peptides originally synthesized used the widely cited21 variant Ile218,5 which is now thought to have resulted from a sequencing error.22 Therefore, only data obtained with the variant Met218,6 as shown here, are included in this report.
To ensure purity, the peptide panel was synthesized with fluorenylmethoxycarbonyl chemical techniques on resin using a base-labile linker rather than with conventional pin technology, and randomly selected peptides were screened by high-performance liquid chromatography and amino acid analysis. For some experiments, a panel limited to 42 of the peptides was selected to span the entire RhD protein with 5 amino acid overlaps. A similar peptide panel was used previously to map the epitopes on the RhD protein that are recognized by autoreactive T cells in AIHA.17 All peptides were used to stimulate cultures at 20 μg/mL, although the responses of the cultures were previously shown to be similar in magnitude and kinetics at peptide concentrations between 5 and 20 μg/mL.
The control antigen Mycobacterium tuberculosispurified-protein derivative (PPD; Statens Seruminstitut, Denmark) was dialyzed extensively against phosphate-buffered saline (PBS; pH 7.4) and filter sterilized before it was added to cultures (10 μg/mL). PPD readily provokes recall T-cell responses in vitro23 24because most citizens of the United Kingdom have been immunized with bacille Calmette-Guérin vaccine. The mitogen concanavalin A (Con A) was obtained from Sigma (Poole, Dorset, United Kingdom) and used to stimulate cultures (10 μg/mL).
Antibodies
Fluorescein isothiocyanate–conjugated or phycoerythrin-conjugated mAbs against human CD3, CD19, CD45, and CD14 were obtained from Dako UK Ltd. Blocking mAbs specific for HLA-DP, HLA-DQ, and HLA-DR (Becton Dickinson, Oxford, United Kingdom) were dialyzed thoroughly against PBS before being added to cultures at the previously determined optimum concentration of 2.5 μg/mL.17,24 Allotype-specific murine mAb against DR15 molecules was a gift from Dr K. Gelsthorpe (Sheffield, United Kingdom). Tissue-culture supernatants containing the murine mAbs UCHL1, specific for human CD45RO,25 or SN130, specific for human CD45RA,26 were gifts from Professors P.C.L. Beverley and G. Janossy (London, United Kingdom), respectively.
Isolation of peripheral blood mononuclear cells and T cells
Peripheral blood mononuclear cells (PBMCs) were separated from fresh blood samples by density gradient centrifugation (Lymphoprep; Nycomed, Denmark). The viability of PBMCs was greater than 90% in all experiments, as indicated by trypan blue exclusion. Where required, T cells were isolated from PBMCs by passage through glass-bead affinity columns coated with human IgG and sheep antihuman IgG immune complexes.23 24 Flow cytometry (FACScalibur; Becton Dickinson) demonstrated that typical preparations contained more than 95% T cells.
Preparation of PBMCs depleted of CD45RO+ and CD45RA+ cells
PBMCs depleted of CD45RO+ or CD45RA+ T cells were prepared by using a modification of a previously described method.24 27 PBMCs were incubated with UCHL1 or SN130 cell-line supernatant, and cells that bound mAb were removed with magnetic beads coated with antibody specific for mouse IgG (Biomag; PerSeptive Biosystems, Framingham, MA). Each depleted population contained fewer than 10% cells expressing the respective CD45 isoform, as determined by flow cytometry (FACScalibur; Becton Dickinson).
Cell-proliferation assays
PBMCs were cultured in 100-μL volumes in microtiter plates at a concentration of 1.25 × 106 cells/mL in the α modification of Eagle medium (Gibco, Paisley, United Kingdom) supplemented with 5% autologous serum, 4 mmol/Ll-glutamine (Sigma), 100 U/mL sodium benzylpenicillin G (Sigma), 100 μg/mL streptomycin sulfate (Sigma), and 20 mmol/L HEPES (pH 7.2; Sigma). All plates were incubated at 37°C in a humidified atmosphere of 5% carbon dioxide and 95% air. The cell proliferation in cultures was estimated from the incorporation of tritiated thymidine in triplicate wells 5 days after stimulation with antigen, as described previously.17 Purified T cells were cultured under similar conditions23,24 (1.25 × 106 cells/mL), together with unfractionated PBMCs that had been irradiated (20 Gy [2000 rad]) to prevent their proliferation and that were added to the wells at a final concentration of 0.6 × 106cells/mL to act as APCs. Proliferation results are presented as either the mean counts per minute (± SE) of the triplicate samples or a stimulation index (SI) expressing the ratio of mean counts per minute in stimulated compared with unstimulated control cultures. An SI greater than 3 with counts per minute above 1000 was interpreted as representing a significant positive response.25
Statistical analysis
A nonparametric test, the Spearman rank correlation, was used for the statistical analysis, and P < .05 was considered to represent significance.
Results
Proliferative responses of PBMCs to the panel of peptides spanning the RhD protein sequence
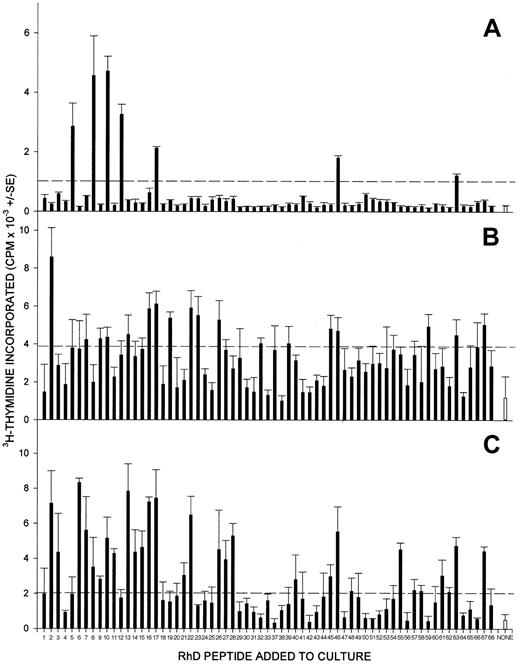
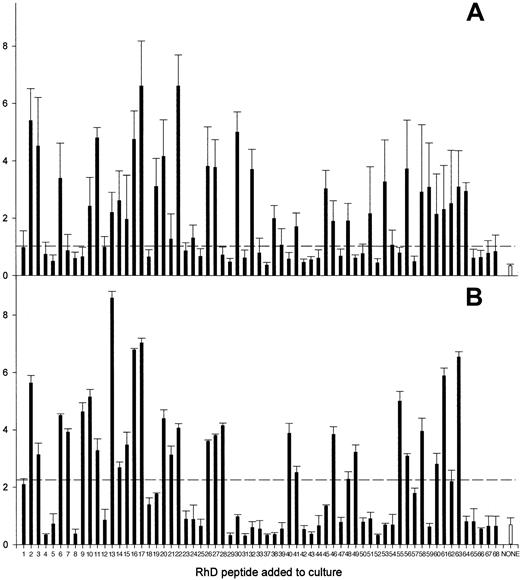
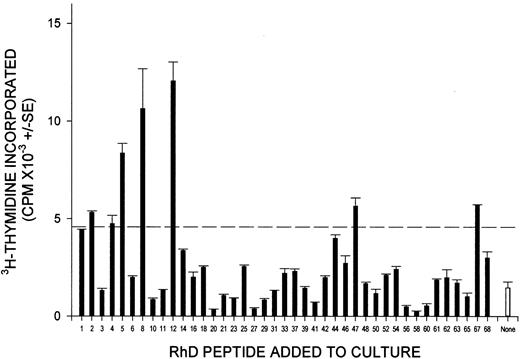
The full panel of up to 68 synthetic peptides corresponding to the sequence of the RhD protein was screened for the ability to stimulate the proliferation of PBMCs from each of the 13 D-negative donors who had generated anti-D alloantibodies after deliberate immunization. The results are summarized in Table 1, and representative data obtained from 3 of these donors are shown in Figure1. One or more peptides from the RhD protein elicited significant proliferation by PBMCs from each of the deliberately alloimmunized donors and, typically, multiple peptides were stimulatory. Longitudinal studies demonstrated that the pattern of response to the peptide panel was reproducible during a period of several months (Figure 2); although the profile of stimulatory peptides showed some changes, this variation was minor compared with that observed among different donors.
PBMCs from all RhD-negative donors alloimmunized with RhD-positive RBCs proliferate in response to peptides from the sequence of the RhD protein.
PBMCs were obtained from representative alloimmune donors 2 (A), 8 (B), and 13 (C) and tested for their ability to proliferate against the panel of peptides spanning the RhD protein. In each case, all the peptides were tested on a single occasion, and the broken line indicates the level of proliferation assumed to represent a positive response (the higher of SI = 3 or cpm = 1000).
PBMCs from all RhD-negative donors alloimmunized with RhD-positive RBCs proliferate in response to peptides from the sequence of the RhD protein.
PBMCs were obtained from representative alloimmune donors 2 (A), 8 (B), and 13 (C) and tested for their ability to proliferate against the panel of peptides spanning the RhD protein. In each case, all the peptides were tested on a single occasion, and the broken line indicates the level of proliferation assumed to represent a positive response (the higher of SI = 3 or cpm = 1000).
The pattern of RhD peptides that stimulate PBMCs from alloimmune donors to proliferate is reproducible.
Proliferative responses of PBMCs from alloimmune donor 12 against the panel of peptides spanning the RhD protein were compared on 2 occasions (A and B) 2 months apart. The broken line indicates the level of proliferation assumed to represent a positive response (the higher of SI = 3 or cpm = 1000).
The pattern of RhD peptides that stimulate PBMCs from alloimmune donors to proliferate is reproducible.
Proliferative responses of PBMCs from alloimmune donor 12 against the panel of peptides spanning the RhD protein were compared on 2 occasions (A and B) 2 months apart. The broken line indicates the level of proliferation assumed to represent a positive response (the higher of SI = 3 or cpm = 1000).
PBMC samples were also obtained from 6 plasma donors and 3 whole-blood donors who had anti-D antibodies as a result of RhD-incompatible pregnancy. Table 2 shows that PBMCs from all these donors proliferated in response to at least one peptide from the full RhD protein panel. These peptide responses were observed even though particular donors had no record of exposure to D-positive RBCs for up to 29 years and are thus comparable with the persistence of serum anti-D alloantibody.
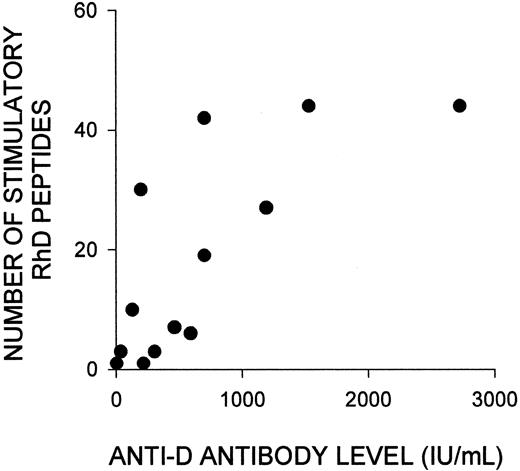
Although the number of peptides that elicited responses by PBMCs varied markedly among the alloimmune donors, Tables 1 and 2 show that this variation was not obviously related to either the length of time since deliberate or natural immunization with RhD-positive RBCs, or the number of such exposures. However, there was a strong positive correlation (Rs = 0.75; P < .003) between the number of stimulatory peptides in deliberately alloimmunized donors and the concentration of anti-D antibodies in their serum (Figure3). No such relation was apparent in the naturally alloimmunized women, but this was a smaller group, with a narrower range of relatively low anti-D antibody levels.
Variation in the number of RhD peptides that stimulate PBMCs from alloimmune donors to proliferate is strongly correlated with the level of anti-D antibody in their serum.
Shown is the relation (Rs = 0.75; P < .003) between the number of RhD peptides that elicited proliferation of PBMCs from each of the deliberately alloimmunized donors (1-13) and the level of anti-D antibody in their serum. If antibody concentrations were measured more than once during the period in which PBMC samples were obtained, the highest level was recorded.
Variation in the number of RhD peptides that stimulate PBMCs from alloimmune donors to proliferate is strongly correlated with the level of anti-D antibody in their serum.
Shown is the relation (Rs = 0.75; P < .003) between the number of RhD peptides that elicited proliferation of PBMCs from each of the deliberately alloimmunized donors (1-13) and the level of anti-D antibody in their serum. If antibody concentrations were measured more than once during the period in which PBMC samples were obtained, the highest level was recorded.
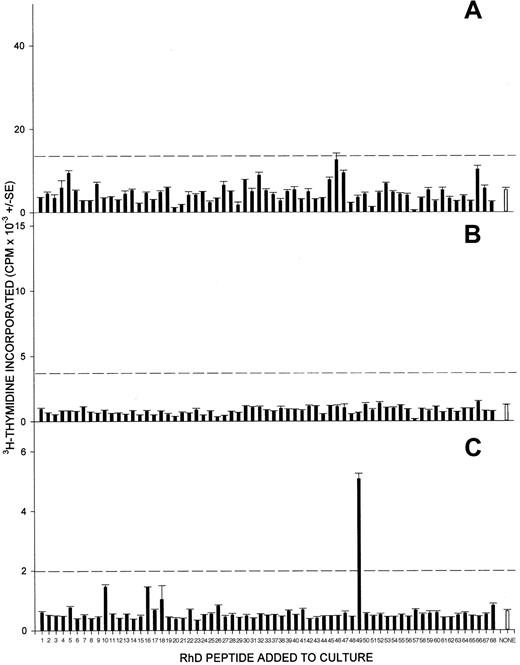
In contrast to the responsiveness of PBMCs from alloimmunized donors, PBMCs from only 2 of the 8 negative-control volunteers with no anti-D alloantibodies proliferated under similar conditions against any RhD peptide from the panel, and any responses observed were weak. Figure4 shows representative results from nonresponsive (4A and 4B) and responsive (4C) control donors. It should be noted that the failure of control donors' PBMCs to respond was specific to the RhD peptides, since in every case, significant proliferation was elicited by both the T-cell mitogen Con A and the recall foreign antigen PPD (results not shown).
PBMCs from control RhD-negative donors, who have not been alloimmunized, rarely proliferate when stimulated with peptides from the RhD protein sequence.
Shown here are the proliferative responses of PBMCs from 3 control unimmunized RhD-negative donors (A, B, and C) against the panel of peptides spanning the RhD protein. The broken line indicates the level of proliferation assumed to represent a positive response (the higher of SI = 3 or cpm = 1000).
PBMCs from control RhD-negative donors, who have not been alloimmunized, rarely proliferate when stimulated with peptides from the RhD protein sequence.
Shown here are the proliferative responses of PBMCs from 3 control unimmunized RhD-negative donors (A, B, and C) against the panel of peptides spanning the RhD protein. The broken line indicates the level of proliferation assumed to represent a positive response (the higher of SI = 3 or cpm = 1000).
Characterization of cells that proliferate in response to peptides from the RhD Protein
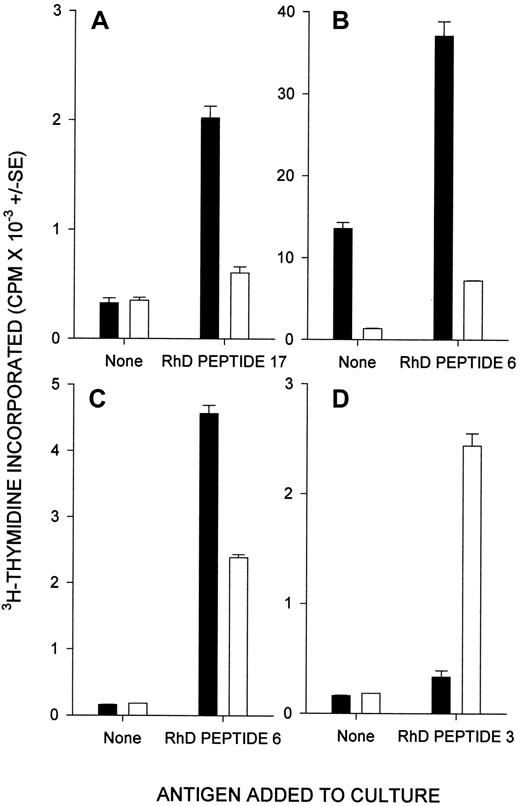
It is generally accepted17,24 27 that PBMC proliferation in vitro in response to peptides is mediated by T cells, and this was demonstrated for the RhD peptides by experiments to stimulate purified T cells. The availability of 450-mL blood donations from the naturally immunized donors allowed purification of sufficient peripheral blood T cells to test their responsiveness to the smaller panel of up to 42 peptides spanning the RhD protein. The strong proliferation of the purified T cells responding to stimulatory RhD peptides is illustrated in Figure 5.
The proliferation of PBMCs from alloimmune donors against RhD peptides is mediated by T cells.
Shown are the proliferative responses of purified peripheral blood T cells from a representative alloimmune donor (22) against the smaller panel of peptides spanning the RhD protein. The apparent discrepancy with the sequences that stimulated PBMCs from this donor (Table 2) was due to differences in the peptide panels screened and the greater responsiveness of the purified T cells. Thus, the T cells were not tested against 2 (17 and 28) of the 3 peptides (2, 17, and 28) that elicited proliferation of PBMCs from the same donor, and PBMCs did respond to 4 of the peptides (5, 8, 12, and 47) that stimulated the T cells, but the response was not strong enough to represent a significant difference.
The proliferation of PBMCs from alloimmune donors against RhD peptides is mediated by T cells.
Shown are the proliferative responses of purified peripheral blood T cells from a representative alloimmune donor (22) against the smaller panel of peptides spanning the RhD protein. The apparent discrepancy with the sequences that stimulated PBMCs from this donor (Table 2) was due to differences in the peptide panels screened and the greater responsiveness of the purified T cells. Thus, the T cells were not tested against 2 (17 and 28) of the 3 peptides (2, 17, and 28) that elicited proliferation of PBMCs from the same donor, and PBMCs did respond to 4 of the peptides (5, 8, 12, and 47) that stimulated the T cells, but the response was not strong enough to represent a significant difference.
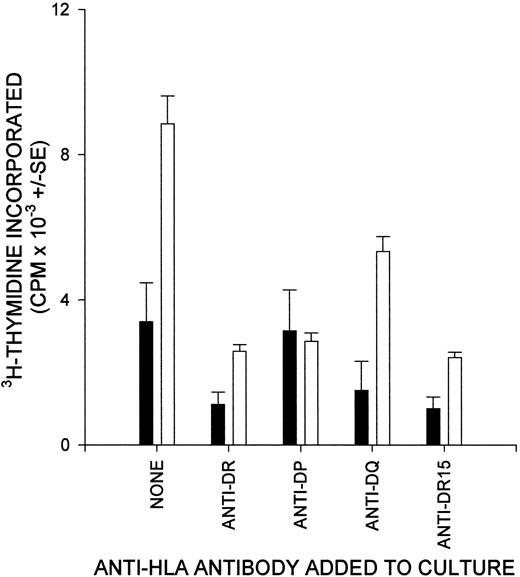
To determine whether the T cells responding to RhD peptides came from the helper subset, which is restricted by MHC class II molecules, blocking antibodies specific for HLA-DP, HLA-DQ, and HLA-DR were tested for their ability to inhibit the RhD peptide–induced proliferation of PBMCs from alloimmunized donors. At least one of the antibodies inhibited proliferation markedly when a total of 17 responses to stimulatory peptides was examined in 6 of the donors. Anti-DR was the most potent blocking antibody for 13 of these responses, whereas anti-DP abrogated 3 responses and anti-DQ abolished only 1 response. Anti-DP and anti-DQ also appeared to inhibit several responses weakly. Figure 6 illustrates the ability of anti-DR in cultures to block proliferative responses to RhD peptides; in this example of a donor homozygous for HLA-DRB1*15, an allele-specific mAb against DR-15 molecules caused identical inhibition. The effect of anti-DP in inhibiting proliferation against particular peptides is also shown.
The proliferation of T cells from alloimmune donors against RhD peptides depends on HLA class II molecules.
Cultures of PBMCs from a representative donor (6) were stimulated with RhD peptide, and class II–restricted responses were blocked by the addition of antibody specific for DP, DQ, DR, or DR15. ▪, no antigen; ■, RhD peptide 17.
The proliferation of T cells from alloimmune donors against RhD peptides depends on HLA class II molecules.
Cultures of PBMCs from a representative donor (6) were stimulated with RhD peptide, and class II–restricted responses were blocked by the addition of antibody specific for DP, DQ, DR, or DR15. ▪, no antigen; ■, RhD peptide 17.
If the helper T cells that proliferated in vitro were responsible for driving anti-D antibody production in the donors, they must have been activated previously in vivo. Therefore, we assessed PBMC proliferation relatively early (on day 5 after stimulation) and cells were cultured in microtiter plates—conditions that strongly favor recall, rather than primary, T-cell responses.17,23,24 To confirm that the T cells proliferating against RhD peptides in vitro had been activated in vivo, depletion experiments determined the isoform of the CD45 molecules they expressed, since primary and recall responses are mediated by T cells bearing CD45RA and CD45RO, respectively.23-27 Responses against at least 2 stimulatory peptides by PBMCs from each of 4 alloimmune donors were analyzed. Except for the response to one peptide in one donor, the proliferating cells were from the CD45RO+ subset containing activated T cells, rather than the CD45RA+ fraction of previously inactive T cells. Representative results are shown in Figure7.
T cells from alloimmune donors that proliferate against RhD peptides in vitro were, with few exceptions, previously activated in vivo.
Shown here are the proliferative responses of CD45RO+ (▪, previously activated) and CD45RA+ (■, previously inactive) T cells from alloimmune donors 12 (A), 17 (B), and 4 (C,D) against selected RhD peptides.
T cells from alloimmune donors that proliferate against RhD peptides in vitro were, with few exceptions, previously activated in vivo.
Shown here are the proliferative responses of CD45RO+ (▪, previously activated) and CD45RA+ (■, previously inactive) T cells from alloimmune donors 12 (A), 17 (B), and 4 (C,D) against selected RhD peptides.
Distribution of stimulatory peptides in the RhD protein
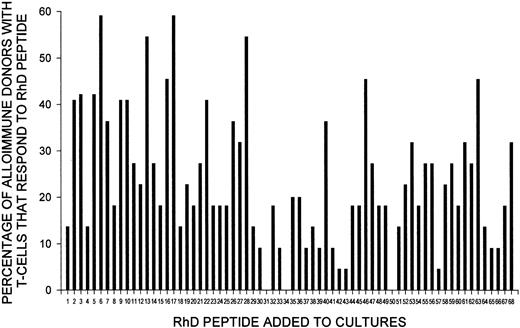
As shown in Tables 1 and 2, the RhD peptides that stimulate T cells to proliferate in vitro can vary among alloimmune individuals. However, an analysis of the proportion of alloimmune donors with T cells that responded to each of the 68 RhD peptides (Figure8) revealed that particular peptides predominated. Thus, peptides 6, 13, 17, and 28 stimulated T-cell proliferation in at least 50% of the alloimmune donors. Notably, peptides 6 and 17 were recognized most often; they elicited proliferative responses by T cells from more than 65% of these donors. The sequences of the 4 dominant peptides that commonly elicited proliferation are included in Table 3, which shows that each of these peptides contains polymorphisms that characterize the RhD protein. Similarly, most, but not all, of the minor stimulatory peptides (those to which T cells from fewer donors responded) carry RhD-specific residues.
Particular peptides from the RhD protein stimulate T cells from most alloimmune donors to proliferate.
Shown here is the proportion of alloimmune donors (n = 22; for peptides 34, 35, and 36, n = 5, since original results obtained using sequence Ile218 were omitted) with T cells that proliferated in response to each of the 68 peptides from the panel spanning the RhD protein.
Particular peptides from the RhD protein stimulate T cells from most alloimmune donors to proliferate.
Shown here is the proportion of alloimmune donors (n = 22; for peptides 34, 35, and 36, n = 5, since original results obtained using sequence Ile218 were omitted) with T cells that proliferated in response to each of the 68 peptides from the panel spanning the RhD protein.
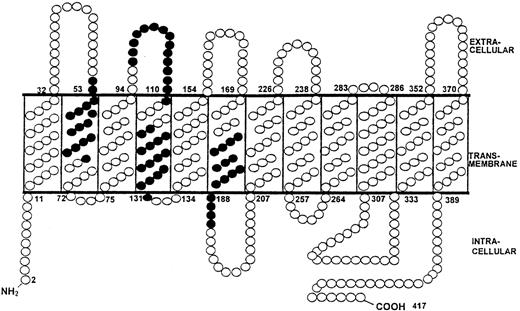
A bias toward recognition of polymorphic sites from the RhD protein is clearly illustrated by Tables 1 and 2, which show that one or more such sequences were stimulatory in each alloimmune donor, even when their T cells responded to a single peptide. Responsiveness to peptides 6, 7, 8, 9, 12, 13, or 14, which bear both RhD and RhC polymorphisms, could theoretically be caused by exposure of donors to Rh protein of either type; however, because it was frequently observed after immunization with RhC-negative R2 cells, it was most likely due to the RhD protein. Figure 9shows the predicted topographic features of the dominant RhD peptides and demonstrates that sequences recognized by alloreactive T cells are derived not only from extracellular and intracellular loops of the Rh proteins but also from membrane-spanning domains.
RhD protein sequences that stimulate T cells from alloimmune donors to proliferate are not limited to extracellular loops.
The predicted topographic features of the RhD protein5 6illustrate the location of the 4 dominant RhD peptides that elicited proliferation by T cells from most of the alloimmune donors. Residues within dominant peptides, ● (peptide 6 aa52-66, peptide 13 aa97-111, peptide 17 aa117-131, and peptide 28 aa177-191).
RhD protein sequences that stimulate T cells from alloimmune donors to proliferate are not limited to extracellular loops.
The predicted topographic features of the RhD protein5 6illustrate the location of the 4 dominant RhD peptides that elicited proliferation by T cells from most of the alloimmune donors. Residues within dominant peptides, ● (peptide 6 aa52-66, peptide 13 aa97-111, peptide 17 aa117-131, and peptide 28 aa177-191).
Discussion
This report describes the first mapping of alloreactive helper T-cell epitopes on the RhD protein. Characterization of the helper response that drives anti-D antibody production by B cells, which has previously been neglected, is the first step in the development of improved or alternative strategies to prevent alloimmunization that are based on immunomodulation of T cells. Such approaches are urgently required because of the growing problems associated with the current prophylactic program for HDN.1-3
Depletion experiments demonstrated that the cells responding in vitro to the Rh peptides were T cells, and the ability of anti-MHC class II antibodies consistently to inhibit the proliferation indicated that these lymphocytes were from the helper subset. The differences in the profile of peptide responses among alloimmune donors may be related to the variations in the donors' HLA type and reflect both the ability of particular class II molecules to bind and present each peptide,14 and the role of the MHC in shaping the T-cell repertoire.28 The relations among the identity of sequences from the RhD protein that contain alloreactive and autoreactive T-cell epitopes, HLA restriction, and peptide affinity for class II molecules are clearly complex and are currently being investigated. The volunteers studied here carry the most common RhD-negative genotype found in Caucasians (homozygous deletion of theRHD gene), and similar studies will be necessary to map the relevant helper determinants in donors who are RhD negative because of an allelic variation, such as that recently described in black Africans.29
Because the aim of the current work was to map the epitopes recognized by T cells that had been activated by the RhD protein in vivo, it was necessary to minimize the primary responses to RhD peptides that we previously observed in unimmunized donors.24 This was achieved by performing the proliferation assays in microtiter plates during a short time course, conditions designed to favor recall proliferation by primed T cells rather than primary responses of naı̈ve T cells.17,23,24 The success of this approach in detecting T cells in vitro that had been primed by the RhD protein in vivo was verified in 2 ways. The first method differentiated between the in vitro proliferative responses of previously stimulated and unstimulated T-helper cells by identifying the CD45 isoform of the cells that responded.23-27 Primary and recall responses in vitro are mediated by T cells bearing CD45RA and CD45RO, respectively,23-27 and we showed that the T cells from alloimmunized donors that proliferated against the RhD peptides were almost invariably drawn from the CD45RO+, previously activated, memory subset. In contrast, previous studies found that, with few exceptions, any T cells from unimmunized donors that respond to Rh peptides bear the CD45RA marker of previously inactive or naı̈ve T cells.24
The second demonstration that the cultures were biased toward supporting recall responses was provided by the observations that none of the peptides from the RhD protein stimulated T-cell proliferation in 6 of the 8 unimmunized donors tested, and that any responses observed were weak and against relatively few peptides. The weak proliferation occasionally observed when control donor T cells were challenged with RhD peptides was likely to have been the result of exposure of donors to environmental antigens that cross-react with the RhD protein,30 particularly given the limited sequence homology between different peptides necessary for T-cell cross-reactivity31 and the large number of peptides screened. The control donors in this study do not express the RhD protein, but T cells from a similar small proportion of healthy, RhD-positive volunteers were previously shown to mount recall responses against some RhD peptides,17 presumably because thymic deletion of T cells reactive with self-Rh proteins is incomplete.24
One striking result of the current study is that primed T cells responsive to epitopes on the RhD protein were detected in the peripheral blood of donors many years after either deliberate or pregnancy-associated alloimmunization. This finding demonstrates that T-cell memory is maintained virtually indefinitely in the absence of known re-exposure to the antigen. Furthermore, the strength of the T-cell response, in terms of the number of RhD peptides that elicit proliferation, does not decline as the length of time since immunization increases. Such long-term maintenance of memory may be due to either persistence of antigen,32 particularly on follicular dendritic cells, or repeated stimulation of memory T cells, which have a lower threshold for activation than naı̈ve T cells,33 by cross-reactive antigens in the environment.30
Another important finding is that the wide variation in the total number of RhD peptides recognized by T cells from different deliberately alloimmunized donors, although not related to the timing or frequency of boosting, was strongly correlated with the level of anti-D antibodies. Our interpretation of these results is that the rate of alloantibody production is determined by the level of help available, which in turn depends on how many epitopes from the RhD protein can stimulate the T-cell repertoire. The factors that govern this variation among donors in the breadth of their T-cell response are under investigation but are likely to include the relative affinity of peptides derived from RhD protein for different HLA class II molecules.14 The correlation we identified may form the basis for a method to predict the ability of individuals to produce high levels of anti-D antibody. It also raises the possibility that an established anti-D antibody response could be boosted or inhibited by manipulating the number of epitopes that elicit T-cell help.
Alignment of the peptides that elicited responses with the sequence and predicted topographic features of the RhD protein5,6reveals that stimulatory epitopes are located throughout the molecule, in extracellular, intracellular, and membrane-spanning domains. This contrasts with the requirement for RhD B-cell epitopes to be exposed on the outer surface of RBCs9,10 but is not unexpected given that antigen must be processed within APCs before recognition by T cells.14 It was also not surprising that most of the RhD peptides capable of eliciting proliferation by T cells from alloimmune donors contain sequences not shared with the closely related RhCc/Ee protein,15 16 which was expressed by these donors. Indeed, we observed that T cells from all alloimmune donors responded to at least one peptide containing RhD-specific polymorphisms, even when only one sequence was recognized. Although T cells that responded to peptides bearing RhD polymorphisms shared with RhC may have the potential to provide help for production of antibody of either specificity, the history of donor immunization indicated that many such responses were the consequence of exposure to RhD, not RhC.
Responses to some RhD peptides that are identical to the corresponding sequence of the RhCc/Ee protein were also observed. Responsiveness to such nonpolymorphic peptides is most likely to be the result of exposure to the RhD protein, since it is relatively rare in control RhD-negative donors who have not been alloimmunized. This raises the question of how T cells that have been primed by a foreign antigen (the RhD protein) can be specific for sequences that are shared with a self-antigen, the RhCc/Ee protein. It is recognized that many peptide sequences in any self-antigen are cryptic, or not presented to T cells after processing from the protein within APCs, and that the corresponding T cells can therefore escape the processes of self-tolerance and remain in an unstimulated, but potentially responsive, state in repertoire.24,34-36 Furthermore, previously cryptic epitopes can be presented to T cells if the conditions under which the antigen is processed change.34-36 Thus, we propose that the sequence differences between the RhD and RhCc/Ee proteins cause them to be differentially processed by APCs so that particular stimulatory peptides, although not containing RhD-specific polymorphisms, are only generated from the RhD protein. The response to such peptides would therefore be specific for the RhD protein and not break tolerance to the RhCc/Ee protein.
Identification of T-helper epitopes on the RhD protein, particularly those that are dominant in most of the population, is the first step toward evaluation of peptide immunotherapy18-20for preventing HDN without the growing problems associated with the current approach.1-3 Characterization of helper epitopes on the RhD protein will facilitate the design of a safe, synthetic immunogen as an alternative to RBCs for boosting anti-D antiserum production by volunteers. In particular, the relation we identified between the strength of the T-cell response to the RhD protein and the level of anti-D alloantibody suggests that immunization with peptides corresponding to helper epitopes, even without the corresponding B-cell determinants, could boost titers. More radical strategies would include the specific control of alloimmunization in RhD-negative women with peptide antigen tolerogens. Studies in animal models have shown that once the dominant T-cell epitopes on an antigen have been identified, it is possible to control the response in vivo by immunization with synthetic peptides corresponding to substituted analogues of the epitopes (including antagonists) or by mucosal administration of the wild-type sequences.18-20Such a strategy would not only be free from ethical concerns relating to the collection of human anti-D antibody for the current prophylactic program but may also have further advantages, such as conferring long-term protection from primary alloimmunization or abrogating an established anti-RhD response. Thus, the use of peptide immunotherapy to render RhD-specific helper T cells tolerant may not only replace the current method of preventing HDN but may offer the possibilities of conferring lifelong unresponsiveness to RhD on RhD-negative girls at puberty and treating women who have already been alloimmunized to prevent HDN in their infants from subsequent pregnancies.
Acknowledgments
We thank the plasma and blood donors from Aberdeen and Edinburgh who participated in this study and the donor-center staff for their help in collecting blood samples, particularly Dr M. A. Greiss and Sister Helen Clark (Aberdeen) and Dr J. Gillon (Edinburgh). HLA genotyping of donors was done by Mr D. W. L. Wilson (Tissue Typing Laboratory, Aberdeen).
Funded by grants from the Scottish National Blood Transfusion Service, the Wellcome Trust, and the Cunningham Trust, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stanislaw J. Urbaniak, Academic Transfusion Medicine Unit, Department of Medicine and Therapeutics, Regional Transfusion Centre, Foresterhill, Aberdeen AB25 2ZW, United Kingdom; e-mail: s.j.urbaniak@abdn.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal