Abstract
Nonrandom interstitial deletions and monosomy of chromosomes 5, 7, and 17 in refractory myelodysplasia (MDS) and acute myelogenous leukemia (AML) suggest a multistep pathway that culminates in aggressive clinical course. Because cytogenetic studies frequently identify chromosome 5 and 17 deletions within a single clone, we searched for allele loss for 5q loci and TP53 gene mutations in the same leukemic samples. Cosegregating deletions of chromosomes 5 and 17 were found to specifically include the 5q13.3 interval between the lociD5S672 and D5S620/D5S626, a locus hypothesized to harbor a tumor suppressor gene1 and the TP53 gene on 17p. A rare patient with secondary refractory MDS and an unbalanced translocation [der(5;17)], which resulted in deletions of the 5q13.3-qter and 17p loci, provided clues on the sequence of genetic alterations. Serial molecular analysis of this patient revealed a dysplastic clone with der(5;17), which gave rise to a leukemic clone on acquiring an inactivating mutation of TP53. Our findings are consistent with functional cooperation between a putative tumor suppressor gene at 5q13.3 that contributes toward the progression of early stages of MDS, and the TP53 gene when mutated, causes transformation to AML.
The significance of cytogenetic alterations in predicting therapeutic outcome was recognized more than a decade ago.2,3 In refractory anemia with excess blasts (MDS-RAEB), refractory anemia with excess blasts in transformation (MDS-RAEB-t), and acute myelogenous leukemia (AML), nonrandom interstitial deletions 5q,7q or monosomy of 5 and 7 are markers for poor prognosis and short remission periods. Similarly, mutations of the tumor suppressor gene,TP53, are associated with resistence to therapy and poor prognosis in hematologic malignancies.4 This is in stark contrast to simple reciprocal translocations such as inv(16) in AML, for which the outcome is more favorable.
Typically, RAEB, RAEB-t, and AML patients with deletions or monosomy of either 5q, 7q, or 17p have numerous additional karyotypic changes, the significance of which has not been extensively investigated. Preferential combination of 1 or more abnormalities may provide clues on the genetic alterations leading to the progression of AML.
Deletions of 5q result in loss of large segments of the chromosome, which translates into physical loss of approximately 2% of the haploid genome. In a previous report, we identified a patient with de novo AML, who harbored a deletion at 5q13, and allowed the characterization of a critical locus between proximal markers, AFMb347yf9/D5S672, and distal markers D5S620/D5S626.1 The importance of this interval was further confirmed in an AML cell line, ML3, in which the entire chromosome 5q sequences are grossly intact, except at 5q13.3 and qter.
Subsequently, several studies on solid tumors support a critical locus at 5q13, which undergoes loss of heterozygosity (LOH) or allelic imbalance. These include malignancies of the lung,5prostate,6 ovary,7 stomach,8pancreas,9 and bladder.10 More importantly, in each of these studies, LOH of 5q13 loci is evident but not 5q23 (APC locus) or 5q31.1 (AML/MDS locus), indicating a distinct tumor suppressor locus at chromosome 5q13.3. The deletions of 5q13 in ovarian cancer are particularly striking in that the 5q13 deletions correlate with mutations of the TP53 gene in 78% of tumors.7
In AML and MDS, mutations of the TP53 gene are infrequent, less than 10%. However, in a subset of patients with “17p- syndrome,” the TP53 gene is frequently mutated.11 A recent report on this distinct clinical entity demonstrated loss of theTP53 gene by 17p deletion in 14 of the 16 cases analyzed by fluorescent in situ hybridization (FISH). Enhanced expression or mutation of the remaining allele was observed in 11 of 14 cases.12 Interestingly, 15 of 17 patients in this study also had karyotypic deletions of chromosome 5. The primary mechanism of loss of chromosome 5 and 17 sequences was by an unbalanced translocation between the 2 chromosomes (9/15 cases). Unbalanced translocations or dicentrics involving chromosomes 5 and 17 have been recognized as recurring abnormalities in myeloid neoplasms by other investigators as well.13 The nonrandom participation of chromosome 5 raises the possibility of an unidentified gene on chromosome 5 whose loss synergizes with the inactivation of theTP53 gene. Because most of these patients have numerous cytogenetic abnormalities at presentation, it is difficult to determine whether the mutation in the TP53 gene occurs first and the unbalanced 5;17 translocation results from the ensuing genomic instability, or whether the der(5;17) is an initial, critical step.
In this report, we show that a putative tumor suppressor locus at 5q13.3 may be the target of a critical gene because patient samples with a mutated TP53 allele also have invariant loss or disruption of 5q13.3. Thus, loss of chromosome 5q13.3 sequences confers a proliferative advantage to a dysplastic clone that can be fully transformed upon acquiring a mutation in the TP53 gene.
Materials and methods
Patient samples
All patient information and samples were obtained through approved protocols. Routine cytogenetic analysis were performed on patients when they were seen at the University of Texas M. D. Anderson Cancer Center. Patient 1 was treated at the University of Texas M. D. Anderson Cancer Center for acute promyelocytic leukemia with a typical t(15;17)(q22;q21) in January 1994. She went into complete remission by July 1994 in response to all trans retinoic acid and idarubicin. An abnormal clone with 45, XX,-5, add(17)(p11.2) was first identified by conventional G-banding during routine follow-up in July 1996. The -5, add(17p) anomaly was later determined to be a der(5;17) by FISH. In December 1996, she presented with 9% blasts in her bone marrow, an abnormal karyotype with multiple cytogenetic changes, including the der(5;17), and was diagnosed with MDS.
Patient database
Information on patients treated at the M. D. Anderson Cancer Center, Houston, TX, in the Department of Hematology since 1985 is available in a departmental database (Microsoft Access). Karyotypic data are available for 2718 patients that have been diagnosed with AML or MDS. Routine cytogenetic analysis was performed on patients at presentation as part of the diagnostic procedure. Patients were identified as having specific chromosomal abnormalities by searching for the following strings in the database: “-5”, “*del(5*", “*5q*”, *(5;*”, “*;5)”, “-7", “*del(7*”, “*7q*”, “*(7;*”, “*;7)*”, “*,-17*”, “*del(17*”, “*17p*”, “*(17;*”, “*;17)*”, “*add(17*”, “*,-16*”, “*del(16*”, “*(16;*”, “*16;)*”, “*inv(16)*”, or “*inv16*”. Searches explicitly excluded t(15;17). Searches were performed for each chromosome alone and then in various combinations. Each karyotype was reviewed to ensure that the string search accurately identified chromosomal changes involving 5q, 7q, and 17p.
Loss of heterozygosity (LOH) analysis
Information for the polymorphic markers that were used in this study was obtained from the Whitehead Institute (http://www.genome.wi.mit.edu) or the Stanford Human Genome Center (SHGC) (http://www-shgc.stanford.edu). Primers and polymerase chain reaction (PCR) amplification conditions for chromosome 5 and 17 markers were obtained from the Genome Database (http://www.gdb.org), the Whitehead institute, or the SHGC. The technique for determining LOH is detailed elsewhere.14
Physical map of 5q13.3 loci
Our previous report on the delineation of the critical 5q13.3 locus relied solely on the radiation hybrid mapping data available at that time from the Whitehead Institute. The current study was facilitated by second generation radiation hybrid maps from the SHGC, Sanger Center. UK, and a Yeast Artificial Chromosome (YAC) contig from the Whitehead Institute. All the markers were verified independently against the YACs. Only those markers that tested positive in 2 or more screens were considered truly positive. Orientation and overlap of the YACs were further confirmed by screening for ends of YACs rescued by inverse PCR.15 The current order is most consistent with centromere-AFMb347yf9-D5S672-D5S620/D5S626-D5S641-GATAP18 104-D5S401-telomere, rather than the previous order of centromere-D5S672-AFMb347yf9-GATAP18 104-D5S620/D5S626-D5S641-D5S401-telomere. Inclusion of additional sequence tagged site (STS) markersD5S1464, D5S806, D5S2029 enhances the resolution of the map.
Somatic cell hybrids
Somatic cell hybrids were generated by fusing the peripheral blood leukemic blasts from patient 1 and LMTK- murine fibroblasts. Ficoll Hypaque separated mononuclear cells in suspension were fused with polyethylene glycol (PEG 1000) and selected in hypoxanthine, aminopterin, and thymidine (HAT) containing medium.16Colonies were screened by PCR to identify those with human chromosome 5 and 17 sequences.
Cell lines
The AML cell line ML3 was cultured in RPMI 1640 medium (Life Technologies, Rockville, MD) containing 10% fetal calf serum. LMTK- mouse fibroblasts were cultured in Dulbecco's Modified Eagle medium (DMEM) (Life Technologies, Rockville, MD) containing 10% fetal calf serum.
Fluorescence in situ hybridization
Alu PCR probes were made by PCR amplifying against YAC DNA. Primers and amplification conditions are described by Liu et al.17Probes were biotinylated using Bionick Labeling System (Life Technologies, Rockville, MD) according to the manufacturer's instructions. Painting probes were purchased from Oncor (Gaithersberg, MD). Hybridization procedures are detailed elsewhere.18
Mutation screening for the TP53 gene
The TP53 exons 4-8 were amplified with intronic primers and the products were sequenced in an ABI 377 automated DNA sequencer using standard dye terminator chemistry. Mutations were verified by at least 2 independent PCR amplifications.
Results
Deletions of chromosomes 5 and 17 cosegregate in RAEB, RAEB-t, and AML patients
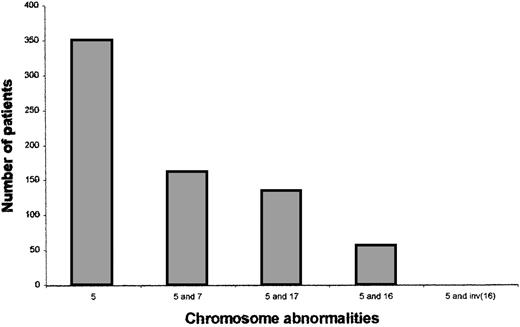
We noted that 6 of the 12 patients who allowed the initial delineation of the critical 5q13.3 locus also had deletions of 17p or monosomy for chromosome 17.1 To determine whether this was due to a selection bias or a nonrandom correlation, we used our institutional database that contains karyotypic data for AML and MDS patients. The overall frequency of loss of 5, 7, and 17 were 11.3%, 13.4%, and 5.97%, respectively. Of those 350 patients with chromosome 5 abnormalities, 134 (38%) had a chromosome 17 abnormality and 161 (46%) had a chromosome 7 abnormality. The correlation between chromosomes 5 and 17 is similar to that seen between chromosomes 5 and 719 (Figure 1). No other chromosomal abnormality showed such a strong cosegregation. For example, anomalies of the long arm of chromosome 16, which is reported to show allelic imbalance in solid tumors,20 are seen only in 55 (16%) of the 350 patients with chromosome 5 anomalies. Additionally, none of the 55 patients with the anomaly inv(16) that is associated with the M4 subtype of AML had loss of chromosome 5. Thus, there is a nonrandom cosegregation of deletions of chromosome 5q and 17p loci in AML and MDS.
Chromosomal abnormalities of patients with AML or MDS.
Patients were identified as having chromosome 5, 7, 17, or 16 abnormalities by reviewing the karyotypes. Each patient has 1 or more cytogenetic changes and karyotypic abnormalities are not exclusive to these 4 chromosomes. The last bar includes only those patients with a 5q abnormality and inv(16) and no other 16 abnormality.
Chromosomal abnormalities of patients with AML or MDS.
Patients were identified as having chromosome 5, 7, 17, or 16 abnormalities by reviewing the karyotypes. Each patient has 1 or more cytogenetic changes and karyotypic abnormalities are not exclusive to these 4 chromosomes. The last bar includes only those patients with a 5q abnormality and inv(16) and no other 16 abnormality.
On the basis of the correlations shown in Figure 1, we examined whether there was a critical target locus on chromosomes 5q, and whether theTP53 gene was indeed the target on 17p. Therefore, the results are presented below in 3 distinct parts: (1) characterization of a putative critical locus at 5q13.3, (2) delineation of the target locus at 17p, and search for TP53 mutations, and (3) the sequence of genetic alterations in a case of secondary MDS with der(5;17).
Delineation of the critical 5q13.3 locus from patients with der(5;17)
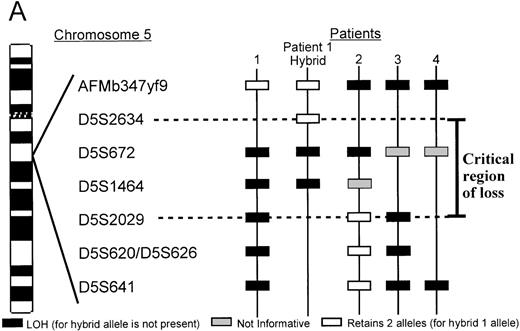
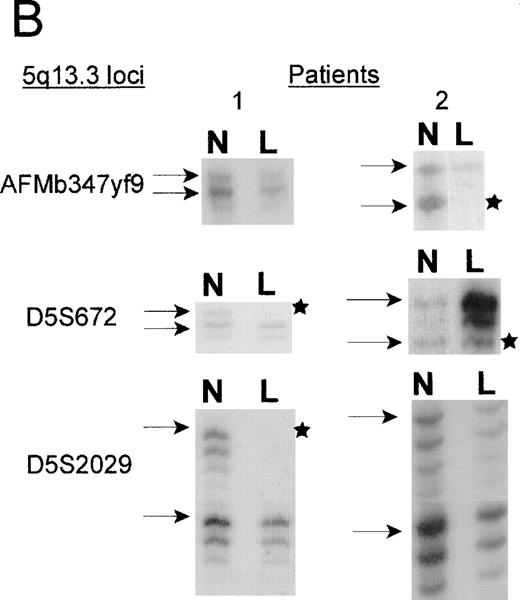
Two patients with unbalanced translocations between chromosomes 5q and 17p facilitated the search for the smallest region of loss on 5q. The der(5;17) in both patients resulted in deletions of 5q13.3 loci, but were in opposite orientations. Patient 2, with de novo AML, retained heterozygosity for all the 5q31.1 loci and the distal 5q13.3 markers D5S2029, D5S620, and D5S626, yet showed a minimal deletion from 5q13.1-q13.3, which included the 5q13.3 markersAMFb347yf9 and D5S672 (Figure2). A similar patient with der(5;17) (patient 1) showed a deletion which apparently overlapped with that of patient 2. As shown in Figure 2B, the informativeness of theAFMb347yf9, D5S672, and D5S2029 loci in both these patients is revealed by the presence of 2 major alleles in the normal sample. The leukemic cells of patient 1 retains 2 alleles ofAFMb347yf9 and a single allele of both D5S672 andD5S2029 loci. Patient 2, however, retains 2 alleles ofD5S2029 and reveals allele loss for both D5S672 andAFMb347yf9. In addition, the deletion pattern in patient 1 confirms the revised order of markers, namely, centromereAFMb347yf9-D5S672-telomere.
A. Delineation of the critical region of loss between in patients 1 and 2.
Marker D5S2634 maps within 30-kb of marker D5S672 (SHGC radiation hybrid map) and is placed centromeric, based on results from our somatic cell hybrid. The ordering of markers is supported by the YAC contig in Figure 3. B. Loss of heterozygosity analysis of polymorphic markers at 5q13.3. Paired samples of DNA from peripheral blood granulocytes or bone marrow blasts and mononuclear cells were PCR amplified for each of the 5q13.3 polymorphic markers. Patient 1 mononuclear cells were short term cultured to select for the leukemic population. The presence of 2 alleles in the control sample rendered a given patient informative for the specific locus. N = normal sample. L = leukemic sample. Arrows mark the location of alleles. Stars indicate allele loss.
A. Delineation of the critical region of loss between in patients 1 and 2.
Marker D5S2634 maps within 30-kb of marker D5S672 (SHGC radiation hybrid map) and is placed centromeric, based on results from our somatic cell hybrid. The ordering of markers is supported by the YAC contig in Figure 3. B. Loss of heterozygosity analysis of polymorphic markers at 5q13.3. Paired samples of DNA from peripheral blood granulocytes or bone marrow blasts and mononuclear cells were PCR amplified for each of the 5q13.3 polymorphic markers. Patient 1 mononuclear cells were short term cultured to select for the leukemic population. The presence of 2 alleles in the control sample rendered a given patient informative for the specific locus. N = normal sample. L = leukemic sample. Arrows mark the location of alleles. Stars indicate allele loss.
Further refinement of the proximal limit of the 5q13.3 critical locus was facilitated by somatic cell hybrids, generated by fusing leukemic blasts of patient 1 and LMTK− murine fibroblasts. A hybrid clone that contained the der(5;17) chromosome but not the normal chromosome 5 was identified and characterized. STS markers localized to this interval by the SHGC were tested against the hybrid by PCR amplification. The der(5;17) chromosome contained markersAFMb347yf9 and D5S2634 but not D5S672. D5S2634, which is retained in the hybrid is within 30 kb (less than 1 cR) of D5S672, according to the SHGC radiation hybrid map. Therefore, the proximal breakpoint in patient 1 resides between the loci D5S672 and D5S2634 (Figure 2A). Lack of informativeness for the D5S1464 locus in patient 2 diminishes finer definition of the telomeric limit of the critical region. Taken together, patients 1 and 2 delineate a critical region of loss between the D5S2634 and D5S2029 loci.
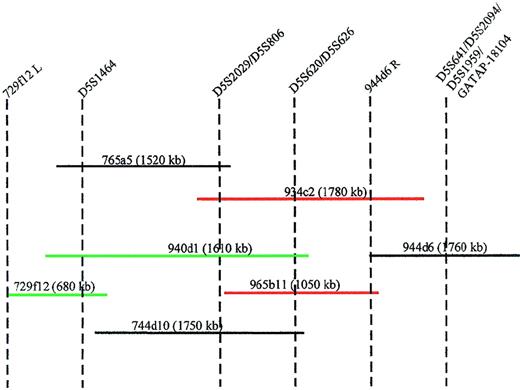
Generation of a YAC contig spanning the D5S1464-D5S641interval
To physically link the markers within the critical 5q13.3 region, we initiated a chromosomal walk from the telomeric end of this locus. The YAC 944d6 served as the starting point and the end rescued fragment from the right arm tested positive by PCR for the YACs 965b11 and 934c2; thus, orienting the D5S641, D5S2094,D5S1959, and GATA-P18104 loci telomeric of theD5S620 and D5S626 loci (Figure3). Screening of the YACs 729f12, 765a5, 940d1, 934c2, and 744d10 with the additional markers yielded an order that is most consistent with centromere-D5S1464-(D5S2029/D5S806)-(D5S620/D5S626)-(D5S641/GATA-P18104)-telomere.
Minimal tiling path of YACs between markers D5S1464 and GATAP-18104.
STS content of each YAC is indicated by vertical dotted lines. The insert size of the YACs is shown in parentheses. YACs in green hybridize to the der(5) chromosome and YACs in red hybridize to the der(3) chromosome. 944d6R = end rescued fragment from the right arm of YAC 944d6. 729f12L = end rescued fragment from the left arm of YAC 729f12.
Minimal tiling path of YACs between markers D5S1464 and GATAP-18104.
STS content of each YAC is indicated by vertical dotted lines. The insert size of the YACs is shown in parentheses. YACs in green hybridize to the der(5) chromosome and YACs in red hybridize to the der(3) chromosome. 944d6R = end rescued fragment from the right arm of YAC 944d6. 729f12L = end rescued fragment from the left arm of YAC 729f12.
Delineation of the 5q13.3 chromosomal breakpoint in the AML cell line, ML3
The AML cell line, ML3, has a der(3;5) and retains heterozygosity for most of the chromosome 5q loci. The translocation disrupts the 5q13.3 region and so facilitated further delineation of the critical region. Inter-alu PCR products from 5q13.3 YACs (Figure 3) were used as FISH probes on metaphases from these cells. YACs 745a9, containing the marker AFMb347yf9, hybridized to the der(5) and YAC 965b11, containing markers D5S620 and D5S626, hybridized to the der(3).1 YACs 848c4 and 765a5, containing markersD5S672 and D5S1464 respectively, hybridized to the telomeric end of the der(5) chromosome. YAC 940d1, containing the markers D5S1464 as well as D5S620, also hybridized to the der(5) chromosome. YAC932c2, with markers D5S2029/D5S806and D5S620/D5S626 hybridized to the der (3) chromosome. Figure3 demonstrates that the 5q13.3 locus disrupted in the ML3 cell line resides between D5S1464 and D5S620/D5S626, a breakpoint at the telomeric end of the critical region of loss (Figure 2).
Absence of a split signal between the der (5) and der (3) chromosomes with either YAC 934c2 or YAC 940d1 could be due to the small region of overlap between the 2 YACs that may represent less than 20% of the insert in either case.
The TP53 gene is deleted in patients with 5q13.3 translocations and deletions
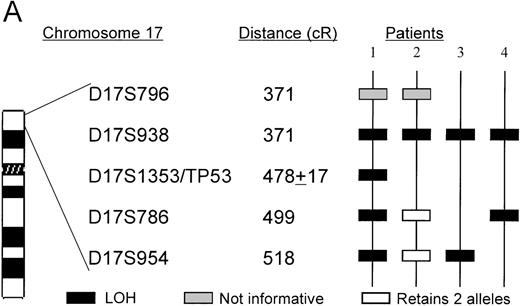
Microsatellite analysis was used to determine whether patients who show allele loss for 5q loci and anomalies of chromosome 17p also show a region of loss that included the TP53 gene. Two pairs of markers that are centromeric (D17S954 and D17S786) and telomeric (D17S938 and D17S796) of the TP53gene were selected. The microsatellite marker D17S1353 maps within the same YAC 728b11 as the TP53 gene, but its orientation with respect to the TP53 gene is unclear. In total, 15 patients with chromosome 5 deletions, including 2 patients whose interstitial deletions were entirely telomeric of the 5q13.3 locus, were analyzed. Surprisingly, we found that the 4 patients who had allele loss for 17p loci had the 5q13.3 deletion in common (ie, contiguous loss of loci within the D5S672-D5S626 interval). The karyotypes and diagnoses of these 4 patients are shown in Table1.
Characteristics of patients with loss of heterozygosity for 5q13.3 loci
| Patient Number . | Karyotype . | Age/Gender . | Diagnosis . | Previous Malignancy . |
|---|---|---|---|---|
| 1 | 45 XX, −5, add(17)(p11.2)[5] | 37/F | Secondary RAEB-t | Yes |
| 42-45 XX, −3, −5, add(6)(p25), −7, −14, −15, add(17)(p11.2), +2 mar [15] | ||||
| 2 | 45 XX, del(5)(q14), del(7)(q21q31), del(9)(q11q22), add(11)(p15), −16, add(17)(p11) | 39/F | De novo AML (M1) | No |
| 3 | 45 XX, various karyotypic changes including ins(3;5)(q31;q31), del(5)(q11q31), dup(8q), t(12;13)(p13;q14) | ??/M | Secondary AML | Yes |
| 4* | 45-48 XY, −2, −5, t(5;12), del(7)(q?), +8, −16, +1-2 r, +1-4 mar[16] 46 XY[4] | 82/M | RAEB-t | Yes |
| Patient Number . | Karyotype . | Age/Gender . | Diagnosis . | Previous Malignancy . |
|---|---|---|---|---|
| 1 | 45 XX, −5, add(17)(p11.2)[5] | 37/F | Secondary RAEB-t | Yes |
| 42-45 XX, −3, −5, add(6)(p25), −7, −14, −15, add(17)(p11.2), +2 mar [15] | ||||
| 2 | 45 XX, del(5)(q14), del(7)(q21q31), del(9)(q11q22), add(11)(p15), −16, add(17)(p11) | 39/F | De novo AML (M1) | No |
| 3 | 45 XX, various karyotypic changes including ins(3;5)(q31;q31), del(5)(q11q31), dup(8q), t(12;13)(p13;q14) | ??/M | Secondary AML | Yes |
| 4* | 45-48 XY, −2, −5, t(5;12), del(7)(q?), +8, −16, +1-2 r, +1-4 mar[16] 46 XY[4] | 82/M | RAEB-t | Yes |
RAEB-t = refractory anemia with excess blasts in transformation; AML = acute myelogenous leukemia.
No cytogenetic abnormality of chromosome 17.
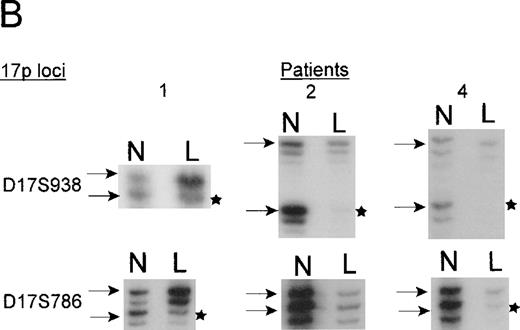
Patients 1, 3, and 4 have deletions, including markers that flank theTP53 gene. Patient 2 has a telomeric deletion that includes markers up to the TP53 region, but because there is a lack of polymorphic markers in this region, a more a precise breakpoint could not be achieved. The TP53 mutation analysis, however, suggests that patient 2 carried only 1 copy of TP53(see below). Interestingly, patient 4 who did not show any cytogenetic anomaly of chromosome 17 appears to have harbored a small deletion (Figure 4).
A. The ordering of markers within the region is based on published mapping data from the SHGC.
Distances in centirays (cR) are from the p-arm telomere as determined by the SHGC. B. Loss of heterozygosity analysis of polymorphic markers at 17p13. Paired samples of DNA from peripheral blood granulocytes or bone marrow blasts and mononuclear cells were PCR amplified for each of the 17p13 polymorphic markers. The presence of 2 alleles in the mononuclear cells rendered a given patient informative for the specific locus. LOH = loss of heterozygosity. N = normal sample. L = leukemic sample. Arrows mark the location of alleles. Stars indicate allele loss.
A. The ordering of markers within the region is based on published mapping data from the SHGC.
Distances in centirays (cR) are from the p-arm telomere as determined by the SHGC. B. Loss of heterozygosity analysis of polymorphic markers at 17p13. Paired samples of DNA from peripheral blood granulocytes or bone marrow blasts and mononuclear cells were PCR amplified for each of the 17p13 polymorphic markers. The presence of 2 alleles in the mononuclear cells rendered a given patient informative for the specific locus. LOH = loss of heterozygosity. N = normal sample. L = leukemic sample. Arrows mark the location of alleles. Stars indicate allele loss.
TP53 gene is mutated in patients with allele loss for 17p and the cell line, ML3
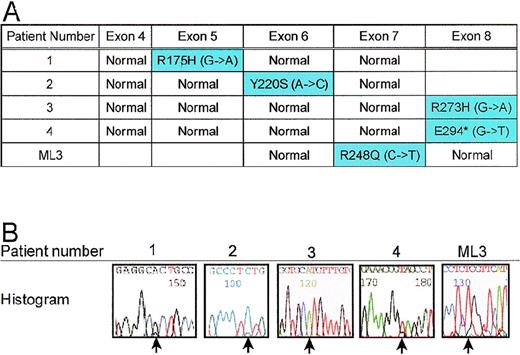
To determine whether the remaining allele of the TP53 gene was mutated, we sequenced exons 4 through 8 in 15 patients with 5q deletions. As expected, all 4 patients with LOH for 5q13.3 and 17p loci also had a mutation of the remaining TP53 allele (Figure5A). Patients 1 and 3 had the commonTP53 mutations, R175H and R273H, respectively; whereas, patients 2 and 4 had more rare mutations Y220S and E294*, respectively. Although the ML3 cells do not appear to have chromosome 17 abnormalities by G-banding (JC Liang, unpublished results), we tested for TP53 mutation given the 5q13.3 abnormality. ML3 cells have a mutation R248Q, which has been reported in solid tumors and Li Fraumeni Syndrome families.21 Patient 2, for whom LOH for the TP53 gene could not be verified, shows only residual evidence of the normal TP53 allele. For patient 3 and ML3, no normal allele could be detected by sequencing (Figure 5B).
A. Mutations in remaining allele of TP53.
Exons 4 through 8 of the TP53 gene were amplified by PCR using intronic primers and sequenced to identify mutations. Patients 1, 2, 3, 4, and ML3 each had a mutation in the coding region of the gene. B. Histograms of mutant TP53 alleles from patients. Point mutations are designated by arrows.
A. Mutations in remaining allele of TP53.
Exons 4 through 8 of the TP53 gene were amplified by PCR using intronic primers and sequenced to identify mutations. Patients 1, 2, 3, 4, and ML3 each had a mutation in the coding region of the gene. B. Histograms of mutant TP53 alleles from patients. Point mutations are designated by arrows.
The der(5;17) precedes mutation of TP53 that is involved in leukemic progression
Patient 1 presented with secondary MDS within 3 years after therapy for her primary malignancy of acute promyelocytic leukemia (APL). As part of the follow-up, several routine cytogenetic studies were performed. An abnormal clone with -5, add(17p11.2) as the sole abnormality was first identified 29 months after her therapy. Retrospective FISH analysis on serial bone marrow samples revealed that the -5, add(17p) was actually a der(5;17). Metaphase FISH also showed that the der(5;17) clone expanded over the previous 8 months (Table2).
Progression of myelodysplasia patient 1
| Time from Primary Diagnosis . | Karyotype . | FISH Results . | Clinical Status . | TP53 Status . |
|---|---|---|---|---|
| 0 mo | 46, XX, t(15;17)(q22;q21) | APL t(15;17) | Presentation | Normal |
| 6 mo | 46, XX | Complete Remission | Normal | |
| 21 mo | ND | 13% der(5;17) | Complete Remission | Normal |
| 24 mo | ND | 33% der(5;17) | Complete Remission | Normal |
| 29 mo | 45, XX, −5, add(17)(p11.2)[6] | 43% der(5;17) | Complete Remission | Normal |
| 32 mo | 45, XX, −5, add(17)(p11.2)[19] | 74% der(5;17) | Complete Remission | Normal/Mutant |
| 35 mo | 45, XX, −5, add(17)(p11.2)[5] 42-45, XX, chromosomes exhibiting shared abnormalities consisting of −3, −5, add(6)(p25), −7, −14, −15, add(17)(p11.2), +2mar[15] | 95% der(5;17) | Relapse MDS 9% blasts | Mutant |
| Time from Primary Diagnosis . | Karyotype . | FISH Results . | Clinical Status . | TP53 Status . |
|---|---|---|---|---|
| 0 mo | 46, XX, t(15;17)(q22;q21) | APL t(15;17) | Presentation | Normal |
| 6 mo | 46, XX | Complete Remission | Normal | |
| 21 mo | ND | 13% der(5;17) | Complete Remission | Normal |
| 24 mo | ND | 33% der(5;17) | Complete Remission | Normal |
| 29 mo | 45, XX, −5, add(17)(p11.2)[6] | 43% der(5;17) | Complete Remission | Normal |
| 32 mo | 45, XX, −5, add(17)(p11.2)[19] | 74% der(5;17) | Complete Remission | Normal/Mutant |
| 35 mo | 45, XX, −5, add(17)(p11.2)[5] 42-45, XX, chromosomes exhibiting shared abnormalities consisting of −3, −5, add(6)(p25), −7, −14, −15, add(17)(p11.2), +2mar[15] | 95% der(5;17) | Relapse MDS 9% blasts | Mutant |
ND = not done; APL = acute promyelocytic leukemia; MDS = myelodysplasia.
We used fixed cells from serial samples to determine the stage at which the mutation in the TP53 gene occurred. The first sign of mutation was seen 32 months after the APL diagnosis, suggesting that the mutation occurred in the der(5;17) clone. Consistent with aTP53 inactivation mutation, numerous cytogenetic changes characteristic of genomic instability, were seen at 35 months (Table2).
Discussion
This study is a combinatorial analyses of 2 distinct cytogenetic alterations, namely, deletions of 5q and 17p. The investigations allow us to conclude that unbalanced translocations between chromosomes 5 and 17 target a critical locus at 5q13.3 and the TP53 gene (patients 1 and 2). Both of these loci may also be deleted by separate events as seen in ML3 and patients 3 and 4.
Cosegregation of deletions of chromosomes 5 and 17
The evidence provided in this report and that of Soenen et al12 suggests that unbalanced chromosomal translocations may be a common mechanism that hematopoietic cells use to cause the physical loss of the TP53 gene. There is a strong indication for cosegregation of loss of chromosome 5 and 17 sequences (Figure 1). The higher frequency of chromosome 5 deletions than 17p deletions suggests it being an earlier genetic lesion. Inversely, 65% (134/205) of those patients with loss of 17p sequences also have deletions of chromosome 5, which is consistent with data reported by others.12 13
Deletions of chromosome 5 occur in a broad range of clonal myeloid anomalies
An eclectic group that includes the indolent, transfusion-dependant refractory anemia (RA), the highly refractory and aggressive RAEB-t, and AML harbor large deletions of chromosome 5. Very little is known about the relationship between the basis of these diverse genetic entities besides the 2 distinct critical regions of loss identified at 5q31.22,23 A rare case of RAEB may show noncontiguous deletions of both 5q31 loci,24 whereas other cases of RA have also been reported with del(5)(q11q13).25 The evidence provided here identifies a distinct refractory subset in which theD5S2634-D5S2029/D5S806 interval is consistently lost and theTP53 gene is mutated.
Nature of TP53 mutations
All the mutations we identified were in the DNA binding domain ofTP53. Of the 5 TP53 mutations that we identified, 2 were not typical. Codon 220 is a common site for mutation, but is usually a change from Tyr to Cys. Codon 294 is rarely mutated.26 Only 3 of the 5 mutations that we identified were transition mutations indicating that the common C to T transition mutation seen in solid tumors may not be the only mechanism of mutation in leukemia. A larger survey of TP53 mutations in secondary AML and MDS will be useful in understanding the mechanism as the mutational spectra in MDS and AML may be different from other cancers.27
Loss of 5q13.3 locus precedes mutations in the TP53 gene
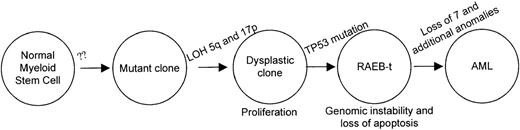
Patient 1 allows us to conclude that deletions of 5q and 17p loci precede the TP53 inactivation although clonal expansion of cells harboring der(5;17) is clearly evident, as shown in Table 2. She was asymptomatic during the evolution of the der(5;17) clone suggesting that the unbalanced translocation conferred a proliferative advantage and did not affect the differentiation potential. Deletion of chromosome 5q is also seen as an acquired anomaly in familial AML and MDS without genetic linkage to chromosome 5q.28 Taken together, the early events leading to the initiation of a dysplastic clone may be independent of loss of chromosome 5q; whereas, deletions in 5q may allow an expansion of the clone that can then be transformed by mutations on TP53 or other genetic alterations. Figure6 illustrates a proposed order and functional consequences of the somatic mutations during the progression of MDS and AML. As TP53 mutations are infrequent in refractory MDS and AML, there are likely to be other genetic alterations that would also facilitate transformation in cells with deletions of chromosome 5. To support this model, additional patients with chromosome 5 and 17 abnormalities will be studied in the future. Furthermore, the placement of TP53 mutation late in the sequence of events is reminiscent of the colon cancer model of transformation.29
Sequence of genetic alterations in patient 1.
A normal myeloid stem cell that acquires loss of 5q13 and 17p loci confers a prolifertaive advantage. The clone accumulates additional chromosomal alterations after inactivation of the TP53 gene. The resulting acute leukemic clone with the numerous cytogenetic alterations is highly refractory to therapy.
Sequence of genetic alterations in patient 1.
A normal myeloid stem cell that acquires loss of 5q13 and 17p loci confers a prolifertaive advantage. The clone accumulates additional chromosomal alterations after inactivation of the TP53 gene. The resulting acute leukemic clone with the numerous cytogenetic alterations is highly refractory to therapy.
Acknowledgments
Our special thanks are due to Dr Elihu Estey for helpful discussions on patient 1. We thank Drs Miloslav Beran, David Claxton, Steven Kornblau, and Susan Roberts for patient material and Dr Hong Liang for helpful discussions. We also thank Rui Yu Wang, Xiuying Lina Wu, and Lisa Chu for assistance with FISH analysis and Rashmi Pershad for the automated DNA sequence analysis. The enthusiastic assistance of Gisela Sanchez William and Ellen Jackson with the patient database is gratefully acknowledged. We thank Dr Walter Hittelman for critical reading of the manuscript.
Supported by grants from Ladies Leukemia League, Metairie, LA, CA66982 and CA55164 from the National Institutes of Health.
Reprints:Lalitha Nagarajan, Department of Molecular Genetics, Box 45, M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: lalitha@odin.mdacc.tmc.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal