Abstract
The ETV6/TEL gene has been reported to fuse toPDGFRβb MDS1/EVI1, BTL, ACS2,STL, JAK2, ABL, CDX2, TRKC,AML1, and MN1. Among them, PDGFRβ,ABL, JAK2, and TRKC are tyrosine kinases (TK). We identified a novel ETV6 partner gene, ARG(ABL-related gene or ABL2), another TK gene in a cell line established from a patient with acute myelogenous leukemia (AML-M3) with a t(15;17)(q22;q11.2) and a t(1;12)(q25;p13), which has the remarkable feature to differentiate to mature eosinophils in culture with all-trans retinoic acid and cytokines. TheETV6/ARG transcripts consisted of exon 1 to 5 of ETV6and the 3′ portion of ARG starting from exon 1B or exon 2, resulting in an open reading frame for a fusion protein consisting of the entire PNT oligomerization domain of ETV6 and all of the functional domains of ARG including the TK domain. This is the same protein structure as identified in the other ETV6 TK fusion proteins. The reciprocal ARG/ETV6 transcript was not expressed, and the normal ETV6 allele was not deleted or rearranged. Although the ABL is known to be involved in various human malignancies, ARG has not been involved in human malignancies despite its high homology to ABL. Thus, this is the first report showing involvement of ARG in human leukemia. The ETV6/ARG protein may be involved in the unique differentiation capacity of this cell line.
ETV6 (also known as TEL), initially cloned as a partner gene of platelet-derived growth factor receptor β (PDGFRβ) in a t(5;12)(q33;p13) translocation, is a member of the ETS family of transcription regulators.1 It contains 2 functional domains, a 5′ pointed (PNT) domain and a 3′ ETS DNA binding domain,1 and fuses to a number of different partner genes such as MDS1/EVI1(3q26),2BTL(4q11),3ACS2(5q31),4STL(6q23),5JAK2(9p24),6,7ABL(9q34),7,8CDX2(13q12),9TRKC(15q24),10,11AML1(21q22),12,13 andMN1(22q11).14 Among them, PDGFRβ,JAK2, ABL, and TRKC are tyrosine kinase (TK) genes.
In the fusion proteins, the PNT domain of ETV6 activates the TK domain of the TK genes,1,7,15 or modifies the properties of the transcription factor as AML1.12 Moreover, it is revealed that not only the functional domains of ETV6/TK proteins but also other sequences may contribute to the properties of the fusion proteins. The same ETV6/TRKC transcript was found in a benign neoplasm (familial fibrosarcoma)10 and in a malignancy (acute myelogenous leukemia [AML]-M2 with an aggressive clinical course).11 Although the 2 putative proteins contained the same functional domains (the PNT domain of ETV6 and the TK domain of TRKC), the sequences were different: ETV6/TRKC protein in familial fibrosarcoma contained an additional 14 amino acids just downstream of the TRKC TDYYR motif,11 whereas the protein in acute leukemia did not contain ETV6 exon 5,11 which may explain the difference.
Here, we report the identification of a novel partner of ETV6,ARG (ABL-related gene or ABL2), another TK, in a cell line established from a patient with AML-M3 with a t(1;12)(q25;p13) and a t(15;17)(q22;q11.2). This translocation resulted in the fusion of the PNT domain of ETV6 to all of the functional domains of ARG. Although ABL is known to be involved in various human malignancies and ARG shows high homology to ABL, ARG is not yet reported to be involved in human malignancies. This is the first report that shows ARGinvolvement in human leukemia.
Materials and methods
Cell lines
The patient was a 66-year-old man with AML-M3. He was successfully induced to the first and second remission with combined chemotherapy. At the second relapse, he was referred to the Niigata University Hospital to be treated with all-trans retinoic acid (ATRA). However, he returned to the previous hospital without receiving ATRA therapy due to his family circumstance. The HT93A cell line was established by one of us (K.K.) from peripheral blood mononuclear cells obtained at the second relapse.At the second relapse, the patient's blood cells showed the following karyotype: 46, XY,1q−, inv(3),4q−,6q+,11p+q−,12p+,15q+,17q−, 21q+,22q+. When the cell line was established, the karyotype was 46, XY, t(1;12)(q25;p13),2q+, t(4;6)(q12;q13), t(15;17)(q22;q11). Further cytogenetic studies revealed that an additional part of 2q+ was derived from the long arm of chromosome 1, that is, the der(2)t(1;2)(q25;q33). We also determined the breakpoint of t(15;17) more precisely as t(15;17)(q22;q11.1). Thus, the corrected karyotype was 46, XY, t(1;12)(q25;p13), der(2)1(1;2)(q25;q33), t(4;6)(q12;q13), t(15;17)(q22;q11.1). With reverse transcriptase-polymerase chain reaction (RT-PCR) using the primers described elsewhere,16a short type PML-RARA transcript was detected. The characteristics of the HT93A cell line are described elsewhere.17 In brief, this cell line has the remarkable capacity to differentiate into eosinophils in culture with ATRA and granulocyte macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3) or interleukin-5 (IL-5), although in culture with ATRA alone, the cells differentiate predominantly into neutrophils. HL60 and K562 were used as controls.
Fluorescent in situ hybridization
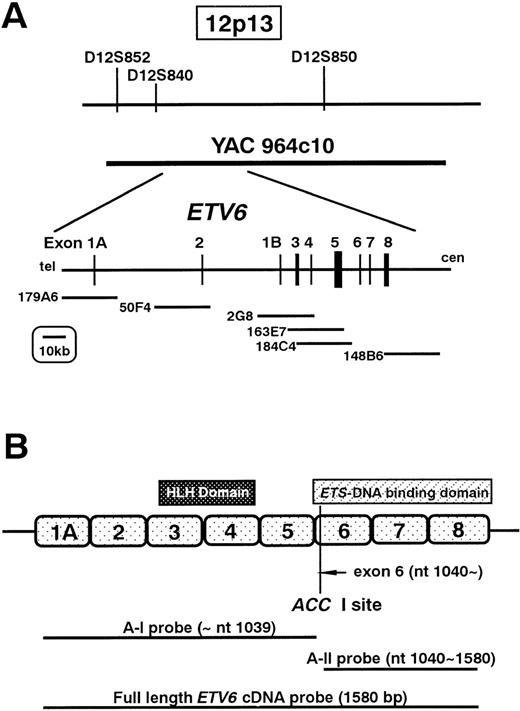
For fluorescent in situ hybridization (FISH) probes, we used the YAC964c10 (kindly provided by Dr Janet D. Rowley, Section of Hematology/Oncology, the University of Chicago, Chicago, IL), which contains the entire ETV6 gene, and the LL12NCO1 cosmid probes (kindly provided by Dr Peter Marynem, University of Leuven, Leuven, Belgium) located within the ETV6 in the following order: 5′—-179A6-50F4-2G8-163E7-184C4-148B6—-3′ (Figure1A). The probes were labeled with biotin-11-dUTP (Boehringer Mannheim, Mannheim, Germany) using nick translation and were hybridized to the HT93A metaphase cells. The probes were detected with Cy-3-Avidin (Amersham Pharmacia, Upsala, Sweden), and then counterstained with 4′, 6-diamindino-2-phenylindole dihydrochloride (DAPI). The presence or absence of the FISH signals was scored on an average of 18 cells (range, 15-21) per probe. Images of the hybridized signals and the DAPI were captured with a liquid-cooled, charge-coupled device camera (Quantix: Photometrics, Tucson, Ariz).
FISH analysis and Southern and Northern hybridizations.
(A) Physical map surrounding ETV6 and the location of the cosmid and YAC probes used in FISH analysis. (B) Schematic representation of ETV6 and the probes used for Southern and Northern hybridizations.
FISH analysis and Southern and Northern hybridizations.
(A) Physical map surrounding ETV6 and the location of the cosmid and YAC probes used in FISH analysis. (B) Schematic representation of ETV6 and the probes used for Southern and Northern hybridizations.
Southern blot analysis
High-molecular weight DNA for Southern hybridization was extracted from each cell line by the standard procedure.18 DNA (10 μg) was digested to completion either with Bam HI, EcoRI, or Sac I and size-fractionated on a 0.7% agarose gel. The DNA was transferred to Hybond-N nylon membrane (Amersham Pharmacia). Probes used for hybridization were full length ETV6complementary DNA (cDNA; kindly provided by Dr Todd Golub, Division of Hematology/Oncology, Brigham and Women's Hospital, University of Harvard, Boston, Mass), ETV6 cDNA fragment of exons 1 to 5 (A-I probe: nt 1-1039), and ETV6 cDNA fragment of exons 6 to 8 (A-II probe: nt 1040-1580). A-I and A-II probes were obtained by digestion of full length ETV6 cDNA by Acc I (Figure 1B). The probes were labeled with α32P-dCTP by random priming and used for hybridization. Hybridization was performed in 0.5 mol/L NaH2PO4/Na2HPO4, pH7.2, 7% sodium dodecyl sulfate (SDS), 1 mmol/L EDTA, with radiolabeledETV6 probes at 65°C. Washes were performed in 40 mmol/L NaH2PO4/Na2HPO4, 1% SDS at 65°C.
Northern blot analysis
Total RNA was extracted by the acid guanidinium phenol-chloroform method using ISOGEN reagent (Nippon Gene, Toyama, Japan). Total RNA (10 μg) was size-fractionated on a 1% agarose/formaldehyde gel and transferred to Hybond-N membrane (Amersham Pharmacia). Probes used for hybridization were the same 3 probes as used in Southern blot analysis,ETV6 1A, ETV6 1B, and an ARG 3′ probe (nt 3001-3753). The latter 3 probes were prepared from total RNA of the HT93A cells by RT-PCR using the following primers: E1AF (ETV6exon 1A nt 25-44: 5′-ATGTCTGAGACTCCTGCTCA-3′) and E2R (ETV6 exon 2 nt 186-167: 5′-CAGGTGCGCAGGCAGGC GGA-3′) for ETV6 1A probe, E1BF (ETV6 exon 1B nt 328-347: 5′-TGTGTCACAGCAGTCTGCCC-3′) and E2R forETV6 1B probe, and A3001F (ARG nt 3001-3020: 5′-GTCCTTATCTCACCCACTCT-3′) and A3753R (ARG nt 3753-3734: 5′-CTACCTCTGCACCACATCAC-3′) for the ARG3′ probe. Hybridization was performed as described above. The same filter was rehybridized with β-actin-specific probe to check the amount of RNA loaded in each lane.
3′-RACE
Total RNA was isolated from each cell line using the ISOGEN reagent. The first strand cDNA was synthesized from 2.5 μg of total RNA using the Super Script Pre-amplification System (Gibco-BRL, Gaithersburg, Md) according to the standard procedures with the primer BamTX (5′-CCCGGATCCT15(A/C/G)-3′). The first PCR was performed with primers E4F (ETV6 exon 4 nt 353-372: 5′-GTGATGTGCTCTATGAACTC-3′) and BamTX using the first strand cDNA as the template. Diluted product of the first PCR was used for the second, semi-nested PCR with primers E5F (ETV6exon 5 nt 510-529: 5′-CAGGCCATCCGTGGATAATG-3′) andBamTX. The thermal cycling profile was as follows: 94°C for 3 minutes, followed by 30 cycles of 94°C for 30 seconds, 55°C for 1 minute, 72°C for 2 minutes, and a final extension at 72°C for 10 minutes. Products were analyzed on an agarose gel, cloned using the pT7 Blue-Vector (Novagen, Madison, Wis) and subjected to DNA sequencing.
RT-PCR
Two micrograms of total RNA isolated from each cell line were subjected to cDNA synthesis using the Super Script II-Reverse Transcriptase (Gibco-BRL). Primers E5F and A590R (ARG nt 590-571: 5′-GTGATGCTGAGTGTGTTATC-3′) were used to confirm the ETV6/ARG transcripts. Primers E1AF and E1BF in combination with A3753R were used to confirm the full-length ETV6/ARGtranscripts. To detect the reciprocal ARG/ETV6 transcripts, primers A11F (ARG nt 11-30: 5′-TCTGTGAGTCGCCTGGAGGC-3′) and A211F (ARG nt 211-230: 5′-CAGCAGGTGGGCCGCGTCGG-3′) in combination with E7R (ETV6 exon 7 nt 1277-1258: 5′-CTGAACAAAAGCCTTTGTCC-3′) and E8R (ETV6 exon 8 nt 1383-1364: 5′-TCAGCATTCATCTTCTTGGT-3′) were used. The thermal cycling parameters were described above. Amplified products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide.
DNA sequencing and homology search
Cycle sequencing was performed using fluorescein-labeled primers derived from the pT7 Blue-vector (T7: 5′-CTAATACGACTC ACTATAGGG-3′ and U19: 5′-GTTTTCCCAGTCACGACGT-3′) and the Vistra systems premixed cycle sequencing kit (Amersham Pharmacia). The reaction products were separated on an automated laser fluorescence sequencer (Amersham Pharmacia). DNA sequences were compared to entries of GenBank using the NCBI Blast server.
Results
FISH study
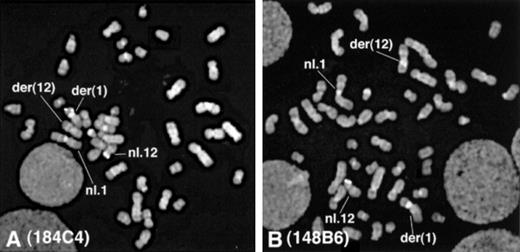
The FISH study indicated that the breakpoint of t(1;12) was located between exon 5 and exon 8 of ETV6. FISH analysis using the YAC964c10 probe showed split signals between the der(1) and the der(12), suggesting the involvement of ETV6 in this t(1;12) translocation.19 Further FISH studies using ETV6cosmid probes showed definite involvement of ETV6. The signals from the cosmids containing exons 1 to 5 (179A6, 50F4, 2G8, 163E7, and 184C4) were found on the der(1) (Figure2A), whereas the signals from the cosmid containing exon 8 (148B6) were found on the der(12) (Figure 2B). These results indicated that the 12p13 breakpoint was located between exon 5 and exon 8 of ETV6. The signals on the normal 12p were always observed with YAC probe and every ETV6 cosmid probe, suggesting that the non-translocated allele of ETV6 was intact.
FISH analysis of the HT93A cells.
(A) The signals were found on the der(1) and on the normal 12p with 184C4 probe, which hybridizes to ETV6 exons 3-5. (B) The signals were found on the der(12) and on the normal 12p with 148B6 probe, which hybridizes to ETV6 exon 8.
FISH analysis of the HT93A cells.
(A) The signals were found on the der(1) and on the normal 12p with 184C4 probe, which hybridizes to ETV6 exons 3-5. (B) The signals were found on the der(12) and on the normal 12p with 148B6 probe, which hybridizes to ETV6 exon 8.
Southern blot hybridization
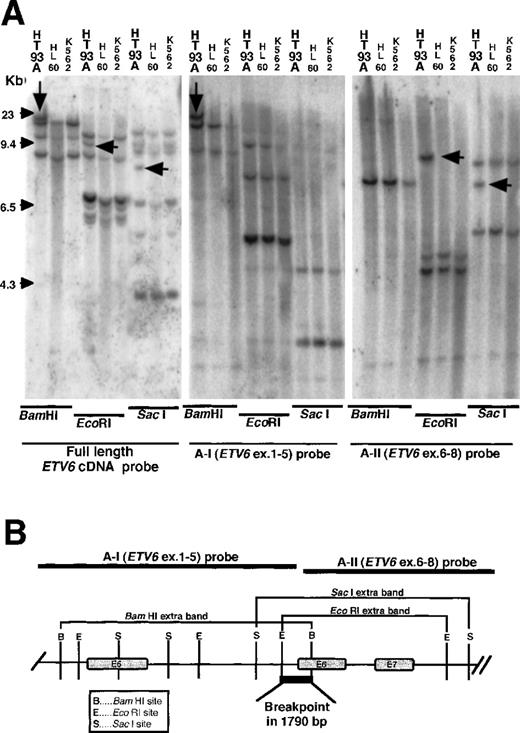
From an additional band detected in each restriction enzyme digest and the restriction map, the breakpoint was speculated between intron 5 and exon 6 of ETV6. Southern blot hybridization using the full length ETV6 cDNA detected a novel band in genomic DNA from the HT93A cells digested with BamHI, EcoRI, andSacI, but not in the DNA from the control cells. The novel band found in the BamHI digest was detected with the A-I probe containing exons 1 to 5 of ETV6, and those found inEcoRI and SacI digests were detected by the A-II probe containing exons 6 to 8 of ETV6 (Figure3A). From the restriction map ofETV6, the breakpoint of ETV6 was speculated to be located within a 1790 bp region between intron 5 and exon 6 ofETV6 (Figure 3B).
Genomic Southern hybridization with the ETV6probes.
(A) With the full length ETV6 cDNA probe, additional bands were found only in the HT93A cells, but not in control cells. The additional bands on Bam HI, EcoRI, and SacI digests were hybridized with A-I probe and A-II probe, respectively. (B) Schematic representation of genomic breakpoint in the HT93A cells. Taking into account the restriction map of ETV6, the breakpoint is deduced to localize in a 1780 bp region that encompasses intron 5 and exon 6 ofETV6.
Genomic Southern hybridization with the ETV6probes.
(A) With the full length ETV6 cDNA probe, additional bands were found only in the HT93A cells, but not in control cells. The additional bands on Bam HI, EcoRI, and SacI digests were hybridized with A-I probe and A-II probe, respectively. (B) Schematic representation of genomic breakpoint in the HT93A cells. Taking into account the restriction map of ETV6, the breakpoint is deduced to localize in a 1780 bp region that encompasses intron 5 and exon 6 ofETV6.
Northern blot hybridization
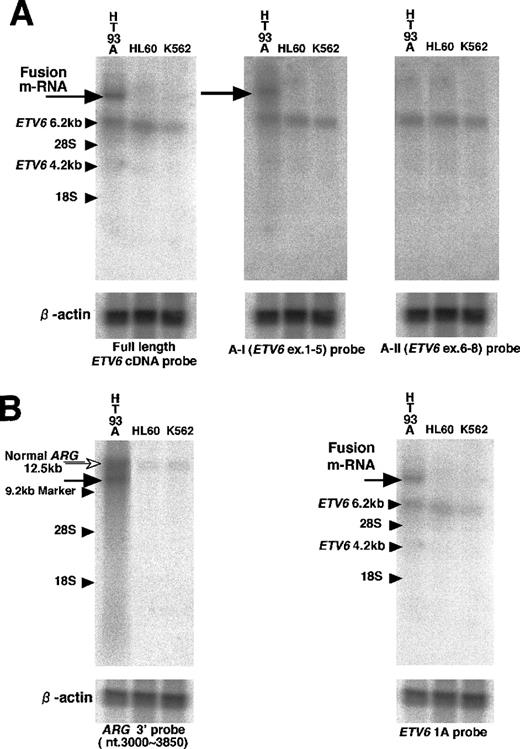
An abnormal transcript band detected in Northern blot hybridization contained exons 1 to 5 of ETV6 and a novel fusion sequence. Northern blot hybridization, using the full length ETV6 cDNA probe, detected an abnormal transcript of more than 10 kb only in the HT93A, which was hybridized with the A-I probe but not with the A-II probe (Figure 4A). This suggested that the abnormal transcript contained exons 1 to 5 of ETV6 and an additional sequence encoded by the unknown fusion partner gene. NormalETV6 transcripts of 2 to 6 kb were also detected in the HT93 A and control cells.
Northern hybridization.
(A) The first hybridization. With full length ETV6 cDNA probe, 6.2 kb transcript (small arrow) was detected in the HT93A, HL60, and K562 cells, but an additional longer transcript (large arrow) was detected only in the HT93A cells. This abnormal transcript was hybridized with A-I (ETV6 exons 1-5) probe but not with A-II (ETV6 exons 6-8) probe. (B) The second hybridization. With theARG 3′ probe, an abnormal transcript (black arrow) was detected only in the HT93A cells at the same position as the abnormalETV6 transcript in the first hybridization. The 12.5 kb normal transcript of ARG (white arrow) was also detected in HT93A cells and control cells. This abnormal band was hybridized only withETV6 exon 1A probe.
Northern hybridization.
(A) The first hybridization. With full length ETV6 cDNA probe, 6.2 kb transcript (small arrow) was detected in the HT93A, HL60, and K562 cells, but an additional longer transcript (large arrow) was detected only in the HT93A cells. This abnormal transcript was hybridized with A-I (ETV6 exons 1-5) probe but not with A-II (ETV6 exons 6-8) probe. (B) The second hybridization. With theARG 3′ probe, an abnormal transcript (black arrow) was detected only in the HT93A cells at the same position as the abnormalETV6 transcript in the first hybridization. The 12.5 kb normal transcript of ARG (white arrow) was also detected in HT93A cells and control cells. This abnormal band was hybridized only withETV6 exon 1A probe.
3′-RACE
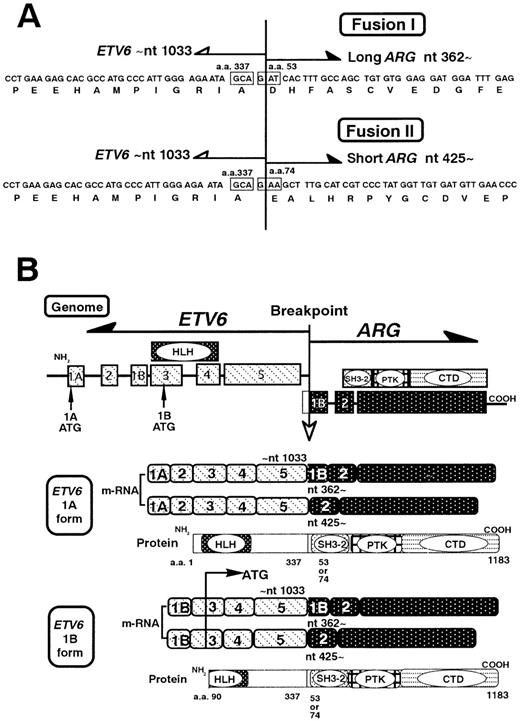
3′-RACE and the BLAST search identified that the novel sequence was ARG. To identify the unknown fusion partner ofETV6, 3′-RACE was performed using primers located in exon 4 and exon 5 of ETV6. Sequence analysis of amplification products detected a novel sequence fused to ETV6. BLAST database searching revealed that the novel sequence was identical toARG, which maps to 1q25. Further sequence analysis revealed 2 types of ETV6/ARG transcripts, 1 containing exon 5 ofETV6 (up to nt 1033) fused to ARG at nt 362 and the other containing exon 5 of ETV6 fused ARG at nt 425 (Figure 5A). The sequence of ETV6exon 5 was fused in-frame to the ARG sequence up to nt 599, and then followed by an Alu repeat sequence, which provided the priming site for the BamTX primer. Careful examination suggested that the Alu sequence was derived from an intron ofARG, suggesting that these 3′-RACE products were amplified from unspliced ARG transcripts. The ARGsequence from nt 362 or 425 to nt 599 is not known to encode any functional domains.
Schematic representation of the ETV6/ARG fusion transcripts.
(A) The nucleotides and deduced amino acid sequence of the ETV6/ARGfusion transcripts. Two fusion breakpoints of the ARG were detected, at nt 362 and nt 425. Both fusions contain the ETV6up to nt 1033, and are in-framed. (B) ETV6 has alternative exon 1A and 1B. Four kinds of the ETV6/ARG transcripts were found. The 1A form ETV6 encodes a complete PNT domain, whereas the 1B form ETV6 encodes a truncated PNT domain, because the translation starts at ATG in exon 3.
Schematic representation of the ETV6/ARG fusion transcripts.
(A) The nucleotides and deduced amino acid sequence of the ETV6/ARGfusion transcripts. Two fusion breakpoints of the ARG were detected, at nt 362 and nt 425. Both fusions contain the ETV6up to nt 1033, and are in-framed. (B) ETV6 has alternative exon 1A and 1B. Four kinds of the ETV6/ARG transcripts were found. The 1A form ETV6 encodes a complete PNT domain, whereas the 1B form ETV6 encodes a truncated PNT domain, because the translation starts at ATG in exon 3.
RT-PCR
Use of RT-PCR detected 4 types of ETV6/ARG transcripts, which contained exons 1 to 5 of ETV6 and the sequence from exon 1B or exon 2 to the end of ARG. To determine whether or not theETV6/ARG transcripts contain the sequences coding functional domains of ARG, we performed RT-PCR with primers inETV6 exon 1A and in the end of ARG, demonstrating 4 kb full length ETV6/ARG transcripts in the HT93A cells. We confirmed that these transcripts contained the sequence from exon 1B or exon 2 to the end of ARG, which encode the Src homology 3 (SH3) domain, SH2 domain, the TK domain of ARG. As forETV6 sequence, we detected 2 types of amino termini of transcripts, 1 containing exon 1A (ETV6 1A form) and the other containing exon 1B (ETV6 1B form). Because exon 1B ofETV6 has no in-frame ATG start codon in a 233 bp sequence, translation starts at in-frame ATG start codon (nt 289-291) in exon 3, resulting in formation of a shortened protein with a truncated PNT domain. To confirm the breakpoint of ETV6 and ARG, we repeated the RT-PCR with E5F as sense primer and A590R as antisense primer using 4kb ETV6/ARG transcript as the template, and detected 2 types of ARG breakpoint, nt 362 and nt 425. Thus, in the HT93A, 4 types of ETV6/ARG transcripts were detected,ETV6 1A/long ARG (nt 362∼), ETV6 1A/shortARG (nt 425∼), ETV6 1B/long ARG (nt 362∼), and ETV6 1B/short ARG (nt 425∼) (Figure 5B). Besides the abnormal ETV6/ARG transcripts, 2 forms of normalETV6 transcripts, the 1A form and 1B form were also detected. The reciprocal ARG/ETV6 transcript was not detected by RT-PCR, suggesting absence or very low level expression of this transcript.
Second Northern blot analysis
The abnormal transcript detected in previous Northern blot analysis contained 1A form ETV6 and ARG sequence. In previous Northern blot analysis, we could detect an abnormal transcript band with full length ETV6 cDNA probe and confirmed that this band was hybridized with A-I (ETV6 exons 1-5) probe. To determine whether this abnormal transcript contains ARG sequence and which isoform of ETV6 is expressed, we did the second Northern blot analysis using the ARG 3′ probe, the ETV6 1A probe, and the ETV6 1B probe. With the ARG 3′ probe, an approximately 10 kb abnormal transcript band was detected only in the HT93A cells at the same position as the abnormalETV6 transcript in the previous hybridization. In the control cells and HT93A cells, the 12.5 kb normal ARG transcript was also detected. Both normal and abnormal transcript of ARG were overexpressed in HT93A cells compared to the control cell lines. Furthermore, the abnormal ARG band was hybridized withETV6 1A probe but not with the ETV6 1B probe (Figure4B). Thus, the abnormal 10 kb transcript contained both ARGsequence and 1A form ETV6 sequence.
Discussion
ARG was first identified as a closely related gene toABL through hybridization with v-ABL probes to human placenta DNA.20 The 2 proteins are structurally quite similar, containing the SH3-SH2 domains, TK domain, and the extensive carboxyl-terminal domain (CTD).21 However, several lines of evidence show that 2 proteins are functionally different. ABLis known to fuse to other genes forming chimeric oncogenes, such asGag/ABL in murine pre-B lymphoma and BCR/ABL in human acute leukemia and chronic myelogenous leukemia. Recently, ABLwas also found to fuse to EVT6.8 15 However,ARG has not fused to any other genes nor been involved in human neoplasms so far. Thus, this is the first report showing ARGinvolvement in a human leukemia through formation of a fusion gene.
The ABL oncoproteins, BCR/ABL and EVT6/ABL, have been demonstrated to transform growth factor-dependent hematopoietic cell lines to factor-independent ones or to transform Rat-1 cells.8,22This property is dependent on the presence of the PNT domain ofETV6 as well as the active TK domain ofABL.8 Recent studies have shown that oligomerization by the coiled-coil motif of BCR or by the PNT domain of ETV6 is essential for constitutive activation of the TK function of the BCR/ABL and EVT6/ABL oncoproteins.8,23In the other ETV6/TK proteins (ETV6/PDGFRß and ETV6/JAK2), oligomerization by the PNT domain of ETV6 has been also demonstrated to lead to constitutive activation of the TKs.6,24 Considering the high structural similarity between ABL and ARG proteins, it is conceivable that this ETV6/ARG protein has transforming activity through the same molecular mechanism shown for the ETV6/ABL protein and other ETV6/TK proteins. However, it should be noted that several functional differences between ABL and ARG protein have been reported. There is a phosphotyrosine-independent interaction between the BCR and the ABL SH2 domain.25 TheARG SH2 domain, however, exhibited 10-fold weaker affinity for BCR than the ABL SH2 domain.25 In another study to determine whether ARG shares the potential oncogenic properties of ABL, Gag/ARG protein unlike Gag/ABL protein showed no transforming activity in the NIH 3T3 focus formation assay, and the transforming activity was recovered by replacement of the CTD of ARG with that ofABL.26 The further study using engineered Gag/ARG/ABL chimeric molecules revealed that the third SH3 domain-binding site in the CTD of ARG had a suppressive effect on NIH 3T3 transformation.26 We have carefully checked the sequence of ARG in the ETV6/ARG transcripts, especially the sequence of the CTD, but have not found any mutation or deletion. The transforming properties of the ETV6/ARG protein should be examined carefully.
Because the PML/RARA transcript was detected in this patient and this cell line and the leukemic cells showed typical morphology of AML-M3, it is likely that PML/RARA protein played a major role and the ETV6/ARG protein played a secondary role in this leukemia. However, the HT93A cells showed the remarkable feature of differentiating into mature eosinophils in culture with ATRA and cytokines, although we do not know whether the patient's leukemic cells had this capacity. Despite the HL60 cells showing multilineage differentiation to neutrophils, macrophages, eosinophils, and basophils, the true AML-M3 cell line having a t(15;17) shows differentiation only to neutrophils and monocytes/macrophages.27 Surprisingly, other investigators have found the same ETV6/ARG transcript in a mature T-ALL cell line with a t(1;10;12) and observed that their cells differentiated into myeloid cells in culture with IL-3 and GM-CSF (F. Griesinger, personal communication, October 29, 1999). Their ETV6/ARG transcripts had the same breakpoint ofETV6 (at nt 1033) and the same 2 breakpoints of ARG (at nt 362 and nt 425). Among ETV6 fusion proteins reported to date, some are involved in both lymphoid and myeloid leukemias.3,6-8 15 The ETV6/ARG protein may be one of them and be associated with the unique differentiation capacity of this cell line. Now, we are examining whether this protein is found in other leukemias using clinical samples with RT-PCR. Further analysis of ETV6/ARG protein would provide further insight into the role of the ETV6 fusion proteins in cell proliferation and differentiation.
Added in proof. While this article was in production, another case with ETV6/ARG fusion and inv(16) was published (Blood. 94:12) as the first report.
Acknowledgments
We thank Dr Janet D. Rowley for kindly providing the YAC964c10 probe, Dr Todd Golub for ETV6 cDNA probe, and Dr Peter Marynen for the LL12NCO1 ETV6 cosmid probes.
Supported in part by a Grant-in-Aid for the Second Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health and Welfare, Japan.
Reprints:Yuko Sato, Division of Molecular Cytogenetics, Department of Clinical Pathology, Research Institute, International Medical Center of Japan, Toyama 1-21-1, Shinjuku-ku, Tokyo, 162-0052, Japan; e-mail: ysato@ri.imcj.go.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal