Abstract
Angiopoietin-1 (Ang-1) is required for developing vessels, and its absence leads to defects in vessel remodeling. Ang-1 has been identified as the ligand for the tyrosine kinase receptor Tie-2, which is expressed specifically on endothelial cells and early hematopoietic cells. In studying the role of Tie-2 and Ang-1 in megakaryocytopoiesis, 3 alternatively spliced species of Ang-1 mRNA (Ang-1.3 kb, Ang-0.9 kb, and Ang-0.7 kb) were identified in addition to the full-length Ang-1 (Ang-1.5 kb), in the megakaryocyte cell line CHRF by reverse transcription–polymerase chain reaction (RT-PCR), and then cloned and sequenced. The expression of 3 alternatively spliced isoforms of Ang-1 was confirmed by RT-PCR using specific primer pairs derived from junction sites and the 3′ end of Ang-1 cDNA, and it was further demonstrated by nuclease protection assay, Northern blotting, and immunoblotting in CHRF cells. Expression of the Ang-1.3 kb isoform was also detected in human primary fibroblast cell line FS4, breast cancer cell line MDAMB-468, and CD34+CD41+ cells of fetal liver and platelets. The function of the 1.5-kb, 1.3-kb, and 0.9-kb isoforms was examined. Recombinant proteins Ang-1.5 and 0.9 kb bind strongly to the recombinant Tie-2 receptor (Tie-2-Fc), whereas the 1.3-kb isoform does not. The Ang-1.3 kb isoform binds to the 1.5-kb isoform. Ang-1.5 kb, but not the 1.3-kb and 0.9-kb isoforms, induces tyrosine phosphorylation of Tie-2 in human umbilical vein endothelial cells. These data suggest that isoforms 1.3 kb and 0.9 kb could serve as dominant negative molecules for the full-length Ang-1. The possible involvement of the newly identified Ang-1 isoforms in angiogenesis and in growth and differentiation of hematopoietic progenitor cells provides a greater complexity to these processes.
Angiogenesis, the process of new growth of blood vessels, is essential for supporting embryonic development, wound healing, and tumor growth. It also plays a role in atherosclerosis, diabetic retinopathy, and thrombosis. Various growth factors, including fibroblast growth factor (FGF) and vascular endothelial growth factor (VEGF) have been implicated in the regulation of vessel formation. Although FGF is a mitogen for a wide range of cell types, VEGF is a mitogen for vascular endothelial cells,1 2 and it appears to play an important role in regulating the function of endothelial cells in normal and disease states.
Recently, angiopoietin-1 (Ang-1), a ligand for the Tie-2 receptor, was isolated by secretion-trap expression cloning.3 Ang-1 signals through a tyrosine kinase receptor (Tie-2/Tek) that is expressed specifically on endothelial cells and early hematopoietic cells.4-7 Mice engineered to lack Ang-1 or its receptor express severe vascular abnormalities during embryogenesis.8 Unlike VEGF, Ang-1 is incapable of mediating endothelial cell growth or tubule formation. However, in the absence of Ang-1, endothelial cells are poorly associated with the underlying matrix and do not properly recruit and associate with periendothelial supporting cells.3,8 Ang-1 has been shown to be able to induce the migration and sprouting of endothelial cells,9,10 and the overexpression of Ang-1 in transgenic mice induces increased vascularization.11 Thus, Ang-1 appears to be involved in a later stage of vessel development. Angiopoietin-2, a natural antagonist for the Tie-2 receptor that disrupts in vivo angiogenesis, has recently been described.12 Both Ang-1 and Ang-2 could modulate VEGF-induced postnatal neovascularization.13 14 Thus it is likely that angiogenic processes depend on the sequential and cooperative effects of the members of VEGF and angiopoietin families.
The endothelial and hematopoietic lineages are closely linked. Several receptor tyrosine kinase molecules are expressed on both lineage cells, including Flk-1, Flt-1, Tie-1, and Tie-2. Tie receptor kinases are of special interest because they are expressed in very early stages of hematopoietic and endothelial cell development.4-7 Indeed, Tie-1 was first isolated from a human chronic myeloid leukemia cell line K562, and its expression was up-regulated during in vitro megakaryoblastoid differentiation of certain leukemia cell lines.15-17 Tie-1 expression was also found in vivo in megakaryocytes, hematopoietic stem cells, and some B cells of adult bone marrow or umbilical cord blood cells.18,19 Tie-2 is also expressed in bone marrow hematopoietic stem cells in a subset of CD34+ or c-KIT+ cells.20
In studying the role of Tie-2 and its ligand, Ang-1, in megakaryocytopoiesis, we discovered the presence of 4 Ang-1 isoforms in the megakaryocyte cell line, CHRF. In this article we report on their cloning, sequencing, and function (ability to bind and phosphorylate Tie-2), and we provide suggestive evidence that 2 of these isoforms may act as dominant-negative inhibitors.
Materials and methods
Cell culture
CHRF-288 cells were a gift from Dr M. Lieberman, University of Cincinnati Medical Center, and were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and 1% penicillin and streptomycin. Human breast cell lines MDA-MB468, MDA-MB231, and MCF7, and prostate cancer cell line DU145 were obtained from the American Type Culture Collection (Rockville, MD) and grown in Dulbecco's modified essential medium (DMEM) containing 10% fetal calf serum and antibiotics. Human diploid primary culture fibroblasts (FS4) were kindly provided by Dr J. Vilcek (New York University Medical Center, New York, NY) and cultured in DMEM with 10% fetal calf serum and antibiotics. Human umbilical vein endothelial cells were derived from fresh umbilical cord blood. Normal human tissue—fetal liver, cord blood, skin, breast, lung, stomach, kidney—were obtained from discarded surgical pathology specimens. CD34+ cells were isolated by low-density centrifugation, monoclonal antibody isolation with super paramagnetic particles (Miltenyi Biotec GmbH, Sunnyvale, CA) and fluorescence-activated cell sorting as previously described.21
Reverse transcription–polymerase chain reaction
Expression of Ang-1 and Ang-2 was determined by RT-PCR. Total RNA was extracted using the acid guanidinium thiocyanate-phenol-chloroform method. cDNA was generated with 1 μg RNA, 200 μm MuLV reverse transcriptase, 20 μm RNAsin, 0.5 mmol/L each of the deoxynucleotide triphosphates (dNTPs), and 100 pmol random hexamer and buffer (Cetus, Norwalk, CT). The reaction mixture was incubated at 42°C for 30 minutes. One tenth of synthesized cDNA was used as a template in the PCR reaction containing 2.5 μm Taq DNA polymerase, 50 mmol/L each dNTP, 8% (vol/vol) dimethyl sulfoxide, PCR buffer provided by the manufacturer (Cetus), and 50 pm primers. Amplification cycles consisted of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 minute. RT-PCR products were analyzed in a 2% agarose gel.
Primers for reverse transcription–polymerase chain reaction
A series of RT-PCR experiments was conducted using pairs of oligonucleotide primers corresponding to different regions and isoforms of the Ang-1 sequences. The detection of Ang-1 expression was performed using sense primer 5′-GGAAGTCTAGATTTCCAAAGAGGC-3′ and antisense primer 5′-CTTTATCCCATTCAGTTTTCCATG-3′, corresponding to sequences 1306-1326 bp and 1711-1734 bp, respectively, designed to give a 429-bp product numbered according to the Ang-1 cDNA sequence submitted to GenBank, accession number U83 508. Amplification of the full-length coding region of Ang-1 was generated with sense primer 5′-GCTGGCAGTACAATGACAGGT-3′ (identical to 5′ end) and antisense primer 5′-TCAAAAATCTAAAGGTCGAAT-3′ (complementary to 3′ end). Two sense primers designed to be located within the junction sites of alternatively spliced mRNA were 5′-GGAATATAAAATGGTTGTATTTAA-3′ (specific for Ang-1.3 kb), and 5′-GTGGCTGCAAAAAGTGTTTTGC-3′ (specific for Ang-0.9 kb). The sense primer specific for Ang-1.3 kb was paired with an antisense primer complementary to the 3′ end of Ang-1 cDNA to give a 312-bp product (for detection of the 1.3-kb isoform). The sense primer specific for Ang-0.9 kb was paired with an antisense primer 5′-ATCGCTTCTGACATTGCGCTT-3′ (1807-1827 bp of the Ang-1 sequence) to give an amplification fragment of 595 bp. Primers used for Ang-2 were 5′-GGCAGCGTTGATTTTCAGAGGACT-3′ and 5′-TTTAATGCCGTTGAACTTATTTGT-3′, corresponding to 1340-1363 bp and 1746-1769 bp of the published sequence.
Cloning and sequencing analysis
cDNA for individual Ang-1 mRNA species was obtained by RT-PCR using 2 primers derived from either the 5′ or the 3′ end of the translated sequences of human Ang-1. The amplified product was fractionated on an agarose gel. Several DNA fragments, ranging in size from 0.7 to 1.5 kb, were observed. These RT-PCR products were cloned into a pGEM-T Easy vector (Promega, Madison, WI). The cloned cDNA was then sequenced with T7 and SP6 sequencing primers and with Ang-1-specific primers. Sequencing data were compared with the published Ang-1 sequence. Isoform sizes were calculated from the sequence data. In some experiments, specific bands from RT-PCR products were excised. The DNA was then eluted, cloned, and sequenced.
Northern blot analysis
The polyadenylated mRNA fraction was isolated by binding to oligo(dT) cellulose, fractionated by electrophoresis on 1% agarose gel in 6.7% formaldehyde, and transferred to a Genescreen Plus nylon membrane (NEN Life Science Products, Boston, MA). The membrane was hybridized at 42°C overnight with a 32P-dCTP-labeled probe—either the 1.5-kb full-length Ang-1, an approximately 0.5-kb alternatively spliced fragment of Ang-1 absent in the 0.9-kb and 0.7-kb isoforms, or a 0.17-kb alternatively spliced fragment of Ang-1 absent in the 1.3-kb and 0.7-kb isoforms. Blots were sequentially washed with varying dilutions of SSC, the last 0.1 × SSC at 65°C for 30 minutes. Autoradiography was carried out at −70°C with intensifying screen.
Nuclease protection assays
cDNA fragments derived from alternatively spliced sites of the 1.3- or 0.9-kb isoform were cloned into pGEM-T vector (Promega) to generate probes 1 and 2. Antisense probes were made using T7 polymerase and α-32p-UTP. Total RNA (20 μg) extracted from CHRF cells or DU145 cells was used for solution hybridization according to the manufacturer's protocol (Ambion, Austin, TX). Addition of hybridization probe 1 to detect the 1.3-kb isoform, followed by RNase digestion, gave a protected band of 262 bp, whereas addition of hybridization probe 1 to detect isoforms 1.5 or 0.9 kb, followed by RNase digestion, gave bands of 219 and 43 bp. Hybridization probe 2 protected the 0.9-kb isoform from RNase digestion, resulting in a 249-bp band, and protected the 1.5-kb and 1.3-kb isoforms from RNase digestion, resulting in 146-bp and 103-bp bands. See “Results” for detailed strategy.
Immunoblotting
Cells were washed twice with phosphate-buffered saline and lysed in lysis buffer (1% Triton X-100, 150 mmol/L NaCl, 10 mmol/L Tris, pH 7.4, 50 μg/mL pepstatin, 20 μg/mL aprotinin, 0.2 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L EDTA, 0.2 mmol/L sodium orthovanadate, and 0.5% NP-40). One hundred micrograms protein, determined by Bio-Rad protein assay, was run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a membrane, incubated with blocking solution (2% powdered milk, 0.2% Tween 20 in phosphate-buffered saline), and reacted with a specific rabbit anti-Ang-1 antibody, kindly provided by Dr George Yancopoulos (Regeneron Pharmaceuticals, NY) or with a goat anti-Ang-1 antibody specific for the N-terminal end (reactive with anti-1.5 kb and anti-1.3 kb isoforms) or C-terminal end (reactive with 1.5-kb isoform but not 1.3-kb isoform) supplied by Santa Cruz Biotechnology (Santa Cruz, CA). After washing, a peroxidase-conjugated second antibody was applied and chemiluminescence generated by incubation with ECL reagents (Amersham Life Science).
Protein isoform expression and purification
cDNA corresponding to the full-length Ang-1 and various isoforms (without signal peptide) was cloned in frame into the pGE × 4T1 vector (Pharmacia) and then transformed to BL21Escherichia coli. Recombinant GST-Ang-1 fusion proteins were purified from bacterial lysates by affinity chromatography on a glutathione Sepharose 4B column (Pharmacia).
Each cDNA isoform was also cloned to pCDNA3 (Invitrogen). To prepare 35S-methionine-labeled Ang-1 isoforms, the plasmid (1 μg each isoform) was transcribed and translated in vitro using the coupled TNT transcription/translation ribosomal system (Promega). The same plasmid was used for transfection of COS cells.
Pull-down assay for isoform-isoform binding
Two different methods were used. In the first method,35S-radiolabeled Ang-1 isoforms 1.3 kb, 0.9 kb, and control protein luciferase were prepared by transcription/translation in vitro and incubated with the GST-Ang-1.5-kb fusion protein in binding buffer (10 mmol/L Tris, pH 7.6, 100 mmol/L NaCl, 0.1 mmol/L EDTA, 1 mmol/L dithiothreitol, 1% nonfat dry milk, 5 mmol/L MgCl2, 0.05% NP40, and 8% glycerol) at 4°C for 60 minutes. The materials were then incubated with glutathione Sepharose 4B beads for 30 minutes and were shaken at 4°C. After washing with binding buffer, the proteins bound to the beads were solubilized in sample lysis buffer, resolved on 12% SDS-PAGE, and exposed to x-ray film. In the second method, COS cells were transfected with a mixture of 1.5-kb and 1.3-kb isoforms or the 1.5-kb isoform alone. Cells were lysed with sample lysis buffer, and isoforms were precipitated with an anti-Ang-1 antibody reactive with the 1.5-kb isoform, not the 1.3-kb isoform (C-terminal antibody) plus or minus protein G beads. Samples were then analyzed by Western blot with an anti-Ang-1 antibody reactive with both the 1.5- and 1.3-kb isoforms (N-terminal antibody). The specificity of the antibodies was verified independently by Western blot against the specific isoforms.
Ligand–receptor binding assay
Binding of the Ang-1 isoforms to the Tie-2 receptor was also evaluated by ligand–receptor precipitation in the presence of 5% bovine serum albumin. In vitro ribosome-translated35S-labeled Ang-1 isoforms were incubated with recombinant Tie-2-Fc protein, followed by precipitation with protein G beads. After washing, the proteins bound to the resin were solubilized in sample lysis buffer and resolved on 12% SDS-PAGE followed by autoradiography.
Tyrosine phosphorylation assay
Human umbilical vein endothelial cells (HUVEC) were plated in T-75 flasks, cultured to confluence, and serum starved for 4 hours. Conditioned media, collected from COS cells transiently transfected with various plasmids of Ang-1 isoforms, were placed on HUVEC for 10 minutes. Cells were then solubilized with lysis buffer (1% Triton X-100, 10 mmol/L sodium pyrophosphate, 100 mmol/L sodium fluoride, 4 mmol/L EDTA, 2 mmol/L sodium orthovanadate, 1 mmol/L phenylmethylsulfonyl fluoride, 100 mmol/L leupeptin, and 50 mg/mL aprotinin). Tie-2 protein was immunoprecipitated from the cell lysates with a goat anti-Tie-2 antibody (1 μg/mL) and autophosphorylation of tyrosine residues was evaluated with antiphosphotyrosine mAb 4G10 (Upstate Biotechnology, NY) by Western blotting, followed by chemiluminescence using ECL (Amersham).
Results
Detection of Ang-1 expression in human cells by RT-PCR
The expression of Ang-1 mRNA was analyzed in several tumor cell lines and normal tissues. Positive signals were obtained from RNA of CHRF, MDA-MB468, and MDA-MB231 human tumor cell lines and from human FS4 fibroblasts, fetal liver CD34+CD41+ cells, and cord blood CD34+ cells and platelets, whereas none were obtained from the RNA of MCF7 and DU145 human tumor cell lines and several normal human tissue specimens (Table1, Figure 1). Ang-2 was not expressed in the cell lines studied (data not shown).
Presence of angiopoietin isoforms 1.5, 1.3, and 0.9 kb in various cell lines and tissues
| . | 1.5 kb . | 1.3 kb . | 0.9 kb . |
|---|---|---|---|
| CHRF*,† | + | + | + |
| Fetal liver CD34+ CD41+ | + | + | ND |
| Platelets† | + | + | + |
| Cord blood CD34+ | + | − | − |
| FS4* | + | + | − |
| MDAMB468* | + | + | − |
| MDAMB231 | + | − | − |
| MCF7 | − | − | − |
| DU145 | − | − | − |
| Skin | − | − | − |
| Breast | − | − | − |
| Lung | − | − | − |
| Stomach | − | − | − |
| Kidney | − | − | − |
| . | 1.5 kb . | 1.3 kb . | 0.9 kb . |
|---|---|---|---|
| CHRF*,† | + | + | + |
| Fetal liver CD34+ CD41+ | + | + | ND |
| Platelets† | + | + | + |
| Cord blood CD34+ | + | − | − |
| FS4* | + | + | − |
| MDAMB468* | + | + | − |
| MDAMB231 | + | − | − |
| MCF7 | − | − | − |
| DU145 | − | − | − |
| Skin | − | − | − |
| Breast | − | − | − |
| Lung | − | − | − |
| Stomach | − | − | − |
| Kidney | − | − | − |
Plus or minus signs refer to the presence or absence by RT-PCR, using specific primers. All reported values were positive for the internal control, -actin.
Positive by Northern blot analysis.
Positive by immunoblot.
ND, not done.
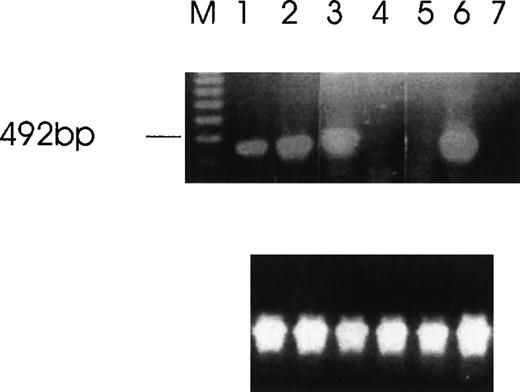
Detection of Ang-1 mRNA expression by RT-PCR with primers designed to give a 429-bp product.
Ethidium bromide-stained agarose gel with RT-PCR products from MDA-MB231 cells (lane 1), MDA-MB468 (lane 2), FS4 (lane 3), MCF7 (lane 4), DU145 (lane 5), CHRF (lane 6), and negative control (absence of cDNA template) (lane 7). The amplification of β-actin was used as an internal control (lower panel).
Detection of Ang-1 mRNA expression by RT-PCR with primers designed to give a 429-bp product.
Ethidium bromide-stained agarose gel with RT-PCR products from MDA-MB231 cells (lane 1), MDA-MB468 (lane 2), FS4 (lane 3), MCF7 (lane 4), DU145 (lane 5), CHRF (lane 6), and negative control (absence of cDNA template) (lane 7). The amplification of β-actin was used as an internal control (lower panel).
Alternatively spliced species of Ang-1 mRNA identified by RT-PCR, RNase protection, Northern blot analysis, and immunoblot
Using RT-PCR to analyze the expression of Ang-1 mRNA in CHRF cells, we noted an extra band migrating at approximately 262 bp with ethidium bromide staining, in addition to the 429 bp-band predicted because of the pair of primers used (Figure2). Another extra band was observed in FS4 cells and MDA-MB468 cells (see below). The extra band was excised from the gel, and the DNA was eluted, cloned, and sequenced. Sequencing indicated that the 262-bp band represented a fragment of an alternatively spliced form of Ang-1 mRNA, later identified as the 1.3-kb isoform. To examine further the expression of multiple isoforms of the Ang-1 coding region in human cells, a pair of primers designed to span the coding region of the Ang-1 gene was applied for RT-PCR analysis using RNA extracted from CHRF cells. Several amplified products were detected. After examining 50 clones and selecting 8 for sequencing, 3 alternative transcripts (Ang-1.3 kb, Ang-0.9 kb, and Ang-0.7 kb) were identified, as was the published Ang-1.5 kb sequence for the coding region (Figures 3 and4). To confirm the presence of these transcripts, new sets of primers were designed with the 5′ end straddling the apparent splice junction site. A set of primers, used to detect the 1.3-kb isoform and designed to give a 312-bp product, is shown in Figure 5A. This isoform was found in FS4, MDA-MB468, and CHRF cells, not in DU145, MDA-MB231, and MCF7 cells (Table 1). The primer pair used to detect the 0.9-kb isoform and designed to give a 595-bp product is shown in CHRF cells (Figure 5B). Also shown is a 428-bp product that represents a double alternatively spliced isoform, Ang-0.7 kb. Neither of these isoforms was found in MDA-MB468, MDA-MB231, or FS4 cells (data not shown).
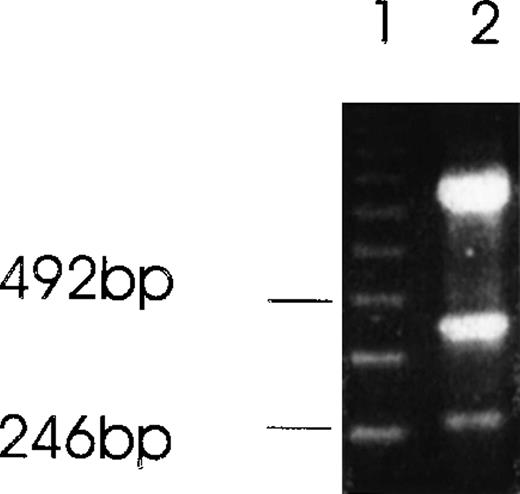
Alternatively spliced species of Ang-1 mRNA in CHRF cells detected by RT-PCR analysis.
Two isoforms of Ang-1 can be detected simultaneously that give rise to single bands at 429 bp and 262 bp (lane 2), representing full-length Ang-1.5 kb and an alternatively spliced 1.3-kb isoform, respectively. The top band is β-actin.
Alternatively spliced species of Ang-1 mRNA in CHRF cells detected by RT-PCR analysis.
Two isoforms of Ang-1 can be detected simultaneously that give rise to single bands at 429 bp and 262 bp (lane 2), representing full-length Ang-1.5 kb and an alternatively spliced 1.3-kb isoform, respectively. The top band is β-actin.
Nucleotide and amino acid sequences of the coding region of the 4 Ang-1 isoforms.
Colinear sequence alignment of Ang-1 cDNA (Ang-1.5 kb) with isoforms Ang-1.3 kb, Ang-0.9 kb, and Ang-0.7 kb cDNA. The amino acid sequence of Ang-1.5 kb is shown above the nucleotide sequence. Dotted lines indicate the spliced sequences. Amino acid and nucleotide residues are numbered to the right of each sequence. The termination codon is indicated by an asterisk.
Nucleotide and amino acid sequences of the coding region of the 4 Ang-1 isoforms.
Colinear sequence alignment of Ang-1 cDNA (Ang-1.5 kb) with isoforms Ang-1.3 kb, Ang-0.9 kb, and Ang-0.7 kb cDNA. The amino acid sequence of Ang-1.5 kb is shown above the nucleotide sequence. Dotted lines indicate the spliced sequences. Amino acid and nucleotide residues are numbered to the right of each sequence. The termination codon is indicated by an asterisk.
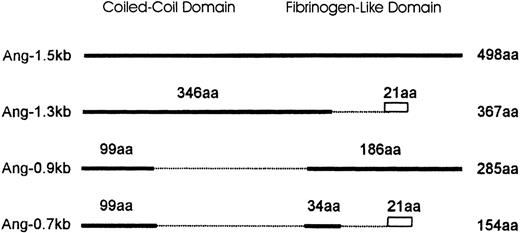
Schematic diagram comparing the isoforms of Ang-1.
Ang-1.5 kb encodes the full-length Ang-1 of 498 amino acids (aa). Isoforms are represented by solid lines. Numbers above the solid line indicate the amino acids identical to the full-length isoform. Amino acids truncated are shown as broken lines. The divergence of predicted amino acids generated by splicing events in Ang-1.3 kb and Ang-0.7 kb (VVFKRSHWDSRKTEQPDLTRC) is shown as a rectangle. The length of each isoform is numbered to the right.
Schematic diagram comparing the isoforms of Ang-1.
Ang-1.5 kb encodes the full-length Ang-1 of 498 amino acids (aa). Isoforms are represented by solid lines. Numbers above the solid line indicate the amino acids identical to the full-length isoform. Amino acids truncated are shown as broken lines. The divergence of predicted amino acids generated by splicing events in Ang-1.3 kb and Ang-0.7 kb (VVFKRSHWDSRKTEQPDLTRC) is shown as a rectangle. The length of each isoform is numbered to the right.
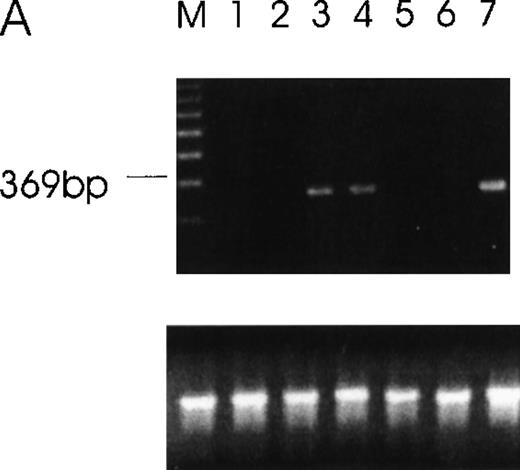
RT-PCR of tumor cells with primers designed to detect specific isoforms using the 5′ primer to straddle the splice junction site.
(A) An alternatively spliced species of Ang-1 mRNA in cells, determined by RT-PCR with a set of primers used to detect Ang-1 isoform 1.3 kb and designed to detect a 312-bp product. Lane 1, negative control without cDNA template; lane 2, DU145; lane 3, FS4; lane 4, MDA-MB468; lane 5, MDA-MB231; lane 6, MCF7; lane 7, CHRF. (B) Primers designed to detect the 0.9- and 0.7-kb isoforms, which give rise to corresponding bands 595 bp and 428 bp, respectively, in CHRF cells.
RT-PCR of tumor cells with primers designed to detect specific isoforms using the 5′ primer to straddle the splice junction site.
(A) An alternatively spliced species of Ang-1 mRNA in cells, determined by RT-PCR with a set of primers used to detect Ang-1 isoform 1.3 kb and designed to detect a 312-bp product. Lane 1, negative control without cDNA template; lane 2, DU145; lane 3, FS4; lane 4, MDA-MB468; lane 5, MDA-MB231; lane 6, MCF7; lane 7, CHRF. (B) Primers designed to detect the 0.9- and 0.7-kb isoforms, which give rise to corresponding bands 595 bp and 428 bp, respectively, in CHRF cells.
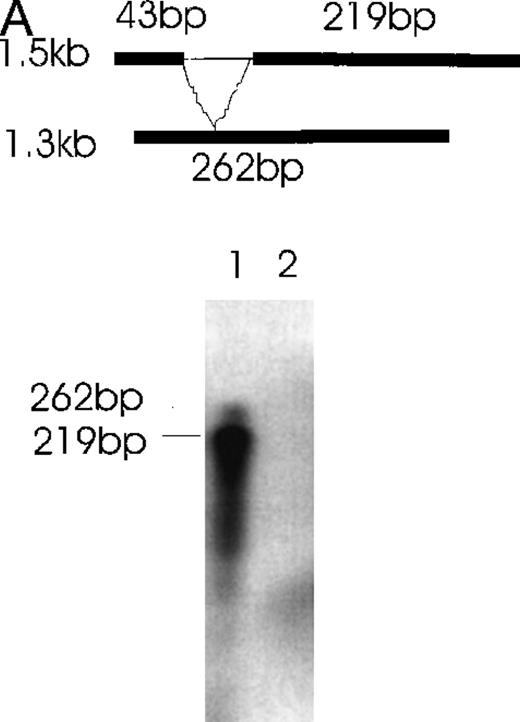
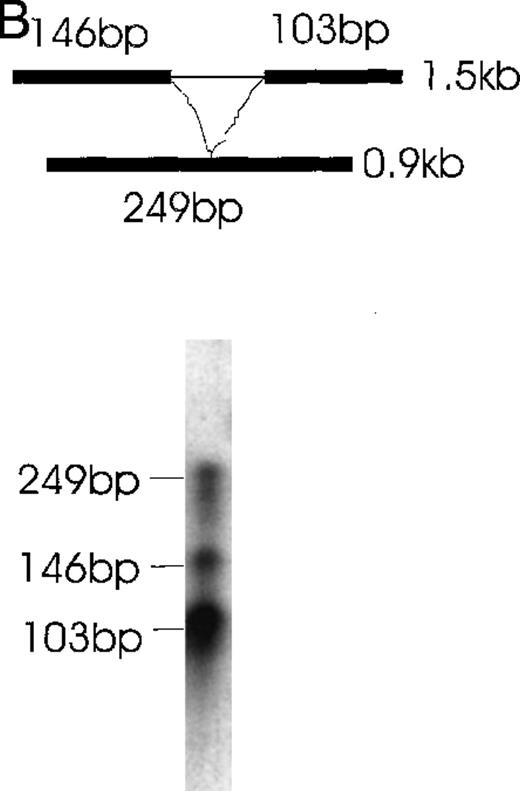
Further confirmation was obtained by the RNase protection assay (Figure6). Alternatively spliced mRNA species of 1.3 kb (Figure 6A) and 0.9 kb (Figure 6B) were clearly detected by designed probes that gave the predicted 262-bp band for the 1.3-kb isoform and the predicted 249-kb band for the 0.9-kb isoform after RNase digestion. In Figure 6A, the 219-bp band and the 43-bp band (not seen) represent the 1.5- and 0.9-kb isoforms. In Figure6B, the 146- and 103-kb bands represent the 1.5- and 1.3-kb isoforms.
Simultaneous detection of the alternatively spliced mRNA species of Ang-1 in CHRF cells by RNase protection assay.
Schematic diagrams of the strategy for determining the alternatively spliced mRNA of Ang-1 are shown on the upper panels of A and B. The thick lines represent fragments derived from various forms of Ang-1 that could be detected by this assay. The alternatively spliced parts of Ang-1 are shown as thin lines. (A) The designed 262-bp antisense probe is derived from the 1.3-kb isoform. Lane 1 shows the RNase-protected 262-bp product, indicating the presence of the 1.3-kb isoform and the 219-bp product representing the 1.5-kb and 0.9-kb isoforms. The 43-bp fragment (not shown) is small and seen only after a longer time exposure. Lane 2 shows the RNA extracted from DU145 cells as a negative control. (B) The designed 249-bp antisense probe is derived from the 0.9-kb isoform. The detected 249-bp band indicates the presence of the 0.9-kb isoform. The 146-bp and 103-bp bands represent the 1.5-kb or 1.3-kb isoforms.
Simultaneous detection of the alternatively spliced mRNA species of Ang-1 in CHRF cells by RNase protection assay.
Schematic diagrams of the strategy for determining the alternatively spliced mRNA of Ang-1 are shown on the upper panels of A and B. The thick lines represent fragments derived from various forms of Ang-1 that could be detected by this assay. The alternatively spliced parts of Ang-1 are shown as thin lines. (A) The designed 262-bp antisense probe is derived from the 1.3-kb isoform. Lane 1 shows the RNase-protected 262-bp product, indicating the presence of the 1.3-kb isoform and the 219-bp product representing the 1.5-kb and 0.9-kb isoforms. The 43-bp fragment (not shown) is small and seen only after a longer time exposure. Lane 2 shows the RNA extracted from DU145 cells as a negative control. (B) The designed 249-bp antisense probe is derived from the 0.9-kb isoform. The detected 249-bp band indicates the presence of the 0.9-kb isoform. The 146-bp and 103-bp bands represent the 1.5-kb or 1.3-kb isoforms.
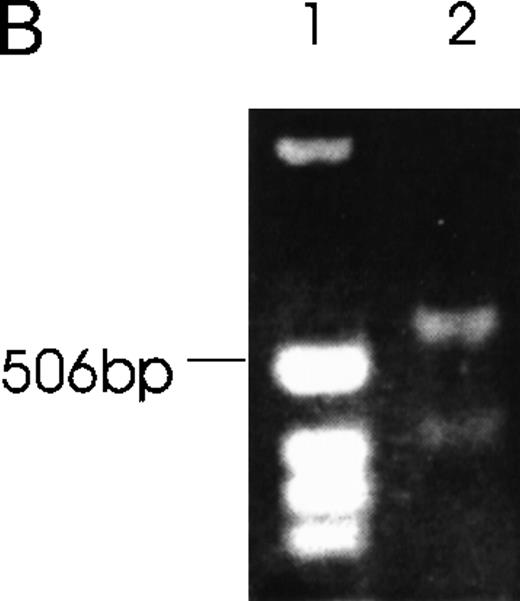
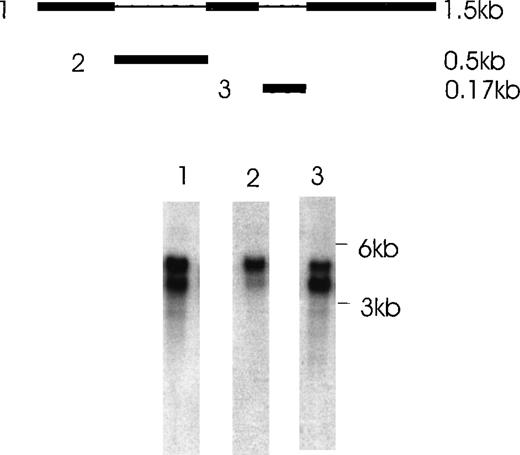
The presence of various isoforms of Ang-1 was also verified by Northern blot analysis of poly A mRNA extracted from CHRF cells. Using the entire coding region 1.5-kb probe (probe 1), 2 positive bands were detected. The upper band was slightly stronger than the lower band, which probably represented the 4 overlapping isoforms—2 merged bands consisting of a 1.5- and 1.3-kb overlap and a 0.9- and 0.7-kb overlap (Figure 7, lane 1). Lane 2 used probe 2, which did not detect the 0.9- and 0.7-kb isoforms but detected the 1.5- and 1.3-kb overlapped isoforms. Lane 3 used probe 3, which detected the 1.5- and 0.9-kb isoforms. Western immunoblot analysis revealed 2 bands that probably represented the 1.5-kb and 0.9-kb isoforms, as calculated from their predicted molecular weights (plus carbohydrate contribution) (Figure 8).
Northern blot analysis of Ang-1 transcripts in CHRF cells.
Polyadenylated RNA (2 μg) was fractionated on a 1% agarose gel and transferred to nylon membranes. Membranes were hybridized with either probe 1,1.5 kb full-length Ang-1 or probe 2 (approximately 0.5 kb of the alternatively spliced part of Ang-1, which is absent in the 0.9-kb and 0.7-kb isoforms), or probe 3 (0.17 kb of the alternatively spliced part of Ang-1, which is absent in the 1.3-kb and 0.7-kb isoforms). A schematic diagram of the probes is shown in the upper panel. The thin lines represent the alternatively spliced parts of Ang-1 that gave rise to the 1.3-, 0.9-, and 0.7-kb isoforms. Lane 1, result from hybridization with probe 1, which detected all 4 mRNA isoforms; lane 2, hybridization with probe 2, which detected the 1.5-kb and 1.3-kb isoforms; lane 3, hybridization with probe 3, which detected the 1.5-kb and 0.9-kb isoforms. Sizes of RNA markers are given on the right.
Northern blot analysis of Ang-1 transcripts in CHRF cells.
Polyadenylated RNA (2 μg) was fractionated on a 1% agarose gel and transferred to nylon membranes. Membranes were hybridized with either probe 1,1.5 kb full-length Ang-1 or probe 2 (approximately 0.5 kb of the alternatively spliced part of Ang-1, which is absent in the 0.9-kb and 0.7-kb isoforms), or probe 3 (0.17 kb of the alternatively spliced part of Ang-1, which is absent in the 1.3-kb and 0.7-kb isoforms). A schematic diagram of the probes is shown in the upper panel. The thin lines represent the alternatively spliced parts of Ang-1 that gave rise to the 1.3-, 0.9-, and 0.7-kb isoforms. Lane 1, result from hybridization with probe 1, which detected all 4 mRNA isoforms; lane 2, hybridization with probe 2, which detected the 1.5-kb and 1.3-kb isoforms; lane 3, hybridization with probe 3, which detected the 1.5-kb and 0.9-kb isoforms. Sizes of RNA markers are given on the right.
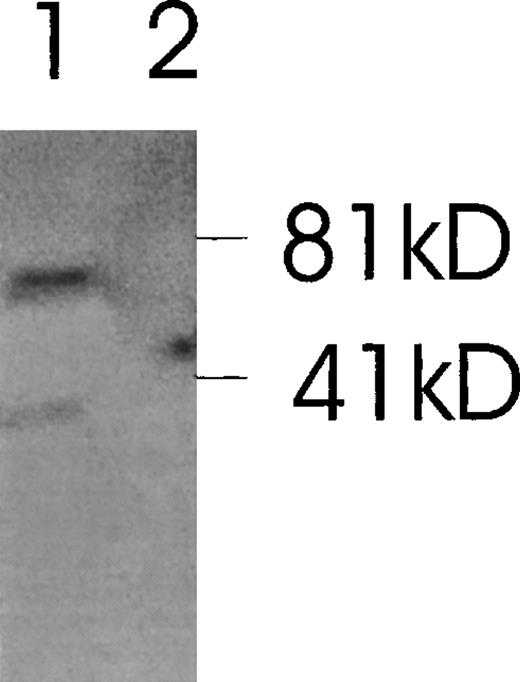
Western immunoblot of 2 Ang-1 isoforms.
CHRF cells were lysed, electrophoresed in 12% SDS-PAGE, transferred to nylon membranes, and immunoblotted with rabbit anti-Ang-1. Lane 1, CHRF cells; lane 2, DU145 cells. Protein marker sizes are shown on the right.
Western immunoblot of 2 Ang-1 isoforms.
CHRF cells were lysed, electrophoresed in 12% SDS-PAGE, transferred to nylon membranes, and immunoblotted with rabbit anti-Ang-1. Lane 1, CHRF cells; lane 2, DU145 cells. Protein marker sizes are shown on the right.
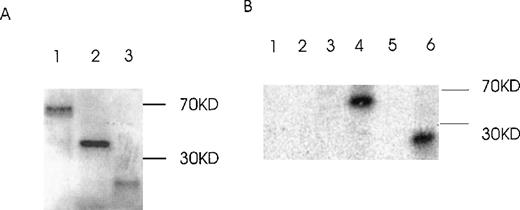
Ang-1.5 kb and Ang-0.9 kb isoforms bind to the Tie-2 receptor
The capability of Ang-1 isoforms 1.5-, 1.3-, and 0.9-kb to bind to the Tie-2 receptor was examined by ligand-receptor precipitation using in vitro translated 35S-labeled isoforms. Figure9A demonstrates in vitro translated isoforms 1.5, 1.3, and 0.9 kb. Figure 9B demonstrates binding of the 1.5- and 0.9-kb isoforms, with absent binding of the 1.3-kb isoform. The absent binding of the 1.3-kb isoform to Tie-2 is compatible with its protein structure, which predicts absent binding.
Binding of Ang-1 isoforms to Tie-2 by ligand–receptor precipitation.
(A) In vitro translated radioactive proteins of isoform 1.5 kb (lane 1), 1.3 kb (lane 2), and 0.9 kb (lane 3) are shown. (B) In vitro translated proteins were incubated with Tie-2-Fc, 1 μg/mL, and precipitated with protein G beads. After washing and elution, samples were discriminated by SDS-PAGE plus autoradiography. Lane 1, 2, and 3 (1.5-, 1.3-, and 0.9-kb isoforms, respectively, with protein G beads alone). Lanes 4, 5, and 6 (1.5-, 1.3-, and 0.9-kb isoforms, respectively, with Tie-2-Fc plus protein G beads. No immunoprecipitation was noted with the 1.3-kb isoform (lane 5) or with control 35S-luciferase (Tie-2-Fc plus protein G beads; data not shown).
Binding of Ang-1 isoforms to Tie-2 by ligand–receptor precipitation.
(A) In vitro translated radioactive proteins of isoform 1.5 kb (lane 1), 1.3 kb (lane 2), and 0.9 kb (lane 3) are shown. (B) In vitro translated proteins were incubated with Tie-2-Fc, 1 μg/mL, and precipitated with protein G beads. After washing and elution, samples were discriminated by SDS-PAGE plus autoradiography. Lane 1, 2, and 3 (1.5-, 1.3-, and 0.9-kb isoforms, respectively, with protein G beads alone). Lanes 4, 5, and 6 (1.5-, 1.3-, and 0.9-kb isoforms, respectively, with Tie-2-Fc plus protein G beads. No immunoprecipitation was noted with the 1.3-kb isoform (lane 5) or with control 35S-luciferase (Tie-2-Fc plus protein G beads; data not shown).
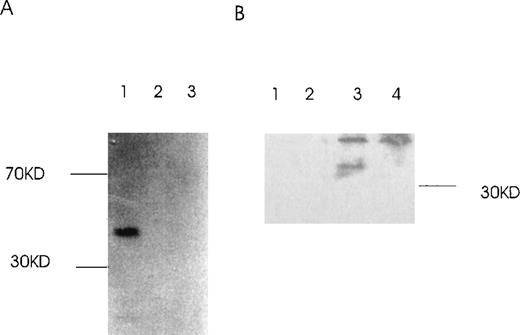
Ang-1.3 kb isoform, but not 0.9 kb-isoform, binds to the 1.5-kb isoform
Because the coiled-coil domain of Ang-1 could be involved in protein multimerization, we examined whether the 1.5-kb isoform could bind to the 1.3- and 0.9-kb isoforms. GST–Ang-1 isoform fusion proteins were constructed and purified (Figure 10). Two different pull-down approaches were used, 1 with glutathione Sepharose 4B beads to pull down GST–Ang-1.5-kb fusion protein with35S-labeled 1.3 kb or 0.9 kb in the mixture (Figure11A), the other with antibody reactive with the 1.5-kb, not the 1.3-kb, isoform and a mixture of cos-transfected 1.5- and 1.3-kb isoforms (Figure 11B). The results indicated that only the 1.3-kb, not the 0.9-kb, protein binds to the 1.5-kb isoform. These data suggested that the 1.3-kb isoform can form multimers with the 1.5-kb isoform.
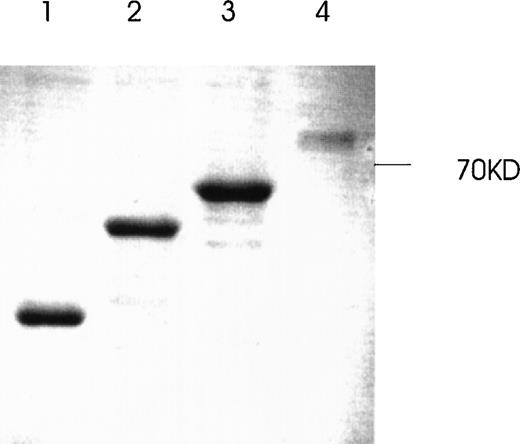
Production and purification of recombinant GST-fusion proteins of Ang-1 isoforms.
Recombinant proteins were purified from bacterial lysates by glutathione Sepharose 4B chromatography and analyzed by SDS-PAGE followed by Coomassie blue staining. The expected molecular weights of fusion proteins were obtained and purified to near homogeneity by this procedure. Lanes 1, 2, 3, and 4 (0.7-, 9.9-, 1.3-, and 1.5-kb isoforms, respectively).
Production and purification of recombinant GST-fusion proteins of Ang-1 isoforms.
Recombinant proteins were purified from bacterial lysates by glutathione Sepharose 4B chromatography and analyzed by SDS-PAGE followed by Coomassie blue staining. The expected molecular weights of fusion proteins were obtained and purified to near homogeneity by this procedure. Lanes 1, 2, 3, and 4 (0.7-, 9.9-, 1.3-, and 1.5-kb isoforms, respectively).
Binding of Ang-1.3-kb isoform protein to 1.5-kb isoform protein in vitro.
(A) The GST–Ang-1.5-kb fusion protein was incubated with35S-labeled 1.3 kb, 0.9 kb, or luciferase prepared by transcription/translation in vitro. Proteins bound to the 1.5-kb fusion protein were identified by incubation with glutathione Sepharose 4B beads, elution, electrophoresis on 12% SDS-PAGE, and autoradiography of the gel. Lane 1 (1.3 kb), lane 2 (0.9 kb), lane 3 (control luciferase protein). (B) cos cells were transfected with either a mixture of 1.5 and 1.3 kb containing vectors or 1.5-kb vector alone. Cells were lysed in lysis buffer, and isoforms were precipitated with or without antibody reactive with the 1.5-kb isoform, not the 1.3-kb isoform, plus protein G beads. Samples were then analyzed by Western blot with an antibody reactive with both the 1.5- and the 1.3-kb isoforms. Lane 1, 1.5- and 1.3-kb transfectant without antibody but with protein G beads; lane 2, 1.5-kb transfectant without antibody but with beads; lane 3, 1.5- and 1.3-kb transfectant with antibody plus beads; lane 4, 1.5-kb transfectant with antibody plus beads.
Binding of Ang-1.3-kb isoform protein to 1.5-kb isoform protein in vitro.
(A) The GST–Ang-1.5-kb fusion protein was incubated with35S-labeled 1.3 kb, 0.9 kb, or luciferase prepared by transcription/translation in vitro. Proteins bound to the 1.5-kb fusion protein were identified by incubation with glutathione Sepharose 4B beads, elution, electrophoresis on 12% SDS-PAGE, and autoradiography of the gel. Lane 1 (1.3 kb), lane 2 (0.9 kb), lane 3 (control luciferase protein). (B) cos cells were transfected with either a mixture of 1.5 and 1.3 kb containing vectors or 1.5-kb vector alone. Cells were lysed in lysis buffer, and isoforms were precipitated with or without antibody reactive with the 1.5-kb isoform, not the 1.3-kb isoform, plus protein G beads. Samples were then analyzed by Western blot with an antibody reactive with both the 1.5- and the 1.3-kb isoforms. Lane 1, 1.5- and 1.3-kb transfectant without antibody but with protein G beads; lane 2, 1.5-kb transfectant without antibody but with beads; lane 3, 1.5- and 1.3-kb transfectant with antibody plus beads; lane 4, 1.5-kb transfectant with antibody plus beads.
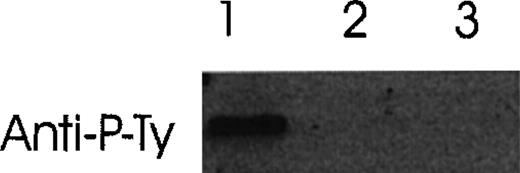
Ang-1.5 kb, but not other isoforms, induces tyrosine phosphorylation of Tie-2
Conditioned media from COS cells transfected with various Ang-1 isoforms were studied for their ability to induce tyrosine phosphorylation. As shown in Figure 12, a positive band was observed in HUVE cells treated with Ang-1.5 kb (full-length), but no band was detected in HUVE cells incubated with the 1.3- and 0.9-kb isoforms.
Induction of autophosphorylation of Tie-2 receptor by Ang-1 isoforms.
HUVE cells were stimulated with conditioned media collected from Ang-1 isoforms (Ang-1.5 kb, lane 1; 1.3 kb, lane 2; or 0.9 kb, lane 3) in transfected COS cells. Cell lysates were immunoprecipitated with anti-Tie-2 antibody. Immunoprecipitates were analyzed by Western blot with antiphosphotyrosine antibody.
Induction of autophosphorylation of Tie-2 receptor by Ang-1 isoforms.
HUVE cells were stimulated with conditioned media collected from Ang-1 isoforms (Ang-1.5 kb, lane 1; 1.3 kb, lane 2; or 0.9 kb, lane 3) in transfected COS cells. Cell lysates were immunoprecipitated with anti-Tie-2 antibody. Immunoprecipitates were analyzed by Western blot with antiphosphotyrosine antibody.
Discussion
These data indicate the presence of Ang-1 in 4 of 6 human cell lines examined (MDA-MB468 and MDA-MB231 breast, CHRF megakaryocyte, and FS4 fibroblast), fetal liver CD34+CD41+ cells, cord blood CD34+ cells and platelets with alternatively spliced mRNA species in 3 of the cell lines (CHRF, MDA-MB468, and FS4), and fetal liver CD34+CD41+ cells and platelets.
Four isoforms of Ang-1 (1.5, 1.3, 0.9, and 0.7 kb) were identified, cloned, and sequenced and then were further confirmed by RT-PCR using specific primers, RNase protection, and Northern blot analysis with specific probes. These 4 isoforms represented amino acid numbers 498, 367, 285, and 154, respectively. Two protein isoforms of Ang-1, Ang-1.5 kb and Ang-0.9 kb, were observed by immunoblot. Ang-1.3 kb and Ang-0.7 kb were not detected by immunoblot, possibly because of the low expression or altered antigenic specificity of the isoforms for the antibody applied.
The structure of Ang-1 contains a secretory signal peptide, a coiled-coil region consisting of residues 100-280 amino acids and a fibrinogen-like region consisting of residues 280-498 amino acids.3 All the isoforms noted could be synthesized with a hydrophobic leader peptide for secretion, indicating that the isoforms described may be of physiologic significance. Isoform Ang-1.3 kb contains the N-terminal coiled-coil region and a truncated second fibrinogen-like domain. In the isoform Ang-0.9 kb, most of the coiled-coil domain is deleted. The isoform Ang-0.7 kb is a truncated form derived from 2 alternatively spliced regions of the full-length Ang-1 in which parts of both the coiled-coil and the fibrinogen-like domains are deleted. The coiled-coil domain is involved in protein assembly into multimeric structures, which could be homomultimers or heteromultimers of related proteins.22-25 Indeed several relatives of Ang-1 have been described that could be candidates for association of the heteromultimer.3 8
It is reasonable to suggest that the various isoforms described in this study may have different functions, depending on their multimeric association. Indeed, the 1.3-kb isoform was found to interact with Ang-1.5 kb. This could be predicted from its structure, eg, the absence of its purported receptor-binding domain but the presence of its coiled-coil multimerization domain. It therefore appears that the Ang-1.3-kb isoform could act as a dominant negative form for the full-length 1.5-kb isoform. In the same context, the 0.9-kb isoform binds to Tie-2 but does not bind to the 1.5-kb isoform, as could be predicted from its intact receptor-binding domain and absent coiled-coil dimerization domains. The 0.9-kb isoform does not activate the receptor and therefore may act as a second dominant negative isoform by interfering with binding of the 1.5-kb isoform to Tie-2. At the molecular level, much remains to be learned about the structure of the isoforms and their mechanism of action in vivo. Will any of the isoforms interact (ie, activate or inhibit) as heteromultimers with related angiogenic species?
The role of Ang-1 and its isoforms in human cells remains to be determined. It is of interest that Ang-1 was present in 3 of 6 malignant tumor cell lines, 1 primary fibroblast cell line, and primitive hematopoietic CD34+ cells from fetal liver, cord blood, and platelets. The 1.3-kb isoform was found in 2 of the 3 positive Ang-1 tumor cell lines, the primary fibroblast cell line, the fetal liver CD34+CD41+ cells, and platelets. The 0.9-kb isoform was only found in CHRF cells and platelets. Thus the role, the pathophysiology, or both of Ang-1 distribution may be related to certain primitive and malignant cells. Regardless of the possible role of Ang-1 and its isoforms in angiogenesis and early hematopoietic cell development, it is clear that 2 of the isoforms, Ang-1.3 and Ang-0.9 kb, theoretically have the possibility to act as dominant negative inhibitors if used therapeutically.
Two families of receptor tyrosine kinases, essential for the normal development of blood vessels, have been identified in endothelial cells. One includes VEGF receptors-1, -2, -3, and -4, which bind members of the VEGF family of growth factors.26 It is of interest that VEGF, like Ang-1, also has at least 4 alternatively spliced mRNA species-encoding isoforms. The interaction of VEGF isoforms with VEGF receptors and extracellular matrix components varies.26 However, it appears that all isoforms of VEGF are capable of inducing endothelial cell proliferation.
The second family of receptor tyrosine kinases includes Tie-1 and Tie-2, together with ligands Ang-1, -2, -3, and -4. Their interaction is likewise involved in vascular development.3-8,27 Blood vessel formation and angiogenesis appear to require the cooperative effects of both families. Angiogenesis is essential for supporting embryonic development and wound healing and plays a pathophysiologic role in diabetic retinopathy, atherosclerosis, thrombosis, and tumor growth. Agents targeting Tie-2 have been shown to inhibit tumor growth and angiogenesis.28 29 Tie-1 and Tie-2 are also expressed in certain cells of the hematopoietic lineage. The possible involvement of the newly identified Ang-1 isoforms in angiogenesis, as well as the growth and differentiation of hematopoietic progenitor cells, provides greater complexity to these processes.
Supported by National Institutes of Health grant HL-13336-26 and by grants from the Helen Polonsky Research Fund and the Dorothy and Seymour Weinstein Research Fund.
Reprints:Yao-Qi Huang, New York University School of Medicine and Kaplan Cancer Center, 550 First Avenue, New York, NY 10016; e-mail: huangy02@mcrcr6.med.nyu.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal