To the editor:
A single tube multiplex PCR method to detect the common + thalassemia alleles
Alpha thalassemias are the commonest single gene disorders in humans. Two different deletions (−α3.7 and −α4.2) in the α globin genes (−α) on the short arm of chromosome 161 are the most common cause of this disorder. Natural selection, by providing protection against a severe form of malaria, is believed to be responsible for elevating and maintaining high frequencies of these heritable genetic defects in various regions of the world where malaria is or has been endemic. This was recently emphasized in an incisive study that suggested that α+ thalassemia may have a major impact on childhood survival in coastal Papua New Guinea by providing continued protection against several infections.2 This very attractive hypothesis needs to be evaluated in different geographical/environmental contexts.
Accurate estimation of the frequency of α+ thalassemia in the population requires DNA analysis. The conventional approach has been to characterize these deletional forms of α+thalassemia by Southern blot hybridization using radioactively labeled probes, a time-consuming, labor-intensive, and expensive procedure. For these reasons, its application to any large-scale screening program was extremely difficult, even though it was very useful for diagnosis of individual patients. With the advent of polymerase chain reaction (PCR)-based methods, detection of α+ thalassemia deletions have become relatively simple and cost-effective.3 4 PCR-based approaches, however, require multiple tube amplifications for precise characterization of the two major, −α3.7 and −α4.2, alleles. Although wieldy for diagnostic purposes, multi-tube testing is cumbersome for large epidemiological screening. In an effort to develop a technique suitable for population screening for α+thalassemia, we have established a multiplex PCR amplification procedure carried out in a single tube for detecting −α3.7 and −α4.2 alleles.
In a previously described PCR-based method,4 selective amplification of the mutant −α3.7 and normal alleles were carried out in separate tubes with distinct primer pairs and experimental conditions. For detecting the deleted allele, oligonucleotide primers flanking the deletion were chosen, and experimental conditions were such that the normal allele did not amplify, as the priming sites are >5 kb apart. Presence of the normal allele in the same sample was confirmed in a separate PCR experiment using one of the primers selected within the sequence of the deletion. For testing the presence of the −α4.2 allele, a single-tube multiplex PCR with three primers was carried out in which the normal and the −α4.2 alleles were recognized by fragment sizes of 2.1 kb and 550 bp, respectively. Overall, three separate PCR reactions had to be carried out to diagnose the presence of two major α+ thalassemic determinants.
We have developed a single-tube multiplex long PCR method to detect the normal, −α3.7 and −α4.2, alleles using primers described earlier.4 This PCR was performed in 25 μL with 0.6 μmol/L primers A, B, D, and E, 350 μmol/L of each dNTP, 2.25 mmol/L MgCl2, 7.5% DMSO, 1.75 units of Expand Long Template DNA polymerase in supplied buffer-3 (Roche GmbH, Mannheim, Germany) and 25 ng of genomic DNA in the PTC-200 thermal cycler (MJ Research, Watertown, MA). After 2 minutes of initial denaturation at 92°C, 10 cycles were performed with denaturation at 92°C for 40 seconds, annealing at 58°C for 1 minute and extension at 68°C for 6 minutes followed by 20 cycles where extension time was increased by 20 seconds after each cycle. PCR products were analyzed in 1% agarose gel in TAE buffer. The expected diagnostic fragment sizes were 1.8 kb, 2.1 kb, and 3.2 kb for −α3.7, −α4.2, and normal alleles, respectively (Figure 1). Under these experimental conditions, fragments of 5.5 kb and 6.3 kb that would be expected for initiations from A+B and D+E primer pairs, respectively, are not amplified in the PCR reaction.
Schematic representation of 1 and 2 genes showing position and orientation of primers.
Primers A and B amplify a 1.8 kb fragment and D and E amplify a 2.1 kb fragment for −α3.7 and −α4.2 deletions, respectively. An amplification product of 3.2 kb for A and E primer pair indicates the presence of normal allele.
Schematic representation of 1 and 2 genes showing position and orientation of primers.
Primers A and B amplify a 1.8 kb fragment and D and E amplify a 2.1 kb fragment for −α3.7 and −α4.2 deletions, respectively. An amplification product of 3.2 kb for A and E primer pair indicates the presence of normal allele.
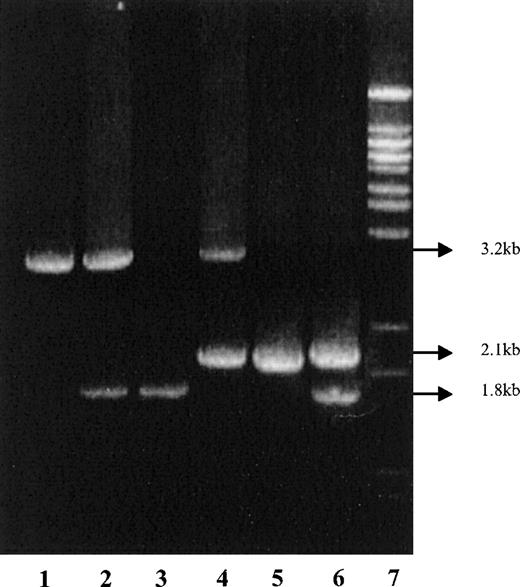
Standardization of the PCR was carried out with DNA samples of known genotypes: −α3.7/−α3.7, −α3.7/αα, −α4.2/−α4.2, −α4.2/αα, and −α4.2/−α3.7 (Figure2).
Identification of amplification products by gel electrophoresis.
Lane 1: normal (αα/αα); 2: heterozygote −α3.7(−α3.7/αα); 3: homozygote −α3.7(−α3.7/−α3.7); 4: heterozygote −α4.2 (−α4.2/αα); 5: homozygote −α4.2 (−α4.2/−α4.2); 5: compound heterozygote −α3.7/−α4.2; and 7: molecular weight marker.
Identification of amplification products by gel electrophoresis.
Lane 1: normal (αα/αα); 2: heterozygote −α3.7(−α3.7/αα); 3: homozygote −α3.7(−α3.7/−α3.7); 4: heterozygote −α4.2 (−α4.2/αα); 5: homozygote −α4.2 (−α4.2/−α4.2); 5: compound heterozygote −α3.7/−α4.2; and 7: molecular weight marker.
We conclude that this single-tube multiplex PCR assay for the common α+ thalassemia determinants is a simple, rapid, and robust method applicable to large population screening for epidemiological purposes. It will need to be used with caution when applied to diagnosis of individuals with compound heterozygous states for large deletions such as −−SEA/−α3.7 or −−SEA/−α4.2, which will be identified as homozygous for the small deletion. Such cases, however, are easily recognized on clinical and hematological features.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal