Abstract
Recently the Belgium–Dutch Hematology–Oncology group initiated a multicenter study to evaluate whether myeloma patients treated with intensive chemotherapy benefit from additional peripheral stem cell transplantation. To determine treatment response accurately, we decided to quantitate malignant cells. To test a consensus quantitation strategy, 5 centers independently determined the immunoglobulin heavy chain sequences of patient tumor cells and developed allele-specific oligonucleotides (ASO) and ASO–polymerase chain reaction (PCR). We compared the reproducibility of real-time quantitation with quantitation using limiting dilutions. We distributed DNA samples with a 4-log range of tumor cell concentrations and found average quantitation values deviating 74% and 42% from the input values with real-time PCR (1 center) and limiting dilutions (4 centers), respectively. Within single centers we found an average variation coefficient of 0.74, with limiting dilutions not significantly different from the average 0.82 center-to-center variation coefficient. Within a single center, real-time quantitation proved more reproducible (average variation coefficient, 0.36). Quantification was confirmed in 3 patients during treatment in the protocol. This report shows that real-time PCR or limiting dilution assays can be used for quantitation in a single multicenter trial. We present a consensus strategy that allows an accurate comparison of quantitation data generated in independent centers.
Multiple myeloma is a malignant disease characterized by an increased number of clonal plasma cells in the bone marrow. Intensive treatment of patients with multiple myeloma results in more complete remission, event-free survival, and overall survival than treatment with conventional doses of chemotherapy. Recently updated results from the French IMF group showed a 43% (median, 57 months) 6-year probability of survival after diagnosis in the myeloma group treated with high-dose chemotherapy versus a 21% probability (median, 42 months) in the group treated with conventional doses. In multivariate analysis the most important prognostic factor was response to treatment.1,2 Based on these results, the Belgium–Dutch Hematology Oncology group (HOVON) initiated a multicenter study (HOVON-24) to evaluate the additional effect of peripheral stem cell transplantation on treatment outcome. In this study myeloma patients received 3 to 4 courses of VAD, followed by cyclophosphamide, stem cell mobilization, and 2 courses of intravenous intermediate-dose melphalan (70 mg/m2). Subsequently, patients were randomized to receive either interferon-α alone or cyclophosphamide/TBI and peripheral stem cell transplantation, followed by interferon-α maintenance. Treatment response was measured using bone marrow plasma cell percentage, plasma cell monoclonality (κ/λ ratio), and serum or urine M-protein concentration as parameters. A reliable assessment of tumor cell fractions below 1%, however, was not possible with morphologic or flow cytometry examinations. The serum or urine M-protein concentration reflected the secreting capability of monoclonal plasma cells rather than the absolute number of tumor cells and was of limited value as a parameter for treatment response. Data generated by Corradini et al3 strongly suggest a correlation between the PCR detection of posttransplant bone marrow tumor cells and treatment outcome. Ninety-two percent of myeloma patients who underwent autologous stem cell transplantation were PCR-positive, and most of these patients had relapses. Among patients subjected to allogeneic bone marrow transplantation, 45% became PCR-negative. Only 1 of 4 PCR-negative patients had relapses. To define treatment response more accurately, we decided to quantitate malignant cells using PCR.
For logistical reasons quantitation data were generated in the participating HOVON centers. Other multicenter studies reporting large center-to-center variations in quantitation data clearly indicate that an accurate comparison of data generated in individual centers is dependent on the standardization of a reproducible method.4The HOVON-24 group, therefore, developed a consensus strategy for the quantitation of malignant cells in myeloma patients. Quantitation was based on amplification of the unique immunoglobulin heavy-chain sequence of the malignant clone using allele-specific oligonucleotides (IgH ASO–PCR). In this report we tested a consensus strategy using a limiting dilution assay in a multicenter setting and compared these data with quantitation results using a recently developed integrated system for thermal cycling, real-time fluorescent detection, and subsequent calculation of PCR product (7700 SDS; ABI-PRISM; Perkin-Elmer, Norwalk, CT).5
Patients, materials, and methods
Patients
Each patient had stage II or III multiple myeloma, according to the staging system of Durie and Salmon.6 Bone marrow cells were obtained by aspiration from the sternum or iliac crest after each patient gave informed consent. Cytospin preparations of bone marrow cells were stained with May–Grünwald–Giemsa, and the morphology of 200 nucleated cells was quantitated by 2 independent investigators. Cells were layered over Ficoll–Hypaque, and the mononuclear layer was collected after density centrifugation and washed in phosphate-buffered saline. These cells were cryopreserved at −196°C in small aliquots.
Nucleic acid extraction
More than 10×106 cryopreserved cells were thawed, washed in phosphate-buffered saline, and counted using a Coulter counter. Five million cells were used for DNA extraction with the QIAamp Blood (Qiagen, Hilden, Germany) kit. The remaining 5×106 cells were used for RNA extraction using the RNAzol B isolation (CAMPRO–Scientific, Veenendaal, The Netherlands) kit. For quantitation of samples containing fewer than 1×106 cells after thawing, whole cell lysates were used.7
cDNA synthesis
One microgram RNA (5 μL) was reversed transcribed in a total volume of 20 μL, containing 50 mmol/L Tris–HCl (pH 8.3), 75 mmol/L KCl, 3 mmol/L MgCl2, 10 mmol/L dithiothreitol, 625 μmol/L dNTP, 5 μmol/L random hexamers (Pharmacia, Uppsala, Sweden), 20 U RNAsin (Promega, Madison, WI), and 200 U Mo-MuLV reverse transcriptase (Life Technologies, Gaithersburg, MD). Reverse transcription was performed at 42°C for 45 minutes.
VH family-specific amplification of IgH using consensus primers
Six independent IgH PCRs were performed using leader region VH family-specific sense primers (Table1) and a constant region antisense primer (cα for IgA-producing tumor clones, cγ for IgG-producing tumor clones). One microliter cDNA was subjected to IgH PCR in a 100-μL PCR solution containing 10 mmol/L Tris–HCl (pH 8.3), 50 mmol/L KCl, 2 mmol/L MgCl2, 250 μmol/L dNTP, 2.5 U Taq DNA polymerase (Life Technologies), and 30 pmol of each primer (Eurogentec, Seraing, Belgium). The PCR was performed for 35 cycles of 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C, and for a final 10-minute extension at 72°C in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany). Products were separated on a 2% agarose gel. As an alternative, Fr1-region VH family-specific sense primers or JHantisense primers were used.
Primers used for PCR and sequencing
| Amplification . | Primers . | |||
|---|---|---|---|---|
| Sense . | . | Antisense . | . | |
| IgH rearrangement tumor clone | VH1-leader VH3-leader VH4-leader VH5-leader VH6-leader | 5′-ATGGACTGGACCTGGAGG-3′ 5′-GAGTTTGGGCTGAGCTGG-3′ 5′-CTGGTGGCAGCTCCCAGA-3′ 5′-CTCGCCCTCCTCCTGGCT-3′ 5′-ATGTCTGTCTCCTTCCTC-3′ | Cγ Cα | 5′-GGGTCTAGACAGGCAGCCCAGGGCCGCTGTGC-3′ 5′-GGCTCAGCGGGAAGACCTTGG-3′ |
| Sequencing or alternative set | VH1/7 VH2 VH3 VH4 VH5 VH6 | 5′-TCTGGGGCTGAGGTGAAGAA-3′ 5′-ACCTTGAAGGAGTCTGGTCCT-3′ 5′-GGGGGTCCCTGAGACTCTC-3′ 5′-GCCCAGGACTGGTGAAGC-3′ 5′-CTGGTGCAGTCTGGAGCAG-3′ 5′-GTACAGCTGCAGCAGTCAGGT-3′ | Jh-cons Jh21 | 5′-GGTCACCGTCTCCTCAGGT-3′ 5′-ACCTGAGGAGACGGTGACC-3′ |
| β-actin reference PCR | β-actin forward | 5′-TCACCCACACTGTGCCCATCTACGA-3′ | β-actin reverse | 5′-CAGCGGAACCGCTCATTGCCAATGG-3′ |
| β-globin reference PCR | PCO5 GH21 | 5′-CAACCTCAAACAGACACCATG-3′ 5′-GGAAAATAGACCAATAGGCAG-3′ | PCO3 | 5′-ACACAACTGTGTTCACTAGC-3′ |
| Amplification . | Primers . | |||
|---|---|---|---|---|
| Sense . | . | Antisense . | . | |
| IgH rearrangement tumor clone | VH1-leader VH3-leader VH4-leader VH5-leader VH6-leader | 5′-ATGGACTGGACCTGGAGG-3′ 5′-GAGTTTGGGCTGAGCTGG-3′ 5′-CTGGTGGCAGCTCCCAGA-3′ 5′-CTCGCCCTCCTCCTGGCT-3′ 5′-ATGTCTGTCTCCTTCCTC-3′ | Cγ Cα | 5′-GGGTCTAGACAGGCAGCCCAGGGCCGCTGTGC-3′ 5′-GGCTCAGCGGGAAGACCTTGG-3′ |
| Sequencing or alternative set | VH1/7 VH2 VH3 VH4 VH5 VH6 | 5′-TCTGGGGCTGAGGTGAAGAA-3′ 5′-ACCTTGAAGGAGTCTGGTCCT-3′ 5′-GGGGGTCCCTGAGACTCTC-3′ 5′-GCCCAGGACTGGTGAAGC-3′ 5′-CTGGTGCAGTCTGGAGCAG-3′ 5′-GTACAGCTGCAGCAGTCAGGT-3′ | Jh-cons Jh21 | 5′-GGTCACCGTCTCCTCAGGT-3′ 5′-ACCTGAGGAGACGGTGACC-3′ |
| β-actin reference PCR | β-actin forward | 5′-TCACCCACACTGTGCCCATCTACGA-3′ | β-actin reverse | 5′-CAGCGGAACCGCTCATTGCCAATGG-3′ |
| β-globin reference PCR | PCO5 GH21 | 5′-CAACCTCAAACAGACACCATG-3′ 5′-GGAAAATAGACCAATAGGCAG-3′ | PCO3 | 5′-ACACAACTGTGTTCACTAGC-3′ |
Direct sequencing
Double-stranded IgH PCR product was sequenced in 2 directions by the dideoxy chain termination method in a 15-cycle PCR using 5 pmol of an end-labeled sequencing primer (Table 1) and a 1-μL (10 ng) template.8 The sequences were probed for homology with published functional VH, DH, and JHgene segments (V-base downloaded from the internet sitehttp://www.mrc-cpe.cam.ac.uk/imt-doc/public/INTRO.html) using DNAPLOT or FASTA software.
Design of allele-specific oligonucleotides
Allele-specific oligonucleotides complementary to the CDRI (sense) and CDRIII (antisense) regions were designed using the Primer Express software program (Perkin–Elmer, Foster City CA; demo version 1.0 ppd). Design criteria for ASO were: temperature between 59°C and 64°C (nearest-neighbor method), no secondary structures with temperature higher than 50°C, no primer dimer formation with δG less than −10 kcal/mol, no stretches of 4 or more Gs, the last 3′ base of the sense primer preferentially complementary to a point mutation in the CDRI region, and the 3 penultimate 3′ bases of the antisense primer in the CDRIII region preferentially in the N region between the VH and D segments.
Tumor clone-specific polymerase chain reaction
An ASO–PCR was performed on 1 μL patient cDNA essentially as described for IgH consensus PCR, except for the use of 30 pmol 5′ and 3′ ASO primers instead of the VH and constant region consensus primers. PCR products were separated on 2% agarose gel. A PCR product of the correct length justified further use of the tested ASO. The specificity of the ASO–PCR (essentially performed as described for IgH consensus PCR) was tested using 0.5 μg patient or 1 μg control DNA (normal bone marrow cells, normal white blood cells [NWBC], and normal tonsillar cells). Three independent allele-specific dilution series were generated by diluting patient marrow DNA in 10-fold decrements into NWBC DNA to yield a concentration of 0.1 μg/μL and 160,000 cells per PCR. Sensitivity of the ASO–PCR was tested using these dilution series as templates. PCR products were separated on 2% agarose gel, transferred to nylon membranes (Hybond N+; Amersham, Roosendaal, The Netherlands), and probed with end-labeled VH family-specific Fr2 or Fr3 probes under stringent conditions. Radioactive signals were visualized on x-ray film (Eastman Kodak, Rochester, NY). Sensitivity of the ASO–PCR was considered satisfactory when PCR product was detectable up to 0.1% tumor cells on agarose gel or 0.01% tumor cells on x-ray film. To increase ASO–PCR sensitivity, the annealing temperature could be optimized by testing at 58°C to 62°C.
Quantitation using limiting dilutions
Patient DNA samples were serially diluted (using /10 decrements) in dH2O.5,9Dilutions were amplified using a 2-step PCR procedure. The initial amplifications were performed using 30 pmol of the Fr1 region or the leader region VH family-specific sense primer and the Jh21 antisense primer. DNA template (10 μL) was subjected to IgH–PCR essentially performed as in the VH family-specific consensus IgH–PCR except for the use of 2 mmol/L MgCl2. Two microliters from the first PCR was used as a template in a nested PCR procedure performed with 30-pmol patient-specific ASO and 2.5 mmol/L MgCl2. PCR products were separated on 2% agarose gel. The most diluted sample that was positive in the above procedure and its neighboring dilutions (2 lower, 2 higher) were subjected to this limiting dilution assay, each sample in 9-fold. Results were corrected for the input of increased DNA, and /108 to /1011dilutions were subjected to a 2-step β-globin reference PCR, each in 10-fold.10 Except for the use of 30 pmol PCO3 and GH21 primers, 5 μL DNA template was subjected to reference PCR performed essentially as tumor-specific ASO–PCR was performed. Two microliters of the first PCR was used as template in a seminested PCR procedure performed with 30 pmol PCO5 and GH21 primers. Precautions necessary to avoid cross-contamination were taken, as described by Kwok and Higuchi.11 The number of tumor cells in a sample was calculated using a program based on Poisson distribution statistics of positive and negative reactions of the PCR at each dilution level.12 As a starting value for Newton's method of iterative approximation, the weighted mean estimate was used. The likelihood maximization and the χ2 minimization were calculated as described by Taswell.13
Quantitation of tumor cells using a calibration curve
As calibrator samples, we used serial dilutions of patient bone marrow taken for the diagnosis containing more than 10% plasma cells in NWBC. The relationship between the fraction of tumor cells and the amount of ASO–PCR product in calibration samples was expressed in a regression equation used to translate the ASO–PCR product (densitometric units, relative reporter group fluorescence, or threshold cycle) in tumor cell fraction of patient samples. Deviations in the logarithmic relationship between tumor cell fraction and amount of PCR product measured using individual calibrator samples were expressed in the correlation coefficient of the regression equation. Consequently, this correlation coefficient expressed the accuracy of the quantitation.
Real-time quantitation of PCR product using the Taqman assay.
Calibrator and unknown samples were subjected to real-time quantitation14-16 using the 5′ nuclease assay17and the ABI/Prism 7700 sequence detector (Perkin–Elmer).16 In this system the tumor-specific PCR products are quantified using a nonextendable, dual-labeled fluorescent probe nested from the PCR primers. This Taqman probe (PE/ABI, Warrington, UK), synthesized according to Lee et al,14 contains a 5′ fluorogenic reporter group (eg, TET or JOE) and a 3′ fluorogenic-quenching group (TAMRA). During laser-induced excitation of the intact Taqman probe (PE/ABI), the 5′ fluorescent reporter dye is quenched by the 3′ quencher dye through Förster-type energy transfer.18,20 During specific amplification, the hybridized Taqman (PE/ABI) probe is hydrolyzed by the 5′ secondary structure-dependent exonuclease activity of Taqpolymerase.15,19,21 The reporter group fluorescence (δRn) caused by hydrolysis of the probe is proportional to the amount of PCR product and is normalized to a passive internal reference signal (ie, the rhodamine derivative ROX). Sample positivity is measured at the cycle number at which emitted fluorescence exceeds the 10× SD of baseline emissions during cycles 3 through 15 (ie, the threshold cycle [Ct]). Ct is proportional to the initial number of target molecules.22 One microgram patient DNA was used as a template. Each sample was amplified in triplicate in the presence of 15 pmol of each specific ASO, 10 pmol Taqman (PE/ABI) probe, 200 μmol/L dNTP, 60 nmol/L passive reference ROX, 1.25 U AmpliTaqGold DNA polymerase (Perkin–Elmer), 4 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 8.3), and 10 mmol/L EDTA in a volume of 50 μL. Samples were heated for 10 minutes at 95°C and amplified for 50 cycles of 0.5 minutes at 95°C and 1.5 minutes at 60°, followed by a final extension of 10 minutes at 72°C. The amount of ASO–PCR product formed is quantitated using the relative reporter group fluorescence (δ Rn) at a given cycle or using the Ct. The ABI/Prism 7700 Sequence Detector System (Perkin–Elmer) computed the fraction of tumor cells in patient samples using the patient-specific linear regression equation as a calibrator.
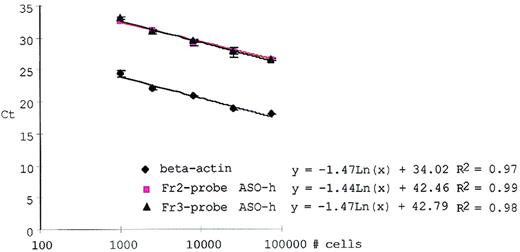
The amount of tumor cells as determined by PCR was dependent on the amount of DNA used in a PCR reaction and on factors affecting amplification fidelity and efficiency. Normalizing the tumor cell–specific ASO–PCR for the input of increased DNA in each sample was therefore requisite. PCR amplification of a reference locus was used for normalization. With real-time PCR the β-actin reference locus was amplified using 15 pmol β-actin forward and reverse primer and 10 pmol β-actin Taqman probe (Perkin–Elmer) using Taqman buffers and cycling parameters as for the ASO–PCR. The amount of increased DNA was computed relative to a dilution curve of 1 μg NWBC DNA in dH2O (5-fold decrements). For each patient sample, the ASO–PCR signal was normalized to the β-actin signal. Figure 1 shows that ASO–PCR for patients and PCR amplification of the β-actin locus had equal amplification efficiencies across a dynamic range of at least 2 log template concentrations (input, 800-80,000 cells).
Quantitating dilution series of tumor cells of patient H in dH2O using real-time ASO–PCR.
Squares and triangles show the average Ct (±SD) of 3 quantitation experiments with the VH3–Fr2 and VH3–Taqman probe, respectively. Quantitation of the absolute number of cells was performed using β-actin real-time PCR (diamonds).
Quantitating dilution series of tumor cells of patient H in dH2O using real-time ASO–PCR.
Squares and triangles show the average Ct (±SD) of 3 quantitation experiments with the VH3–Fr2 and VH3–Taqman probe, respectively. Quantitation of the absolute number of cells was performed using β-actin real-time PCR (diamonds).
Conventional quantitation of PCR product using densitometric imaging.
Calibrator and unknown samples were used as templates in ASO–PCRs, which were performed for 35 cycles on 1 μg DNA essentially as described for IgH consensus PCR except for the use of 30 pmol 5′ and 3′ ASO primers instead of VH and constant region consensus primers. Quantitation was performed as previously described.7
Results
Development of a consensus strategy for quantitation of myeloma tumor cells
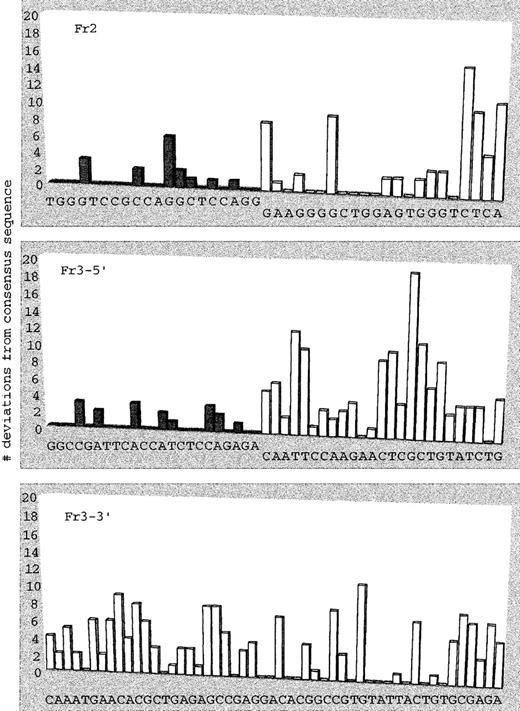
The unique immunoglobulin heavy-chain gene rearrangement (IgH) is generally used as a marker for malignant B-lineage clones. To quantitate myeloma tumor cells by IgH rearrangement, 1 of 2 approaches can be followed: either probe IgH PCR products with a tumor-specific probe23-25 or use tumor-specific IgH primers.26-31 Preliminary experiments showed occasional false-positive control samples in IgH ASO–PCR using only 1 tumor-specific primer (Figure 2). In addition, we observed that mismatched probes were able to hybridize and function in quantitation strategies. To avoid false positivity and the necessity of developing expensive patient-specific fluorogenic Taqman (PE/ABI) probes for real-time PCR, we tested a consensus strategy using 2 tumor-specific IgH primers and confirmation of amplified IgH sequences with consensus probes. Using Clustal W big ‘n’ FAT version 1.4 (EMBL, Heidelberg, Germany),32 we aligned functional VHsequences.33 34 We were unable to find probe sequences complementary to all VH germline sequences and initiated a probe design for the VH3 family (approximately 50% of patient sequences; later, probes for VH1, VH2, and VH4 were designed). Using the consensus VH3 germline sequence, we plotted the deviations we found in a panel of 24 myeloma tumor sequences (Figure 3). Using the Primer Express software program (Perkin–Elmer; demo version 1.0 ppd), we designed VH family-specific Fr2 and Fr3 probes suitable for real-time PCR. Design criteria for probes were: temperature between 66°C and 70°C (nearest-neighbor method); last 5′ base A, T, or C; more C than G bases; no secondary structures with temperature higher than 50°C; and no probe dimer formation with δG less than −10 kcal/mol. The VH3–Fr2 and VH3–Fr3 probes matched 53% and 43% of myeloma tumor IgH sequences with a VH3 rearrangement, respectively. Eighty percent of all tumor VH3 sequences matched with at least 1 of these probes. The remaining 20% of myeloma tumor VH3 sequences showed single mismatches with either the VH3–Fr2 or the VH3–Fr3 probe.
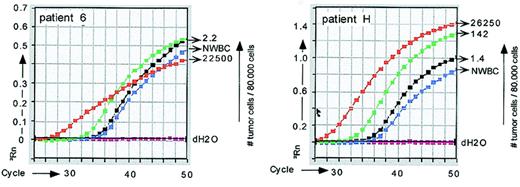
Real-time PCR quantitation of tumor cells from patient 6 and patient H.
Tumor cells were quantitated with IgH ASO–PCR using consensus sense primers (Table 1) and tumor-specific antisense primers (for patient 6) or vice versa (for patient H). Both strategies resulted in false positivity in control NWBC populations for these patients.
Real-time PCR quantitation of tumor cells from patient 6 and patient H.
Tumor cells were quantitated with IgH ASO–PCR using consensus sense primers (Table 1) and tumor-specific antisense primers (for patient 6) or vice versa (for patient H). Both strategies resulted in false positivity in control NWBC populations for these patients.
Deviations from the consensus VH3 germline sequence were found in a panel of 24 myeloma tumor sequences obtained in the HOVON-24 study.
Deviations from the consensus VH3 germline sequence were found in a panel of 24 myeloma tumor sequences obtained in the HOVON-24 study.
Validating the quantitation strategy in a multicenter setting
To test our consensus strategy for the quantitation of myeloma tumor cells, we distributed bone marrow samples from patient H (70% tumor cells) and patient 6 (30% tumor cells) among 5 participating centers. We compared the immunoglobulin heavy-chain sequence of the malignant clone obtained by the individual centers (Figure4). No sequence heterogeneity was observed. Within well-defined restrictions (see “Materials and methods”), each center chose allele-specific oligonucleotides (Figure 4) and developed its own ASO–PCR. On analysis using agarose electrophoresis, each individual ASO–PCR detected at least 1 malignant cell in a background of 1000 NWBC. A blotting procedure and then Fr3 consensus probe of the PCR samples enhanced the sensitivity of the assay by a factor of 10 (results not shown).7 For validation of the consensus strategy, unknown dilutions of DNA from patients H and 6 were distributed, and the quantitation results of 5 participating centers (4 using a limiting dilution assay and 1 using real-time PCR) were compared (Table 2). The average values found with the limiting dilution assay deviated 42% from the input value. Using a normalized real-time PCR, we calculated values that on average deviated 74% from the input value. There was no significant difference from the average quantitation values found using limiting dilutions. Variation coefficients with the limiting dilution assay ranged from 0.36 to 1.06 (average, 0.74) within single centers. We found an average 0.82 center-to-center variation coefficient that was not significantly different from the average variation coefficient found within single centers (Table 2). Center-to-center variation was reduced significantly, dropping from 0.82 to 0.59 with the single limiting dilution assay, when each center used the average of 3 independent limiting dilution assays as a quantitation value. Variation coefficients with real-time quantitation ranged from 0.29 to 0.48 (average, 0.36) and were significantly lower than with the limiting dilution assay.
IgH sequences of the malignant clone in patients 6 and H and the allele-specific oligonucleotides developed in 5 independent centers to quantitate malignant cells.
Deviations from germline sequences are given in superscript. CDR sequences are underlined.
IgH sequences of the malignant clone in patients 6 and H and the allele-specific oligonucleotides developed in 5 independent centers to quantitate malignant cells.
Deviations from germline sequences are given in superscript. CDR sequences are underlined.
Day-to-day and multicenter reproducibility of quantitating malignant myeloma cells using real-time IgH ASO-PCR or a limiting dilution assay
| Patient . | Center . | Quantitation method . | No.analyses . | Type of variation . | Input tumor cells (no.) . | Calculated tumor cells (no.) . | ASO-PCR variation . | Control PCR variation . | Total variation . | Average per patient . | Average total . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variation coefficient | |||||||||||
| 6 | E | Limiting dilution | 3 | Day-to-day | 9.6 | 10.1 | 0.24 | 0.56 | 0.61 | ||
| 6 | C | Limiting dilution | 3 | Day-to-day | 9.6 | 13.7 | 0.32 | 0.92 | 0.97 | ||
| 6 | A | Real time PCR | 4 | Day-to-day | 9.6 | 12.6 | 0.15 | 0.26 | 0.30 | ||
| 6 | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 9.6 | 11 | 0.24 | 0.72 | 0.76 | ||
| 6 | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 9.6 | 10.7 | 0.11 | 0.44 | 0.45 | ||
| 6 | E | Limiting dilution | 3 | Day-to-day | 1600 | 2306 | 0.09 | 1.04 | 1.04 | 0.83 | |
| 6 | C | Limiting dilution | 3 | Day-to-day | 1600 | 695 | 0.65 | 0.84 | 1.06 | 1.02 | |
| 6 | A | Real-time PCR | 4 | Day-to-day | 1600 | 1890 | 0.32 | 0.14 | 0.35 | 0.33 | |
| 6 | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 1608 | 1287 | 0.50 | 0.89 | 1.02 | 0.89 | |
| 6 | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 1608 | 1701 | 0.45 | 0.49 | 0.67 | 0.56 | |
| H | E | Limiting dilution | 3 | Day-to-day | 56 | 54 | 0.64 | 0.49 | 0.81 | ||
| H | C | Limiting dilution | 3 | Day-to-day | 56 | 26 | 0.28 | 0.22 | 0.36 | ||
| H | A | Real-time PCR | 4 | Day-to-day | 56 | 33 | 0.46 | 0.14 | 0.48 | ||
| H | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 56 | 45 | 0.73 | 0.50 | 0.89 | ||
| H | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 56 | 45 | 0.55 | 0.37 | 0.66 | ||
| H | E | Limiting dilution | 3 | Day-to-day | 560 | 866 | 0.40 | 0.33 | 0.52 | 0.67 | 0.75 |
| H | C | Limiting dilution | 3 | Day-to-day | 560 | 956 | 0.56 | 0.04 | 0.56 | 0.46 | 0.74 |
| H | A | Real-time PCR | 4 | Day-to-day | 560 | 1702 | 0.27 | 0.10 | 0.29 | 0.39 | 0.36 |
| H | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 560 | 993 | 0.43 | 0.44 | 0.62 | 0.76 | 0.83 |
| H | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 560 | 990 | 0.34 | 0.44 | 0.56 | 0.61 | 0.59 |
| Patient . | Center . | Quantitation method . | No.analyses . | Type of variation . | Input tumor cells (no.) . | Calculated tumor cells (no.) . | ASO-PCR variation . | Control PCR variation . | Total variation . | Average per patient . | Average total . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variation coefficient | |||||||||||
| 6 | E | Limiting dilution | 3 | Day-to-day | 9.6 | 10.1 | 0.24 | 0.56 | 0.61 | ||
| 6 | C | Limiting dilution | 3 | Day-to-day | 9.6 | 13.7 | 0.32 | 0.92 | 0.97 | ||
| 6 | A | Real time PCR | 4 | Day-to-day | 9.6 | 12.6 | 0.15 | 0.26 | 0.30 | ||
| 6 | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 9.6 | 11 | 0.24 | 0.72 | 0.76 | ||
| 6 | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 9.6 | 10.7 | 0.11 | 0.44 | 0.45 | ||
| 6 | E | Limiting dilution | 3 | Day-to-day | 1600 | 2306 | 0.09 | 1.04 | 1.04 | 0.83 | |
| 6 | C | Limiting dilution | 3 | Day-to-day | 1600 | 695 | 0.65 | 0.84 | 1.06 | 1.02 | |
| 6 | A | Real-time PCR | 4 | Day-to-day | 1600 | 1890 | 0.32 | 0.14 | 0.35 | 0.33 | |
| 6 | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 1608 | 1287 | 0.50 | 0.89 | 1.02 | 0.89 | |
| 6 | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 1608 | 1701 | 0.45 | 0.49 | 0.67 | 0.56 | |
| H | E | Limiting dilution | 3 | Day-to-day | 56 | 54 | 0.64 | 0.49 | 0.81 | ||
| H | C | Limiting dilution | 3 | Day-to-day | 56 | 26 | 0.28 | 0.22 | 0.36 | ||
| H | A | Real-time PCR | 4 | Day-to-day | 56 | 33 | 0.46 | 0.14 | 0.48 | ||
| H | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 56 | 45 | 0.73 | 0.50 | 0.89 | ||
| H | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 56 | 45 | 0.55 | 0.37 | 0.66 | ||
| H | E | Limiting dilution | 3 | Day-to-day | 560 | 866 | 0.40 | 0.33 | 0.52 | 0.67 | 0.75 |
| H | C | Limiting dilution | 3 | Day-to-day | 560 | 956 | 0.56 | 0.04 | 0.56 | 0.46 | 0.74 |
| H | A | Real-time PCR | 4 | Day-to-day | 560 | 1702 | 0.27 | 0.10 | 0.29 | 0.39 | 0.36 |
| H | E, C, G, U | Limiting dilution | Single | Multicenter (4) | 560 | 993 | 0.43 | 0.44 | 0.62 | 0.76 | 0.83 |
| H | E, C, G, U | Limiting dilution | Triplo 5 | Multicenter (4) | 560 | 990 | 0.34 | 0.44 | 0.56 | 0.61 | 0.59 |
The factor-range expresses the quotient of actual maximal and minimal values found in the quantitation assays.
Triplo 5, mean of 5 PCR reactions; whole procedure is repeated 3 times (15 PCR).
Quantification of patient samples in time
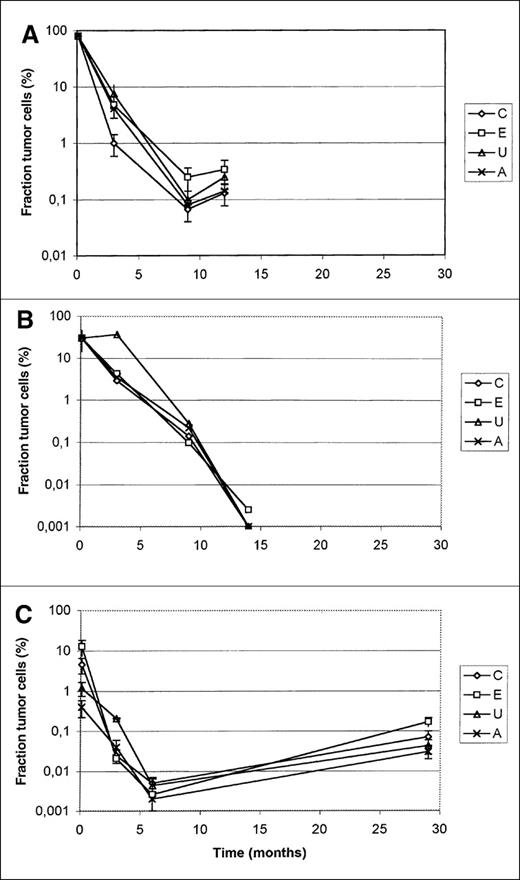
To test further our consensus strategy for the quantitation of myeloma tumor cells, we distributed subsequent bone marrow samples from 3 patients among 4 centers. Three centers used the limiting dilution strategy, and 1 center used real-time PCR (Center A). Figure 5 shows the results. The first data point in Figure 5A was not quantified by PCR but was measured by differential count of the plasma cells. As shown, the limiting dilution assay gives results similar to those for real-time quantitative PCR. Small differences were explained by ASO–PCR and β-globin PCR to measure total DNA.
Quantification of bone marrow sample over time in 3 patients entered in the trial.
The first values in A and B were the number of plasma cells counted in the initial bone marrow slides. In C, this sample was quantified by PCR. Centers C, E, and U applied the limiting dilution approach; center A quantified by real-time PCR.
Quantification of bone marrow sample over time in 3 patients entered in the trial.
The first values in A and B were the number of plasma cells counted in the initial bone marrow slides. In C, this sample was quantified by PCR. Centers C, E, and U applied the limiting dilution approach; center A quantified by real-time PCR.
Performance of the IgH consensus probes in real-time quantitation
Accuracy.
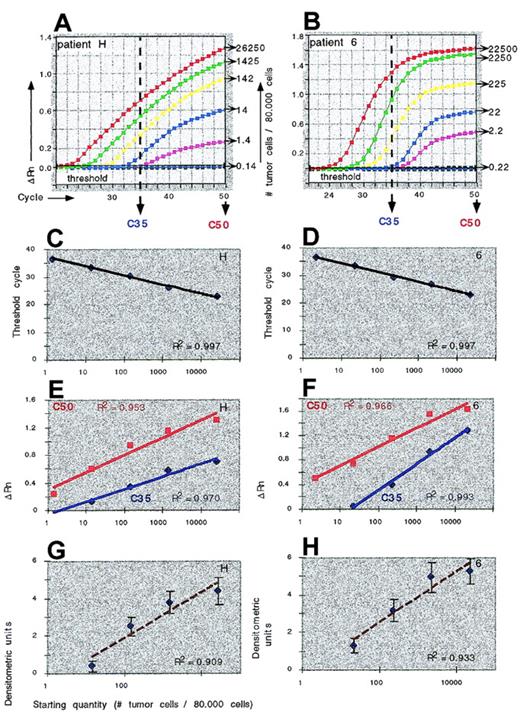
Using a 5-log range of tumor cell concentrations in real-time PCR (Figure 6A), we measured the increase in fluorescence (δ Rn) after 35 cycles and 50 cycles, reflecting the amount of ASO–PCR product generated at 35 or 50 cycles of real-time PCR. We found regression equations (relating the δ Rn to tumor cell concentrations) with correlation coefficients of 0.97 to 0.99 and 0.95 to 0.97 for 35 and 50 cycles, respectively (Figure 6C). Using Ct as a parameter for the amount of PCR product, we consistently found regression equations with correlation coefficients above 0.99 (Figure 6B). In contrast, the detection of blotted ASO–PCR product with labeled probes and densitometric imaging of PCR products resulted in regression equations with correlation coefficients of 0.91 for patient H and 0.93 for patient 6 (Figure 6D). We concluded that using calibration curve equations describing the logarithmic relationship between Ct and the starting quantity (ie, fraction of tumor cells) produced quantitation data with the highest accuracy.
ASO–PCR quantitation of malignant cells.
Quantitation of malignant cells in samples from myeloma patients H and 6 using real-time IgH ASO–PCR (A-C) or conventional ASO–PCR and densitometric scanning (D). Calibration curves were constructed by plotting the initial number of tumor cells against the Ct (B), the amount of emitted fluorescence (PCR product) at 35 and 50 cycles (C), or the density (D).
ASO–PCR quantitation of malignant cells.
Quantitation of malignant cells in samples from myeloma patients H and 6 using real-time IgH ASO–PCR (A-C) or conventional ASO–PCR and densitometric scanning (D). Calibration curves were constructed by plotting the initial number of tumor cells against the Ct (B), the amount of emitted fluorescence (PCR product) at 35 and 50 cycles (C), or the density (D).
Sensitivity.
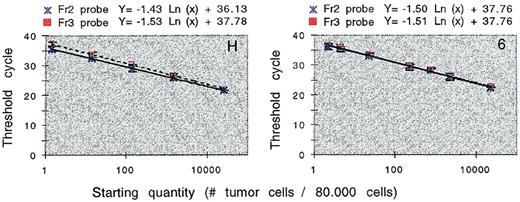
Using dilution series bone marrow of patient H and patient 6, we compared the performance of the VH3–Fr2 Taqman (PE/ABI) probe with the VH3–Fr3 Taqman probe in real-time IgH ASO–PCR. The dilution series from patient 6 had an amplification efficiency of 97%, irrespective of the probe used. The dilution series from patient H was amplified with an amplification efficiency of 100% or 96% with the use of Fr2 or Fr3 probes, respectively (Figure 7). We concluded that real-time ASO–PCR (Figures 5 and 6) was able to detect 1 DNA target or 1 tumor cell in a background of 80 000 normal cells (sensitivity, 1.25×10−5). To achieve this sensitivity, a minimum of 38 amplification cycles was required. Conventional detection of blotted ASO–PCR product with labeled probes and densitometric imaging (Figure 6D) is able to detect 10 DNA targets (sensitivity, 1.25×10−4).
Comparing the performance of the VH3–Fr2 and VH3–Fr3 probe in real-time quantitation.
Bone marrow from patients H and 6 were subjected to real-time IgH ASO–PCR in 5-fold dilutions. Using these results, calibration curves were constructed by plotting the initial number of tumor cells against the threshold cycle (±SD). The VH3–Fr3 probe in patient H has 1 mismatch.
Comparing the performance of the VH3–Fr2 and VH3–Fr3 probe in real-time quantitation.
Bone marrow from patients H and 6 were subjected to real-time IgH ASO–PCR in 5-fold dilutions. Using these results, calibration curves were constructed by plotting the initial number of tumor cells against the threshold cycle (±SD). The VH3–Fr3 probe in patient H has 1 mismatch.
Reproducibility.
With conventional ASO–PCR and densitometric quantitation (Table 2), variation coefficients ranged from 0.68 to 0.88 (average, 0.77). Using a 5-log range of template concentrations in real-time PCR, variation coefficients within a single PCR ranged from 0.04 to 0.37 (average, 0.21) (Table 3). We assessed day-to-day variation of real-time quantitation using identical calibration samples as PCR templates on 5 consecutive days (Table 3). Variation coefficients ranged from 0.23 to 0.57 (average, 0.38). Real-time ASO–PCR performed with the VH3–Fr3 probe and the VH3–Fr2 probe showed no significant differences in reproducibility (Table 3). Alignment of the VH3–Fr3 probe with the tumor VDJ sequence of patient H showed a mismatch on the 3′ position 18 of the probe. We concluded that this mismatch had no consequences for reproducibility and sensitivity of the real-time PCR and that single mismatched probes may be used in real-time quantitation.
Quantitation reproducibility using real-time ASO-PCR or conventional ASO-PCR and densitometric scanning
| Patient . | Quantitation method . | Probe . | Type of variation . | Input tumor cells . | Calculated quantity . | Variation coefficient . | Quantity interval (95%) . | Average variations . | Variation coefficient . |
|---|---|---|---|---|---|---|---|---|---|
| H | Real-time PCR | Fr2 | Intra PCR | 1425 | 1422 | 0.2 | 1141-1703 | ||
| Day-to-day | 1425 | 1634 | 0.23 | 1252-2016 | |||||
| Intra PCR | 1.4 | 0.8 | 0.14 | 0.7-0.9 | Real-time PCR intra PCR | 0.17 | |||
| Day-to-day | 1.4 | 1.7 | 0.41 | 1.0-2.4 | Real-time PCR inter PCR | 0.32 | |||
| H | Real-time PCR | Fr3 | Intra PCR | 1425 | 2026 | 0.1 | 1830-2222 | ||
| Day-to-day | 1425 | 1493 | 0.36 | 958-2028 | |||||
| Intra PCR | 1.4 | 1.2 | 0.37 | 0.8-1.6 | Real-time PCR intra PCR | 0.24 | |||
| Day-to-day | 1.4 | 1.6 | 0.47 | 0.8-2.4 | Real-time PCR inter PCR | 0.42 | |||
| H | Real-time PCR | Fr2 and Fr3 | Real-time PCR intra PCR | 0.21 | |||||
| Real-time PCR inter PCR | 0.37 | ||||||||
| 6 | Real-time PCR | Fr2 | Intra PCR | 2250 | 2601 | 0.04 | 2491-2711 | ||
| Day-to-day | 2250 | 1677 | 0.57 | 715-2639 | |||||
| Intra PCR | 22 | 21.3 | 0.36 | 13.7-28.9 | Real-time PCR intra PCR | 0.2 | |||
| Day-to-day | 22 | 18.4 | 0.37 | 11.6-25.2 | Real-time PCR inter PCR | 0.47 | |||
| 6 | Real-time PCR | Fr3 | Intra PCR | 2250 | 3543 | 0.17 | 2990-4136 | ||
| Day-to-day | 2250 | 3010 | 0.34 | 1994-4026 | |||||
| Intra PCR | 22 | 18 | 0.24 | 13.7-22.2 | Real-time PCR intra PCR | 0.21 | |||
| Day-to-day | 22 | 18.7 | 0.28 | 13.5-23.9 | Real-time PCR inter PCR | 0.31 | |||
| 6 | Real-time PCR | Fr2 and Fr3 | Real-time PCR intra PCR | 0.21 | |||||
| Real-time PCR inter PCR | 0.39 | ||||||||
| H | Densitometric | Fr2 | Intra PCR | 1425 | 5065 | 0.68 | 1609-8521 | ||
| Intra PCR | 1.4 | 6.5 | 0.72 | 1.8-11.2 | Densitometric patient H | 0.7 | |||
| 6 | Densitometric | Fr2 | Intra PCR | 2250 | 11004 | 0.88 | 1318-20690 | ||
| Intra PCR | 22 | 16.5 | 0.78 | 3.6-29.4 | Densitometric patient 6 | 0.83 | |||
| All | Densitometric | 0.77 | |||||||
| Real-time PCR intra PCR | 0.21 | ||||||||
| Real-time PCR inter PCR | 0.38 |
| Patient . | Quantitation method . | Probe . | Type of variation . | Input tumor cells . | Calculated quantity . | Variation coefficient . | Quantity interval (95%) . | Average variations . | Variation coefficient . |
|---|---|---|---|---|---|---|---|---|---|
| H | Real-time PCR | Fr2 | Intra PCR | 1425 | 1422 | 0.2 | 1141-1703 | ||
| Day-to-day | 1425 | 1634 | 0.23 | 1252-2016 | |||||
| Intra PCR | 1.4 | 0.8 | 0.14 | 0.7-0.9 | Real-time PCR intra PCR | 0.17 | |||
| Day-to-day | 1.4 | 1.7 | 0.41 | 1.0-2.4 | Real-time PCR inter PCR | 0.32 | |||
| H | Real-time PCR | Fr3 | Intra PCR | 1425 | 2026 | 0.1 | 1830-2222 | ||
| Day-to-day | 1425 | 1493 | 0.36 | 958-2028 | |||||
| Intra PCR | 1.4 | 1.2 | 0.37 | 0.8-1.6 | Real-time PCR intra PCR | 0.24 | |||
| Day-to-day | 1.4 | 1.6 | 0.47 | 0.8-2.4 | Real-time PCR inter PCR | 0.42 | |||
| H | Real-time PCR | Fr2 and Fr3 | Real-time PCR intra PCR | 0.21 | |||||
| Real-time PCR inter PCR | 0.37 | ||||||||
| 6 | Real-time PCR | Fr2 | Intra PCR | 2250 | 2601 | 0.04 | 2491-2711 | ||
| Day-to-day | 2250 | 1677 | 0.57 | 715-2639 | |||||
| Intra PCR | 22 | 21.3 | 0.36 | 13.7-28.9 | Real-time PCR intra PCR | 0.2 | |||
| Day-to-day | 22 | 18.4 | 0.37 | 11.6-25.2 | Real-time PCR inter PCR | 0.47 | |||
| 6 | Real-time PCR | Fr3 | Intra PCR | 2250 | 3543 | 0.17 | 2990-4136 | ||
| Day-to-day | 2250 | 3010 | 0.34 | 1994-4026 | |||||
| Intra PCR | 22 | 18 | 0.24 | 13.7-22.2 | Real-time PCR intra PCR | 0.21 | |||
| Day-to-day | 22 | 18.7 | 0.28 | 13.5-23.9 | Real-time PCR inter PCR | 0.31 | |||
| 6 | Real-time PCR | Fr2 and Fr3 | Real-time PCR intra PCR | 0.21 | |||||
| Real-time PCR inter PCR | 0.39 | ||||||||
| H | Densitometric | Fr2 | Intra PCR | 1425 | 5065 | 0.68 | 1609-8521 | ||
| Intra PCR | 1.4 | 6.5 | 0.72 | 1.8-11.2 | Densitometric patient H | 0.7 | |||
| 6 | Densitometric | Fr2 | Intra PCR | 2250 | 11004 | 0.88 | 1318-20690 | ||
| Intra PCR | 22 | 16.5 | 0.78 | 3.6-29.4 | Densitometric patient 6 | 0.83 | |||
| All | Densitometric | 0.77 | |||||||
| Real-time PCR intra PCR | 0.21 | ||||||||
| Real-time PCR inter PCR | 0.38 |
The factor-range expresses the quotient of actual maximal and minimal values found in the quantitation assays.
Discussion
Recently, HOVON initiated a multicenter study to evaluate whether myeloma patients treated with intensive chemotherapy benefit from additional peripheral stem cell transplantation. Treatment outcome was measured using the bone marrow plasma cell percentage, plasma cell monoclonality (κ/λ ratio), and serum or urine M-protein concentration as parameters. For patients with follicular non-Hodgkin lymphoma, a significantly reduced disease-free survival rate correlated with the presence of PCR-detectable lymphoma cells after therapy.35,36 The sustained PCR detection of abcr-abl gene rearrangement in patients with chronic myeloid leukemia correlated with a higher probability of relapse.37Although it has not been proven that treatment response is correlated with a decrease in the number of myeloma tumor cells, data generated by Corradini et al3 strongly suggest a correlation between PCR detection of myeloma tumor cells after therapy and treatment outcome. Longitudinal PCR quantitation of myeloma cells in HOVON-24 might confirm that increases in tumor cell number predict clinical relapse. For these reasons we decided to quantitate the fraction of malignant cells as an additional parameter to monitor treatment response.
In this report we compared the reproducibility of several PCR methods to quantitate myeloma tumor cells. With the construction of a tumor-specific PCR, the amplification target of choice is a well-described marker restricted to and ubiquitously present in all malignant cells. Because such markers are unavailable for most patients with multiple myeloma, the unique IgH rearrangement is generally used as a marker for the malignant clone. IgH PCR approaches to quantitate myeloma tumor cells either probe IgH PCR products with a tumor-specific probe23-25 or use tumor-specific IgH primers.26-28,30,31 Because the quantitation of myeloma tumor cells with consensus IgH primers and a tumor-specific probe is generally less sensitive than with the use of allele-specific oligonucleotides24-31 and because we occasionally observed false positivity while using consensus IgH primers, we chose to use 2 IgH ASO and sequence confirmation with consensus IgH probes. Early reports describe quantitation methods using calibration curve equations that relate the amount of ASO–PCR product found in patient samples (with an unknown fraction of tumor cells) with the amount of ASO–PCR product found in dilution series (with a known fraction of tumor cells). Measuring the amount of PCR product was labor intensive and involved Southern blot analysis, screening with labeled probes, and densitometric analysis.26,28,30 We found that in this way accurate measurements were limited to a 3-log range of templates (Willems et al7; this study). In this study, we tested real-time IgH ASO–PCR and a limiting dilution assay as alternatives for the quantitation of tumor cells.5
With real-time IgH ASO–PCR, the amount of PCR product is proportional to the amount of emitted fluorescence (δ Rn) because of probe hydrolysis. In addition, the initial number of PCR target molecules is proportional to the Ct in which a fixed amount of emitted fluorescence (10× base fluorescence) is detected.16,22 This report shows that calibration curve equations relating the amount of tumor cells with δ Rn or Ct allow an accurate quantitation of a 5-log range of myeloma tumor cells (1×10−5 to 1×100). We suggest the use of Ct as a parameter reflecting the amount of PCR product formed in real-time PCR because relating the starting quantity (tumor cells) with the threshold cycle resulted in calibration curve equations with the highest correlation coefficients (consistently above 0.99). Reproducibility within a single real-time PCR was high (average variation coefficient, 0.21). Even day-to-day measurements resulted in good reproducibility (average variation coefficient, 0.38). In contrast, reproducibility within a single ASO–PCR that was quantitated using blotting, probing, and densitometric analysis was much lower (average variation coefficient, 0.77). We conclude that real-time IgH ASO–PCR is a highly reproducible, accurate, and sensitive method to quantitate myeloma tumor cells. We recommend the use of real-time PCR in multicenter trials for a number of additional reasons. Closed tube analysis with real-time PCR reduces contamination risk and false-positive findings. Standardization (a major concern in multicenter studies) is warranted with real-time PCR because standardized equipment, PCR buffers, strict PCR guidelines, and quantitation statistics are available. Recently, we concluded that real-time PCR master mixes with all ingredients (without template) could be stored at −80°C for long periods of time without notable loss of PCR efficiency.38 Distributing ready-to-use master mixtures to individual centers will enhance reaction reproducibility, facilitate standardization, and reduce PCR contamination risk even more.
We tried to develop consensus instead of patient-specific fluorogenic probes to facilitate the standardized quantitation of myeloma tumor cells. Alignment studies of functional VH germline genes showed that a minimum of 2 consensus probes per VH family were required to develop a consensus strategy for quantitation myeloma tumor cells. During this study we initiated a probe design for tumor cell IgH rearrangements involving the VH3 family. Alignment of 24 myeloma tumor cell IgH VH3 rearrangements obtained in the HOVON study with a VH3–Fr2 and a VH3–Fr3 probe suitable for real-time PCR showed that 80% of tumor VH3 sequences were complementary to either probe. Alignment of the VH3–Fr3 probe with the tumor IgH sequence of patient H showed a mismatch on position 3′ of the probe. This mismatch had no consequences for sensitivity, accuracy, or reproducibility of the real-time quantitation. In preliminary experiments, we observed identical results for the quantitation of tumor VH4 sequences using VH4–Fr2 and VH4–Fr3 consensus probes (results not shown). Assuming that all single mismatches are of no consequence for real-time quantitation, 2 probes would be sufficient to quantitate all tumor VH3 sequences. The consensus real-time IgH ASO–PCR strategy presented here circumvented the need to develop a new expensive fluorogenic probe for each patient. In addition, it was a useful strategy for the quantitation of tumor cells in other B-cell malignancies. For many patients with non-Hodgkin lymphoma (NHL), the best available marker to quantitate malignant cells is the IgH rearrangement. For these patients the consensus real-time IgH ASO–PCR strategy is recommended, though ongoing somatic hypermutation of the malignant clone in most patients with follicular NHL might interfere with accurate tumor cell counts.39 In patients with acute lymphoblastic leukemia, frequent VH replacement might obstruct real-time quantitation of acute lymphoblastic leukemia cells with consensus fluorogenic probes.40 41 Quantitation of these cells probably requires the use of consensus IgH sense primers and patient-specific CDR3 fluorogenic probe to avoid false-positive findings.
The equipment to perform real-time PCR was not available to all participating HOVON centers. We therefore distributed patient samples and compared the use of real-time PCR (1 center) and a limiting dilution assay (4 centers) for quantitation of myeloma tumor cells in a multicenter setting. Limiting dilution assays used statistics to translate the highest dilution of patient samples with tumor cells that were ASO–PCR detectable into the number of tumor cells in this sample.5,9 A prerequisite for the statistical procedure with the limiting dilution assay is that a single copy of the CDR3 region of the malignant clone be detected. By diluting the bone marrow of 2 patients with more than 10% plasma cells in NWBC, each center was able to demonstrate that this requirement was met (results not shown). Using the limiting dilution assay, we calculated values that on average deviated 13% from the input value, and we found an average variation coefficient of 0.37 (without correction using a reference PCR). Comparable results with a limiting dilution assay were obtained by Cremer et al,5 who calculated values that on average deviated 32% from the input value and had an average variation coefficient of 0.32. We tested the real-time PCR and the limiting dilution assay in a multicenter setting. The average values found with real-time quantitative PCR were not significantly different from the averages found with the normalized limiting dilution assay. Reproducibility of the real-time quantitation was higher (average variation coefficient, 0.36) than with the limiting dilution quantitation (average variation coefficient, 0.74). In addition, the average variation coefficient inside each center (0.74) was not significantly different from the average center-to-center variation coefficient of 0.82, indicating that standardization of the limiting dilution assay was successful. To reduce center-to-center variation, we tested the use of 3 independent limiting dilution assays to quantitate each sample. The reproducibility of the triple limiting dilution assay (average variation coefficient, 0.59) approached that of the real-time quantitative PCR (average variation coefficient, 0.36). In addition, we distributed the bone marrow samples of 3 study patients. These in-time quantifications during follow-up confirmed reproducibility. We conclude that both quantitation methods result in accurate quantitation values and that both methods can be used in a single multicenter trial.
We present a strategy to quantitate malignant cells in myeloma patients using IgH ASO–PCR. Using the consensus strategy, an accurate comparison of quantitation data generated in independent centers is possible. This strategy can be adapted for the quantitation of tumor cells in other B-cell malignancies and may serve as a template for the construction of consensus protocols for multicenter quantitation studies in general.
Reprints:Reinier Raymakers, Department of Internal Medicine, Division of Hematology, 544 University Hospital Nijmegen, Postbus 9101, 6500HB Nijmegen, the Netherlands.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal