The receptor for interleukin 5 (IL-5) consists of a cytokine-specific chain (IL-5R) and a signaling β chain, which is shared with interleukin 3 (IL-3) and granulocyte-macrophage colony-stimulating factor (GM-CSF). These 3 cytokines can act in eosinophil development and activation in vitro, but gene deletion or antibody blocking of IL-5 largely ablates eosinophilic responses in models of allergic disease or helminth infection. We investigated factors acting in differential IL-5R gene splicing to generate either the membrane-anchored isoform (TM-IL-5R) which associates with the common β chain to allow IL-5 responsiveness, or a secreted, antagonist variant (SOL-IL-5R). In a murine myeloid cell line (FDC-P1), transfected with minigenes allowing expression of either IL-5R variant, IL-5 itself, but not IL-3 or GM-CSF, stimulated a reversible switch toward expression of TM-IL-5R. A switch from predominantly soluble isoform to TM-IL-5R messenger RNA (mRNA) expression was also seen during IL-5-driven eosinophil development from human umbilical cord blood-derived CD34+ cells; this was accompanied by surface expression of IL-5R and acquisition of functional responses to IL-5. IL-3 and GM-CSF also supported eosinophil development and up-regulation of TM-IL-5R mRNA in this system, but this was preceded by expression of IL-5 mRNA and was inhibited by monoclonal antibody to IL-5. These data suggest IL-5-specific signaling, not shared by IL-3 and GM-CSF, leading to a switch toward up-regulation of functional IL-5R and, furthermore, that IL-3 and GM-CSF-driven eosinophil development is dependent on IL-5, providing an explanation for the selective requirement of IL-5 for expansion of the eosinophil lineage.
Selective accumulation of eosinophils is a feature of allergic inflammation, as observed in atopic asthma, where cells are implicated in tissue damage contributing to bronchial hyperresponsiveness.1,2 Interleukin 5 (IL-5) acts in development, activation, and tissue survival of eosinophils.3-5 Increased expression of IL-5 at sites of allergic disease,6-8 which correlates with clinical disease activity,9,10 and data from blocking antibody or gene deletion experiments in animal models11-13 suggest that IL-5 is the major determinant of eosinophilic inflammation.
High-affinity receptors for human IL-5 are formed from cytokine-specific α chain associated with a common signaling receptor β chain shared with interleukin 3 (IL-3) and granulocyte-macrophage colony-stimulating factor (GM-CSF).14,15 Responsiveness of cells to IL-5 appears to be controlled at the level of expression of IL-5Rα.16 Differential splicing of the IL-5Rα gene can generate different messenger RNA (mRNA) isoforms: a membrane-anchored isoform (TM-IL-5), which can associate with the common β chain to form the active receptor complex, or at least 2 secreted variants (SOL-IL-5Rα), which lack the membrane anchor and show receptor antagonist activity.17 We hypothesize that control of this splicing event will determine IL-5 receptor expression, IL-5 responsiveness, and thus eosinophil expansion and activation.
We have previously examined expression of IL-5Rα isoforms in bronchial mucosal biopsies from asthma subjects and found increased relative expression of mRNA for TM-IL-5Rα and decreased SOL-IL-5Rα in those subjects with more active disease.18 Thus, increased IL-5 responsiveness of eosinophils might result in more pronounced inflammation and airway responsiveness. In addition, we have recently detected TM-IL-5Rα expression by CD34+ cells in the airway in asthmatic subjects.19 Together with a report of increased numbers of bone marrow CD34+/IL-5Rα+ cells after allergen inhalation challenge of asthmatic subjects,20 this raises the possibility that IL-5 could act early in eosinophil development from progenitors.
Here, minigene constructs and human primary CD34+ cells were used to examine cytokine regulation of IL-5Rα isoform expression. Our data suggest that IL-5, but not IL-3 or GM-CSF, can switch toward expression of the active TM-IL-5Rα and that eosinophil development, even that driven by IL-3 or GM-CSF, may be, at least in part, IL-5 dependent.
Materials and methods
Materials
Vectors pSV-SPORT (Life Technologies, Rockville, MD) and pBluescriptII (Stratagene, La Jolla, CA) were used to construct minigenes. Restriction enzymes HindIII, BglII, Xho1, Sac1, EcoR1, Not1, and BamH1 were from Gibco BRL. Dulbecco's modified Eagle's medium (DMEM) and RPMI medium were from Gibco BRL, DEAE-dextran from Pharmacia, and chloroquine, puromycin, TPA, and PKH2-GL were from Sigma Chemicals (St Louis, MO). Poly-A+ RNA was extracted using the FastTrack system (Invitrogen Inc, San Diego, CA), and total RNA was extracted for reverse transcriptase-polymerase chain reaction (RT-PCR) by lysis in 4 mol/L guanidine thiocyanate, 25 mM sodium citrate, 0.5% n-lauryl sulfate, and 100 mM mercaptoethanol (all from Sigma). Cell selection and IL-5Rα isoform switching experiments were performed using rmIL-3 (Nippon Roche, Kamakura, Japan), rhIL-5 (purified using a 5A5 immunoaffinity column as described previously22), rmGM-CSF (Biogen SA, Geneva, Switzerland), or rmIL-4 (conditioned medium from transfected CHO cells, a gift from Dr W. Muller). In certain experiments, conditioned medium from phorbol myristate acetate (PMA)-stimulated EL-4 cells was used as the source of cytokines. In all cases, saturating concentrations supporting growth of FDC-P1 cells were used.
Human umbilical cord blood was collected into McCoy's 5A medium (Sigma), with 15% fetal calf serum (FCS; Sigma), and antibiotic, antimycotic (penicillin, streptomycin, and amphotericin mix; Life Technologies, Paisley, Scotland) and 50μM β-mercaptoethanol (Life Technologies), and cultured in Iscove's modified Dulbecco's medium (Life Technologies), in 12-well plates (Nunc, Roskilde, Denmark). Histopaque was from Sigma, and MACS-CD34+ immunomagnetic columns from Miltenyi Biotech, (Bisley, UK). Monoclonal antibodies were anti-CD34 (HPCA2, Becton Dickinson, Oxford, UK), antimajor basic protein (BMK 13, Pharmacia Upjohn, Milton Keynes, UK), anti-CD9 (Serotec, Oxford, UK), anti-hIL-3Rα,6H6 anti-hGM-CSF8G6, and antihuman common β chain (IC1) (all kind gifts from Prof. Angel Lopez, Hanson Institute, Adelaide, Australia) and anti-hIL-5Rα (α16, Van der Heyden et al, unpublished observations). For CD34+ cultures rhIL-3 (Genzyme Corporation, West Malling, UK), rhIL-5 (Pharmingen), or rhGM-CSF (Genzyme) was used. Chromotrope 2R and May Grunwald Giemsa stains, used to identify eosinophils in cultures, were from BDH, Lutterworth, UK.
Generation of minigenes
pSV-SPORT was used as vector frame for the minigene constructions. A Xho1-Sac1 cDNA fragment from pCDM8-hIL-5RαSOL14 was combined with a Sac1-EcoR1 fragment containing exon 11 from λghIL5Rα-623 in pBluescript II. Likewise, a HindIII-BglII fragment containing exon 12 from λghIL5Rα-1123 was combined with a BglII-Xba1 cDNA segment from pCDM8-hIL-5RαTM.17 pSV-MINI-1 was assembled by combining Xho1-Not1 (BamH1 linkered) and HindIII (BamH1 linkered)-Not1 from the previous constructs. pSV-MINI-2 contains an additional SV40 polyadenylation site at the BamH1 fusion point. The minigene constructs are shown in Figure 1C.
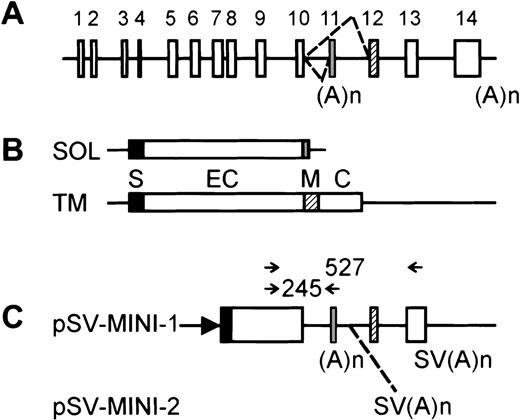
Structure of the hIL-5R minigene and PCR primer design.
(A) hIL-5Rα gene structure. The alternative splicing choice leading to the transcripts encoding the secreted or membrane-anchored isoforms is indicated by dashed lines. Exons are shown as boxes and are numbered on top, with exon 11 corresponding to the “soluble”-specific exon and exon 12 encoding the transmembrane region. Polyadenylation sites are shown below. (B) The 2 major transcripts from the hIL-5Rα gene. SOL encodes the major secreted variant; TM encodes the membrane-associated receptor.8 Boxes are translated regions, with protein domains shown: S indicates signal peptide; EC, extracellular domain; M, membrane anchor; C, cytoplasmic tail. (C) The structure of the minigenes. Boxes are translated regions, either from cDNA segments or from exons. (A)n is the polyadenylation site present in the “soluble”-specific exon; SV(A)n comes from SV40 and is artificially added. The position of the extra SV(A)n in pSV-MINI-2 is shown by a dashed line. The arrow represents the SV40 early promoter. On top, arrows indicate the positions of the primers used in the (competitive) RT-PCR experiments. Sizes of possible reaction products are indicated. Throughout the figure, black, gray, and hatched boxes correspond to exons, protein domains, or coding regions specifying the signal peptide, secreted isoform, or membrane-anchored isoform, respectively.
Structure of the hIL-5R minigene and PCR primer design.
(A) hIL-5Rα gene structure. The alternative splicing choice leading to the transcripts encoding the secreted or membrane-anchored isoforms is indicated by dashed lines. Exons are shown as boxes and are numbered on top, with exon 11 corresponding to the “soluble”-specific exon and exon 12 encoding the transmembrane region. Polyadenylation sites are shown below. (B) The 2 major transcripts from the hIL-5Rα gene. SOL encodes the major secreted variant; TM encodes the membrane-associated receptor.8 Boxes are translated regions, with protein domains shown: S indicates signal peptide; EC, extracellular domain; M, membrane anchor; C, cytoplasmic tail. (C) The structure of the minigenes. Boxes are translated regions, either from cDNA segments or from exons. (A)n is the polyadenylation site present in the “soluble”-specific exon; SV(A)n comes from SV40 and is artificially added. The position of the extra SV(A)n in pSV-MINI-2 is shown by a dashed line. The arrow represents the SV40 early promoter. On top, arrows indicate the positions of the primers used in the (competitive) RT-PCR experiments. Sizes of possible reaction products are indicated. Throughout the figure, black, gray, and hatched boxes correspond to exons, protein domains, or coding regions specifying the signal peptide, secreted isoform, or membrane-anchored isoform, respectively.
Analysis of IL-5R isoform expression and switching
Cos-1 cells maintained in DMEM with 10% FCS were transfected following established procedures using DEAE-dextran and chloroquine. Cells were harvested and mRNA was prepared approximately 72 hours after transfection. FDC-P1 cells were grown in RPMI with 10% FCS and 10% conditioned medium from TPA-stimulated EL4 cells (as a source of murine IL-3), and were transfected by electroporation using settings at 300 V and 1500 μF (Easyject plus electroporator, EquiBio, Kent, UK). A stable transfectant FDC-P1-MINI-1.1 was obtained by cotransfection with a vector conferring puromycin resistance and selection in 1μg/mL puromycin. PolyA+ RNA was prepared using the FastTrack procedure (Invitrogen). Total RNA samples were prepared from isothiocyanate lysed cells essentially as described.24
Expression of mRNA for either soluble or membrane-associated IL-5Rα isoforms was detected by Northern analysis or RT-PCR. For Northern blots, Zeta-probe GT membranes (Biorad, Hercules, CA), and probes corresponding to the extracellular domains, thus hybridizing with both mRNA isoforms, or corresponding to the cytosolic domain only, were used as specified, and products were identified on size criteria (see Figure1). Competitive RT-PCR was optimized allowing rapid estimation of the ratios of expression of both hIL-5Rα isoforms. RT-PCR was performed using a cDNA synthesis kit with RnaseH− MMLV Reverse Transcriptase according to the supplier's instructions, Taq polymerase was from Gibco-BRL. A forward primer was designed on the extracellular part of the receptor at position 1033 to 105617 and 2 reverse primers, specific for the transcripts encoding either the secreted (position 1279-1298, 5′-TCAGATACGGTGTGGGGCAG)22or membrane-anchored isoform (position 1539-1561, 5′-TGAGGCACTGAGGCATGTGTGAG)22 (referred to as SOL-transcripts and TM-transcripts, respectively). Predicted PCR reaction products are 245bp and 527bp for the SOL- and TM-transcripts, respectively. Pilot experiments, using various ratios of plasmids containing the cloned cDNAs for either isoform, indicated an approximately 5-fold bias toward amplification of the SOL-transcript (data not shown). A likely explanation for such bias is the different sizes of the amplified reaction products. In cultured human umbilical cord blood-derived CD34+ cells, IL-5 mRNA expression was also analyzed by RT-PCR (forward: 5′-CAAACGCAGAACGTTTCAAGA; reverse: 5′-CAGGTACCCCCTTGCACAGTT). In certain experiments, fainter bands migrating close to the predicted bands could be observed, but these were found to be artefacts by cloning and sequencing.
Surface expression of IL-5R protein
Cell labeling and analysis of clonal expansion in selection of IL-5R isoform expression
To determine whether IL-5Rα isoform switching represented selective outgrowth of a subpopulation from the cell line, or a switch of the whole population, a number of techniques were used. To avoid rhIL-5 exerting selection pressure in cell expansion, FDC-P1 cells were grown in rmIL-4 together with rhIL-5, or were cotransfected with the pSVZeo-mIL-5Rα vector, which directs the expression of the murine IL-5Rα subunit. Here, a cDNA encoding the mIL-5Rα chain is expressed under control of an SV40 early promoter. These cells were then cultured in murine IL-5.
To measure the plating efficiency of cells growing in either conditioned medium or rhIL-5, FDC-P1-MINI-1.1 cells (FDC-P1 cells transfected with the IL-5Rα minigene) were plated in microtiter plates at 1 or 3 cells per well in medium containing either EL4 conditioned medium alone or in medium containing IL-5. Colonies were counted at day 7 (EL4 CM) or day 12 (hIL-5).
To label cells, the lipophilic chromophore PKH2-GL was purchased from Sigma and used according to the manufacturer's instructions and reference 26. Briefly, 107 FDC-P1-MINI 1.1 cells were collected in 1 mL and were incubated for 7 minutes at room temperature with 4 μM PKH2-GL. Medium with 10% FCS was added and cells were separated from free label by centrifugation through a serum cushion. These cells were then cultured in conditioned medium or rhIL-5 as above and analyzed at 14 days by flow cytometry to examine distribution of the fluorescent dye among daughter cells.
Umbilical cord blood CD34+ cell culture
Human umbilical cord blood was collected from the Maternity Unit, Chelsea and Westminster Hospital (with approval of the local Ethics Committee). Mononuclear cells were isolated by centrifugation over Histopaque (Sigma) followed by red cell lysis in sterile water and adherence at 1 to 2 hours at 37°C. CD34+ cells were isolated from nonadherent cells using MACS-CD34+immunomagnetic columns (Miltenyi Biotech), and checked for purity by staining with anti-CD34 monoclonal antibody HPCA2 (Becton Dickinson). For CD34+ cultures rhIL-3, rhIL-5, or rhGM-CSF was added at 1 ng/mL and cells were grown in Iscove's modified Dulbecco's medium, with 20% FCS, 1% antibiotic/antimycotics, and 50 μM mercaptoethanol in 12-well plates. Cells were counted and fed with fresh medium at days 7 and 14 of culture.
Eosinophil phenotype was identified by chromotrope 2R and May Grunwald Giemsa staining, and by immunostaining with antibodies directed against eosinophil major basic protein (BMK13). Positive cells were enumerated and expressed as a percentage after counting at least 200 total cells. RNA was extracted from cells at days 0, 3, 5, 7, and 14 of culture as above, and flow cytometry was performed at days 0, 3, 5, 7, and 14 for cytokine receptor expression and phenotypic markers, and cells were used for chemokinesis and shape change assays at days 0, 4, 7, and 14 of culture. In some experiments, to determine IL-5 dependence of eosinophil development in IL-3 and GM-CSF, the blocking antibody to IL-5, 5A5,22 was added to cultures at a concentration of 10 μg/mL.
Functional responses to IL-5
For chemokinesis experiments Transwell filters (3 μm pore size) were purchased from Millipore (Watford, UK), and the rhIL-5 used was a kind gift of Dr Tim Wells, Serono, Geneva, Switzerland. Umbilical cord blood CD34+ cells cultured for various periods in cytokines were added to the top well, then exposed to medium alone or IL-5 at 0.1 nM or 1 nM added for 1 hour to both chambers. Cells migrating to the bottom chamber were counted using a flow cytometer (FACScan, Becton Dickinson, San Jose, CA), with relative cell counts obtained by acquiring events for 60 seconds.27 Chemokinetic index was expressed as the number of cells in the bottom well at 1 hour with stimulus divided by that with medium alone.
Results
Minigene constructs allow differential expression of membrane associated (TM-IL-5R) or soluble (SOL-IL-5R) IL-5R mRNA isoforms
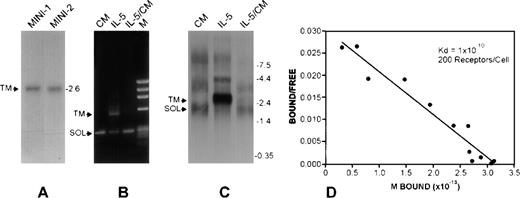
The signaling competent transmembrane or the soluble antagonist isoforms of IL-5Rα are generated by differential gene switching to exclude exon 11 and skip to include exons 12, 13, and 14 (TM-IL-5Rα) or to include and terminate at exon 11 (SOL-IL-5Rα)14 17 (Figure 1A and B). We constructed minigenes combining cDNA and genomic DNA segments of these IL-5Rα isoforms to allow examination of factors regulating relative expression of these 2 isoforms (Figure 1C). Transient transfection of Cos-1 cells led to expression of the membrane-associated IL-5Rα only (TM-transcript, Figure 2A). In contrast, in FDC-P1-MINI-1.1 cells, a stable subclone derived from the murine promyelocytic FDC-P1 cell line, predominant expression of the secreted variant (SOL-transcript, Figure 2B and C, CM lanes) was seen, and the TM-transcript could only be detected by RT-PCR (data not shown).
Transcription patterns from hIL-5R gene and minigenes.
(A) Northern blot hybridization experiments were performed using a probe that detects both membrane associated (TM) and soluble (SOL) IL-5Rα mRNA isoforms, using RNA from Cos-1 cells transfected with the pSV-MINI-1 or -2 constructs. In both cases, only IL-5Rα-TM was detected. Construct pSV-MINI-2 includes an SV40 polyadenylation sequence downstream of exon 11, ruling out the possibility that the absence of the SOL mRNA is due to a deficiency in recognizing the SOL transcript-specific polyadenylation signal. The size and location of the TM-specific transcript is indicated. (B and C) Competitive RT-PCR analysis and Northern analysis of IL-5Rα transcripts from FDC-P1-MINI-1.1 cells grown in EL4-conditioned medium (lanes CM, a source of murine IL-3), or rhIL-5 (lanes IL-5) for 2 weeks, or in rhIL-5 for 2 weeks followed by EL-4-conditioned medium for a further 7 days (lanes IL-5/CM). Representative of 3 independent experiments. (D) Culture in rhIL-5 induces high-affinity IL-5 binding sites on FDC-P1-MINI-1.1 cells. Human IL-5 was radiolabeled using125I by the iodogen procedure and Scatchard plot analysis was performed. No binding was detected to cells grown in EL-4 conditioned medium alone.
Transcription patterns from hIL-5R gene and minigenes.
(A) Northern blot hybridization experiments were performed using a probe that detects both membrane associated (TM) and soluble (SOL) IL-5Rα mRNA isoforms, using RNA from Cos-1 cells transfected with the pSV-MINI-1 or -2 constructs. In both cases, only IL-5Rα-TM was detected. Construct pSV-MINI-2 includes an SV40 polyadenylation sequence downstream of exon 11, ruling out the possibility that the absence of the SOL mRNA is due to a deficiency in recognizing the SOL transcript-specific polyadenylation signal. The size and location of the TM-specific transcript is indicated. (B and C) Competitive RT-PCR analysis and Northern analysis of IL-5Rα transcripts from FDC-P1-MINI-1.1 cells grown in EL4-conditioned medium (lanes CM, a source of murine IL-3), or rhIL-5 (lanes IL-5) for 2 weeks, or in rhIL-5 for 2 weeks followed by EL-4-conditioned medium for a further 7 days (lanes IL-5/CM). Representative of 3 independent experiments. (D) Culture in rhIL-5 induces high-affinity IL-5 binding sites on FDC-P1-MINI-1.1 cells. Human IL-5 was radiolabeled using125I by the iodogen procedure and Scatchard plot analysis was performed. No binding was detected to cells grown in EL-4 conditioned medium alone.
IL-5, but not IL-3 or GM-CSF, up-regulates membrane expression of IL-5R
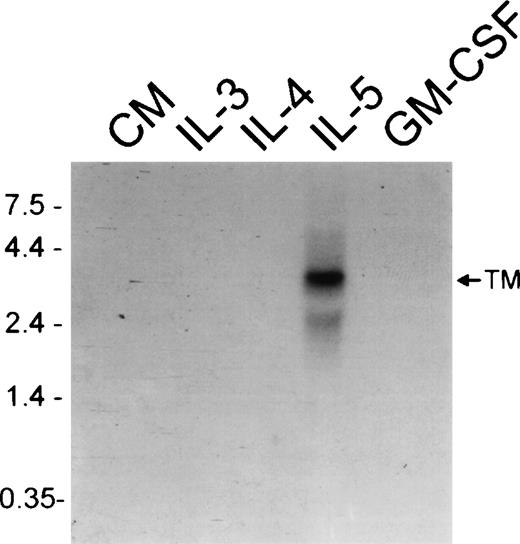
To examine factors regulating relative expression of mRNA for transmembrane and soluble IL-5Rα isoforms, FDC-P1-MINI-1.1cells were cultured with cytokines for 2 weeks, then examined by RT-PCR and Northern analysis. As shown in Figure 2A and B, when the FDC-P1-MINI-1.1 cells were cultured with recombinant human IL-5 there was up-regulation of the TM-IL-5Rα mRNA, which was not seen when cells were cultured with rmIL-3, rmGM-CSF, rmIL-4, or conditioned medium from PMA-stimulated EL-4 cells (Figure3). This IL-5-dependent switch to TM-IL-5Rα mRNA expression was accompanied by acquisition of high-affinity IL-5 binding at the cell surface, detected by binding of radiolabeled IL-5 (Figure 2D). There was no detectable IL-5 binding to FDC-P1 transfectants before culture with IL-5, nor to cells cultured in conditioned medium alone.
Expression of membrane-associated IL-5R.
IL-5 but not IL-3 or GM-CSF induces a switch toward membrane-associated IL-5Rα isoform expression. Northern analysis using a probe that detects the membrane-associated (TM) IL-5Rα mRNA isoform using RNA extracted from FDC-P1-MINI-1.1 cells grown in conditioned medium (CM), rmIL-3, rmIL-4, rhIL-5, or rmGM-CSF for 2 weeks.
Expression of membrane-associated IL-5R.
IL-5 but not IL-3 or GM-CSF induces a switch toward membrane-associated IL-5Rα isoform expression. Northern analysis using a probe that detects the membrane-associated (TM) IL-5Rα mRNA isoform using RNA extracted from FDC-P1-MINI-1.1 cells grown in conditioned medium (CM), rmIL-3, rmIL-4, rhIL-5, or rmGM-CSF for 2 weeks.
IL-5 switching to membrane-associated IL-5R expression in FDC-P1 transfectants is an induced event
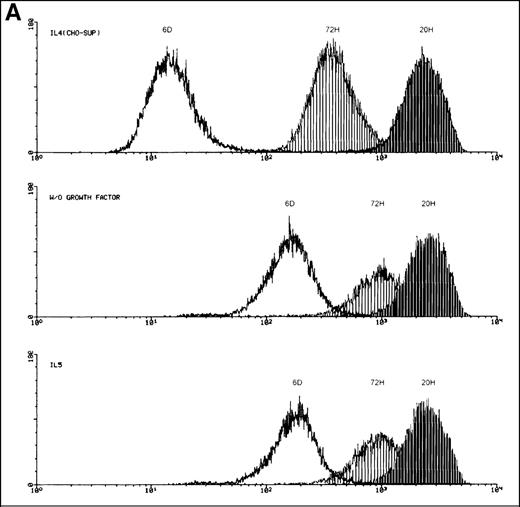
The kinetics of switching to hIL-5Rα TM-transcripts in the FDC-P1-MINI-1.1 system were slow, with a maximal effect after approximately 2 weeks (data not shown). Although TM-IL-5Rα expression by FDC-P1 transfectants at baseline was only detectable by RT-PCR, it remained possible that IL-5 selectively expanded a minority TM-IL-5Rα+ population rather than inducing a switch to TM-IL-5Rα expression. To address this issue we performed a number of experiments. First, as shown in Figure 2A and B, the IL-5-induced switch to TM-IL-5Rα mRNA expression was fully reversible if cells were cultured for a further week in conditioned medium alone, in the absence of IL-5. Second, we labeled FDC-P1-MINI-1.1 cells with the green fluorescent lipophilic chromophore PKH2-GL, which incorporates into the membrane with an elution half-life of 5 to 8 days. On every cell division the fluorescent dye is equally partitioned into the daughter cells, resulting in halving of the fluorescence as measured by flow cytometry. We reasoned that if clonal selection and expansion occurred, a small subpopulation of cells with decreased fluorescence intensity would become apparent. As a control, we took advantage of the fact that FDC-P1-MINI-1.1 cells become factor independent when grown in the absence of any growth factor. Figure4A illustrates that cells grown in human IL-5 (top panel) behaved as a single cell population, and this growth paralleled that seen in medium alone (middle panel). The same conclusion was reached when we measured the plating efficiency of cells growing in either conditioned medium or IL-5. Plating efficiencies were above 70% in both cases, arguing against selection of a minority of the starting cells by IL-5 (data not shown). Third, we examined whether the change in hIL-5Rα isoform expression could also be observed when FDC-P1-MINI-1.1 cells were grown in the presence of murine IL-4 together with human IL-5. Because this cell line can use mIL-4 as a growth factor, no selection pressure on cell survival and proliferation depending on the presence of functional hIL-5Rα subunits can exist under these circumstances. Results are shown in Figure 4B. Panels show results from RT-PCR experiments when cells were grown in medium containing conditioned medium, IL-5 alone, IL-4 alone, or IL-4 and IL-5 combined as indicated. The upper part of each panel shows the products of a competitive PCR; the lower part represents PCR data using primers for the TM-transcript only. TM-transcripts are detected in the presence of IL-5, but also, although at a lower level and with slower kinetics, in the presence of IL-4 plus IL-5. Building on this observation, we transfected the FDC-P1-MINI-1.1 cell-line with the pSVZeo-mIL-5Rα vector, which directs the expression of the murine IL-5Rα subunit. Here, a cDNA encoding the mIL-5Rα chain is expressed under control of an SV40 early promoter. Such cells are now capable of growing in the presence of murine IL-5. Under those conditions, we could again observe the isoform expression switch of hIL-5Rα transcripts from the minigene (Figure 4C). This again underscores the inducible character of the switching, because IL-5 will not exert any growth selection in these conditions.
Induced up-regulation of the membrane-bound hIL-5R isoform by IL-5.
(A) Flow cytometry plots of FDC-P1-MINI-1.1 cells labeled with the lipophilic chromophore PKH2-GL. Cell growth in human IL-5 (bottom panel) showed a single population, which parallels spontaneous autocrine outgrowth (middle panel). If a fast proliferating subpopulation were present, dilution of the fluorescent dye would be much more pronounced and would approach fluorescence intensities as shown for cells grown in IL-4-containing medium (top panel). Representative of 2 independent experiments. (B) RT-PCR analysis of IL-5Rα isoform mRNA expression by FDC-P1-MINI-1.1 cells grown in medium containing EL4 CM, IL-5 alone, IL-4 alone, or IL-4 and IL-5 combined. The upper part of each panel shows the products of a competitive PCR; the lower part represents PCR data using primers for the TM-transcript only. TM-transcripts are detected in the presence of IL-5, but also, although at a lower level, and with slower kinetics in the presence of IL-4 plus IL-5. Representative of 2 independent experiments. (C) FDC-P1-MINI-1.1 cells transfected with the pSVZeo-mIL-5Rα vector, which directs the expression of the murine IL-5Rα subunit, will grow in the presence of murine IL-5, independent of hIL-5. Under these conditions, switch of isoform expression of hIL-5Rα transcripts from the minigene was also observed (arrow). Sampling time points are shown. Representative of 2 independent experiments.
Induced up-regulation of the membrane-bound hIL-5R isoform by IL-5.
(A) Flow cytometry plots of FDC-P1-MINI-1.1 cells labeled with the lipophilic chromophore PKH2-GL. Cell growth in human IL-5 (bottom panel) showed a single population, which parallels spontaneous autocrine outgrowth (middle panel). If a fast proliferating subpopulation were present, dilution of the fluorescent dye would be much more pronounced and would approach fluorescence intensities as shown for cells grown in IL-4-containing medium (top panel). Representative of 2 independent experiments. (B) RT-PCR analysis of IL-5Rα isoform mRNA expression by FDC-P1-MINI-1.1 cells grown in medium containing EL4 CM, IL-5 alone, IL-4 alone, or IL-4 and IL-5 combined. The upper part of each panel shows the products of a competitive PCR; the lower part represents PCR data using primers for the TM-transcript only. TM-transcripts are detected in the presence of IL-5, but also, although at a lower level, and with slower kinetics in the presence of IL-4 plus IL-5. Representative of 2 independent experiments. (C) FDC-P1-MINI-1.1 cells transfected with the pSVZeo-mIL-5Rα vector, which directs the expression of the murine IL-5Rα subunit, will grow in the presence of murine IL-5, independent of hIL-5. Under these conditions, switch of isoform expression of hIL-5Rα transcripts from the minigene was also observed (arrow). Sampling time points are shown. Representative of 2 independent experiments.
Membrane associated IL-5R is up-regulated during development of human eosinophils from umbilical cord blood-derived CD34+cells
Although the data from FDC-P1-MINI 1.1 cells suggested that IL-5 could switch to TM-IL-5Rα isoform expression, it was possible that this was a function of the cell line or transfection system. To examine control of IL-5Rα isoform expression during primary human eosinophil development, cultures of umbilical cord blood-derived CD34+cells were used. When these cells were cultured in IL-3 and IL-5, 84.5% (range 75-88%, n = 10 independent cultures) had eosinophilic granules identified by chromotrope 2R staining and immunocytochemistry for major basic protein (MBP) by day 14 of culture. Eosinophil lineage was confirmed by flow cytometric staining for phenotypic markers, which showed loss of CD34 by day 3 of culture, with acquisition of surface CD9, IL-3Rα, GM-CSFRα, and common β chain increasing to day 14 (data not shown).
The RT-PCR of cells cultured in IL-3 and IL-5 showed up-regulation of TM-IL-5Rα mRNA relative to that of the soluble isoform at days 5 to 7 of culture (Figure 5A). Expression of mRNA for the TM isoform diminished by day 14 of culture, to a pattern of predominant SOL-IL-5Rα mRNA expression, as seen in mature blood eosinophils. Presumably this reflects stage-specific regulation of relative expression of mRNA for IL-5Rα SOL and TM isoforms rather than simple consumption of cytokines, because cytokines were added to cultures at days 7 and 14.
IL-5R expression by developing primary human eosinophils.
(A) The effect of cytokine treatment on IL-5Rα isoform mRNA expression by human cord blood CD34+ cell-derived eosinophil progenitors. RT-PCR using primers specific for the TM-transcript, both TM and SOL transcripts, and β actin mRNA was performed on RNA extracted from cord blood CD34+ cells cultured in IL-3+IL-5 for 0, 3, 5, 7, or 14 days. Results are representative of 5 separate experiments. (B) Interleukin 5Rα surface expression is detected after 7 days of culture of CD34+progenitors in IL-3 and IL-5. Flow cytometry plot shows surface staining with α16 (an IgG1 mouse monoclonal antibody to human IL-5Rα chain) or isotype control IgG1 antibody. CD34+cord blood-derived progenitors were stained at the time of isolation (day 0) and after 7 days of culture in IL-3 and IL-5. The plot shown is typical of 10 independent experiments. (C) CD34+ umbilical cord blood progenitors acquire biologic responsiveness to IL-5 by day 7 of culture in IL-3 and IL-5. Chemokinetic and polarization responses of cord blood-derived CD34+ cells cultured in IL-3 and IL-5 for 4, 7, and 14 days. Cells added to the top well were exposed to medium alone (open bars) or IL-5 at 0.1 nM (hatched bars) or 1 nM (black bars). Chemokinetic index is the number of cells in the bottom well at 1 hour with stimulus divided by that with medium alone. Polarization in response to medium alone or 0.1 nM and 1 nM IL-5 was assessed by change in forward scatter (FSC) signal on flow cytometry (mature peripheral blood eosinophils showed a mean change in forward scatter of 120 units to 1 nM IL-5, n = 3, data not shown). The figure shows a typical response at days 7 and 14. No response to IL-5 was seen at day 3 to 4 in 7 independent experiments. Responsiveness of cells cultured for 7 days to IL-5 was variable and may reflect different rates of eosinophil development in different cultures; however, similar responses to those shown were seen from cells cultured for 7 days in IL-3 and IL-5 in 4 independent experiments.
IL-5R expression by developing primary human eosinophils.
(A) The effect of cytokine treatment on IL-5Rα isoform mRNA expression by human cord blood CD34+ cell-derived eosinophil progenitors. RT-PCR using primers specific for the TM-transcript, both TM and SOL transcripts, and β actin mRNA was performed on RNA extracted from cord blood CD34+ cells cultured in IL-3+IL-5 for 0, 3, 5, 7, or 14 days. Results are representative of 5 separate experiments. (B) Interleukin 5Rα surface expression is detected after 7 days of culture of CD34+progenitors in IL-3 and IL-5. Flow cytometry plot shows surface staining with α16 (an IgG1 mouse monoclonal antibody to human IL-5Rα chain) or isotype control IgG1 antibody. CD34+cord blood-derived progenitors were stained at the time of isolation (day 0) and after 7 days of culture in IL-3 and IL-5. The plot shown is typical of 10 independent experiments. (C) CD34+ umbilical cord blood progenitors acquire biologic responsiveness to IL-5 by day 7 of culture in IL-3 and IL-5. Chemokinetic and polarization responses of cord blood-derived CD34+ cells cultured in IL-3 and IL-5 for 4, 7, and 14 days. Cells added to the top well were exposed to medium alone (open bars) or IL-5 at 0.1 nM (hatched bars) or 1 nM (black bars). Chemokinetic index is the number of cells in the bottom well at 1 hour with stimulus divided by that with medium alone. Polarization in response to medium alone or 0.1 nM and 1 nM IL-5 was assessed by change in forward scatter (FSC) signal on flow cytometry (mature peripheral blood eosinophils showed a mean change in forward scatter of 120 units to 1 nM IL-5, n = 3, data not shown). The figure shows a typical response at days 7 and 14. No response to IL-5 was seen at day 3 to 4 in 7 independent experiments. Responsiveness of cells cultured for 7 days to IL-5 was variable and may reflect different rates of eosinophil development in different cultures; however, similar responses to those shown were seen from cells cultured for 7 days in IL-3 and IL-5 in 4 independent experiments.
Up-regulation of relative expression of mRNA for the TM-IL-5Rα isoform at day 7 of culture of CD34+ cells in IL-3 and IL-5 was accompanied by the acquisition of IL-5Rα expression at the cell surface (Figure 5B).
To determine the functional responsiveness of these developing cells to IL-5 we examined chemokinetic responses and cellular activation by IL-5 as judged by shape change. Chemokinetic responses to IL-5 were not seen at day 4 of culture, but were maximal by day 7 of culture (Figure 5C). In addition, cellular activation in response to IL-5 was maximal by day 7 (see Figure 5C).
Eosinophil development from human CD34+ cells cultured in IL-3 and GM-CSF is preceded by autologous expression of IL-5 mRNA and can be inhibited by a monoclonal antibody blocking IL-5
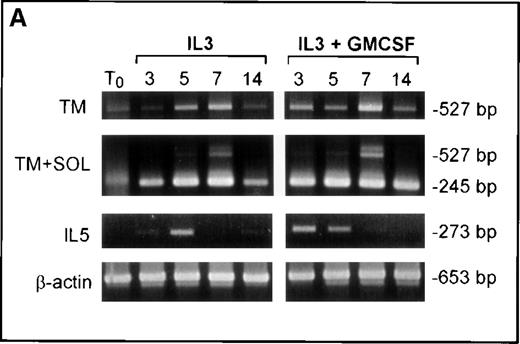
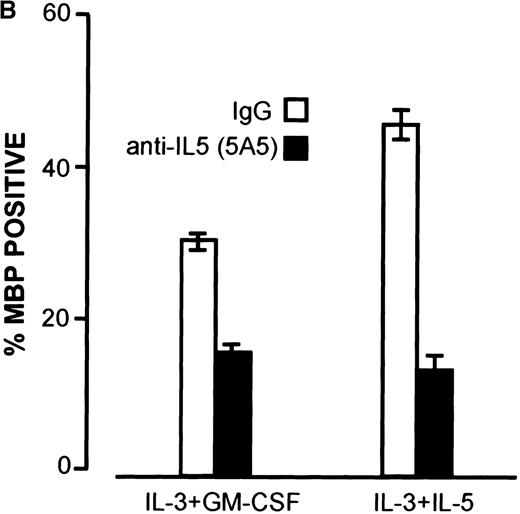
Results from the FDC-P1 cell line suggested that the activity of IL-5 in switching to predominant expression of the TM-IL-5Rα is not shared by IL-3 and GM-CSF. However, previous reports had documented that IL-3 and GM-CSF can support eosinophil development. To examine this further, we cultured human umbilical cord blood-derived CD34+ cells in IL-3 alone, or IL-3+GM-CSF, without addition of exogenous IL-5. In these cultures eosinophil development was also observed, and this was accompanied by up-regulation of mRNA for TM-IL-5Rα also seen at days 5 to 7, as seen when cells were cultured with IL-5 (Figure 6A). To evaluate whether endogenous IL-5 might play a role in this system, we examined IL-5 mRNA expression by RT-PCR, and added a blocking antibody to IL-5. As shown in Figure 6A, IL-5 mRNA was detected in cells grown in IL-3+GM-CSF or IL-3 alone at days 3 to 5 of culture, before the switch to predominant TM-IL-5Rα expression. Furthermore, addition of the blocking anti-IL-5 monoclonal antibody, 5A5, to CD34+ cells cultured in IL-3 or IL-3+GM-CSF inhibited eosinophil development (Figure 6B).
Up-regulation of TM-IL-5R.
Up-regulation of TM-IL-5Rα mRNA expression in umbilical cord blood CD34+ cells grown in IL-3 and GM-CSF is preceded by IL-5 mRNA expression, and anti-IL-5 inhibits eosinophil development from cord blood-derived CD34+ cells grown in IL-3 and GM-CSF. (A) RT-PCR for TM and SOL IL-5Rα mRNA and IL-5 mRNA expression by umbilical cord blood CD34+ cells grown in IL-3 or IL-3 and GM-CSF for 0, 3, 5, 7, and 14 days. Results are representative of 3 independent experiments. (B) A neutralizing anti-hIL-5 monoclonal antibody (5A5, 10 μg/mL) was added to CD34+ cord blood cells cultured at 2 × 104 cells per 100μL in 96-well plates with IL-3 and GM-CSF, as above. Three wells were set up for each time point and cells were harvested at 3, 5, 7, and 14 days of culture for cell counts after May Grunwald Giemsa or major basic protein staining. Data shown are means and standard error for 3 separate experiments.
Up-regulation of TM-IL-5R.
Up-regulation of TM-IL-5Rα mRNA expression in umbilical cord blood CD34+ cells grown in IL-3 and GM-CSF is preceded by IL-5 mRNA expression, and anti-IL-5 inhibits eosinophil development from cord blood-derived CD34+ cells grown in IL-3 and GM-CSF. (A) RT-PCR for TM and SOL IL-5Rα mRNA and IL-5 mRNA expression by umbilical cord blood CD34+ cells grown in IL-3 or IL-3 and GM-CSF for 0, 3, 5, 7, and 14 days. Results are representative of 3 independent experiments. (B) A neutralizing anti-hIL-5 monoclonal antibody (5A5, 10 μg/mL) was added to CD34+ cord blood cells cultured at 2 × 104 cells per 100μL in 96-well plates with IL-3 and GM-CSF, as above. Three wells were set up for each time point and cells were harvested at 3, 5, 7, and 14 days of culture for cell counts after May Grunwald Giemsa or major basic protein staining. Data shown are means and standard error for 3 separate experiments.
Discussion
Expression of IL-5 is increased in atopic allergic disease and helminth parasite infections and is closely implicated in the control of tissue eosinophilia.3,7,11 Control of the relative expression of the signaling, membrane-associated (TM) or soluble, antagonist (SOL) IL-5 receptor α-chain isoforms appears to be at the level of gene splicing, and may determine IL-5 responsiveness of both developing and mature eosinophils.16 17 The present data show that IL-5 itself switches to predominant expression of the TM-IL-5Rα in a minigene reporter system and during primary human eosinophil development, suggesting ligand control of receptor expression at the gene level. Furthermore, this property of IL-5 was not shared by IL-3 and GM-CSF, which share common receptor β chains, suggesting that IL-5 can signal through cytokine-specific pathways not dependent on the common β chain.
One explanation for our findings would be specific signaling via the IL-5Rα chain, not involving the β chain, and thus not shared with IL-3 and GM-CSF. IL-5 signaling has been defined via both IL-5Rα and βc, with activation of STAT1 and 5 via Jaks 1 and 2, the Ras/mitogen activated protein kinase, phosphatidyl inositol 3kinase, JNK/ERK, and SHP-2 pathways.30-33 Whether differential activation of these pathways by different receptor α chains could result in cytokine-specific downstream events remains to be established. To date, evidence for α chain-specific signaling within this cytokine subfamily is limited to selective CREB phosphorylation and subsequent Egr-1 activation in TF-1 cells34 and selective FAK up-regulation in differentiating myeloid cells35 by GM-CSF but not IL-3. Differential phosphorylation effects between IL-3 or GM-CSF and IL-5 have also been described in TF-1 cells.36 Evidence for specific differentiation signals is also suggested from work showing that transfection of FDCP-Mix cells with either the GM-CSFRα or IL-3Rα leads to either macrophage differentiation or expansion of blast cells respectively (Dr T. Whetton, personal communication, 1999). IL-5 specific signaling may also be explained by direct DNA binding of IL-5, which has been shown to possess a nuclear localization sequence and to have the capability of direct nuclear binding, at least in in vitro systems.37 Linear accumulation of translocated IL-5 instead of a receptor-mediated amplification process would also explain the slow kinetics observed in isoform switching. Alternatively, the very low IL-5Rα expression levels in untreated FDC-P1-MINI-1.1 cells may retard a full response. Signaling competence via such low expression levels has been shown to occur for IL-5-induced differentiation of B cells.38
The observed switch to TM-IL-5Rα expression by IL-5 in FDC-P1 cells was shown to result from an induced event in the whole cell population, rather than selective outgrowth of a minority population of cells expressing TM-IL-5Rα at baseline. We have not conclusively demonstrated whether this results from a switch at the level of gene transcription, but detected a similar switch in immature transcripts (data not shown), suggesting that this is indeed a switch in gene splicing. The minigene transfection system used here resulted in predominant SOL-IL-5Rα in FDC-P1 cells, in which IL-5 was shown to induce a switch to TM-IL-5Rα expression, but led to constitutive TM-IL-5Rα mRNA expression in Cos-1 cells. The IL-5-induced up-regulation of TM-IL-5Rα during development of eosinophils from CD34+ cells occurred at around day 7 of culture, but did not persist. Despite continuing exposure of the cells to IL-5, by day 14 cells expressed predominant SOL-IL-5Rα mRNA, a pattern also detected in mature peripheral blood eosinophils. Taken together, the present data suggest cell- and stage-specific signaling events and factors acting in control of IL-5Rα gene splicing. Two IL-5Rα promoter sites have been defined,39 40 although the transcription factors acting to control gene splicing remain to be established.
It is of note that recent data suggest that in mature peripheral blood eosinophils predominant expression of TM-IL-5Rα at the mRNA level is rapidly down-regulated by IL-5, IL-3, and GM-CSF at the level of transcription, although the effects of longer term cultures were not examined.41 The relative expression of IL-5Rα isoforms and its regulation in mature eosinophils remains to defined. It also needs to be mentioned that the SOL-IL-5Rα protein has antagonistic properties in vitro, which may indicate a subtle balance in the IL-5-induced control of its receptor isoform expression. However, it remains unclear which signals are required for production and secretion of this SOL-IL-5Rα isoform by eosinophils.
The data from umbilical cord blood-derived CD34+ cells suggest that IL-5-induced up-regulation of TM-IL-5Rα expression is also relevant in primary human cells, and, in particular, may play an important role in eosinophil development. The demonstration that even when eosinophils develop in culture with IL-3 and GM-CSF, this is dependent on endogenously produced IL-5, may go some way to explaining the IL-5 dependence of eosinophilia seen in vivo when compared to in vitro culture systems. Thus, although IL-3 and GM-CSF could support development of eosinophils from bone marrow cultures,42anti-IL-5 monoclonal antibody treatment or gene deletion of IL-5 largely abrogated eosinophilic responses in both helminth parasitized and inhaled allergen challenged mice.11,13,43 This is encouraging for therapeutic approaches based on selective inhibition of IL-5.12 Transgenic mice expressing IL-5 under the T-cell-dependent CD2 promoter had marked eosinophilia.44 It is of note that mice deficient in IL-5 do have bone marrow and blood eosinophils, albeit in reduced numbers, and that the eosinophil phenotype is similar to that seen in mice lacking both βcand IL-3, where IL-3 and GM-CSF could not substitute for IL-5 in driving eosinophil development.43,45 This suggests that factors other than IL-3, IL-5, or GM-CSF can induce some eosinophil development, but that IL-5 is crucial to eosinophil expansion and mobilization in allergic and parasitic disease.7 46
The current results suggest that the very low expression level of the hIL-5Rα subunit is the initial rate-limiting step in isoform switching. The slow kinetics may represent a built-in protection mechanism against premature eosinophil expansion. Progenitor cells at this stage may be dependent on IL-3 or GM-CSF or both for survival and expansion, explaining the requirement for these cytokines in eosinophil differentiation assays. Indeed, in our CD34+ culture system eosinophil expansion was not seen in IL-5 alone (data not shown). Proliferation of eosinophilic progenitor cells may then first require the up-regulation of the IL-5Rα-subunit, which is only obtained on a prolonged exposure to IL-5. Such a situation is obtained in persistent helminth parasite infections in vivo or in allergic inflammation.1,7,11 It is of note that FDCP-MIX cell lines transfected with IL-5Rα can proliferate to IL-5, but do not show increased differentiation to eosinophils.47 This suggests that either additional signaling machinery via IL-5Rα or additional factors are required for eosinophil differentiation and that IL-5 may act as a growth and survival factor for progenitor cells developing toward the eosinophil lineage via stochastic or programmed mechanisms.
Although the switch to TM-IL-5Rα was slow in transfected cell lines, taking 2 weeks, the kinetics in the CD34+ cord blood system were more rapid, with maximal expression by day 7. This may reflect differences in the cells studied or culture systems, but it is of note that increased numbers of CD34+ cells expressing TM-IL-5Rα were detected in the bone marrow as early as 24 hours after allergen inhalation challenge of asthmatics.20 In addition, CD34+/TM-IL-5Rα mRNA+ cells were recently detected in airway biopsies from atopic asthmatic subjects.19 Taken together, this suggests that switching to TM-IL-5Rα expression may be accelerated in vivo when compared to the culture systems studied and that IL-5 may act with other factors in induction of IL-5Rα on CD34 cells, which may be a key step in eosinophil development. The ability of CD34+ progenitors themselves to produce IL-5 is supported by the recent finding of IL-5 mRNA+ CD34+ cells in bone marrow of sensitized Balb/c mice after allergen challenge,48 and suggests that autocrine (and paracrine) production of IL-5 by CD34+ cells may contribute to IL-5Rα isoform switching.
The specific role of IL-5 in switching mRNA isoforms for its own receptor may have broader significance for many other cytokine receptor systems. It may also represent a specific target for therapeutic intervention in diseases such as asthma.
Acknowledgments
We wish to thank Dr Philippe Lassalle and Tania Tuypens for help in setting up the competitive RT-PCR procedure and generating the minigene constructions, respectively; Dr Geert Plaetinck, Professor Colin Sanderson, and Professor Tim Williams for helpful discussions; Professor Tony Whetton for sharing unpublished data; Professor Angel Lopez for the gift of monoclonal antibodies; and Wim Drijvers for artwork.
X.V.O. and A.V. were supported by a grant from the Verkennend Europees Onderzoek (VEO-011VO797) and the Geconcerteerde Onderzoeks Acties (GOA 96012), respectively. Part of this work was supported by the Wellcome Trust, UK, by the Medical Research Council, UK, and by GlaxoWellcome, UK.
Reprints:Jan Tavernier, Flanders Interuniversity Institute for Biotechnology, University of Ghent, Faculty of Medicine, Department of Medical Protein Research, K.L. Ledeganckstraat 35, B-9000 Ghent, Belgium; e-mail: jan.tavernier@rug.ac.be.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal