The most common chromosomal translocation in acute promyelocytic leukemia (APL), t15;17(q22;q21), creates PMLRAR andRARPML fusion genes. We previously developed a mouse model of APL by expressing PMLRAR in murine myeloid cells. In order to examine the mechanisms by which PMLRAR can initiate leukemia, we have now generated transgenic mice expressingPMLRARm4 and RARm4, proteins that are unable to activate transcription in response to retinoic acid.PMLRARm4 transgenic mice developed myeloid leukemia, demonstrating that transcriptional activation by PMLRAR is not required for leukemic transformation. The characteristics of the leukemias arising in the PMLRARm4 transgenic mice varied from those previously observed in our PMLRAR transgenic mice, indicating that ligand responsiveness may influence the phenotype of the leukemic cells. The leukemias that arose in PMLRARm4transgenic mice did not differentiate in response to retinoic acid therapy. This result supports the hypothesis that a major therapeutic effect of retinoic acid is mediated directly through thePMLRAR protein. However, a variable effect on survival suggested that this agent may be of some benefit in APL even when leukemic cells are resistant to its differentiative effects. Transgenic mice expressing high levels of RARm4 have not developed leukemia, providing evidence that the PML domain ofPMLRAR plays a specific and critical role in the pathogenesis of APL.
Retinoids are signaling molecules with significant roles in development and differentiation.1,2 These biologic effects led to the hypothesis that retinoids might be useful agents in the treatment of human malignancies. Hence, retinoids have been evaluated as possible therapies for a variety of human neoplasms including leukemias, skin cancers, cervical cancer, and neuroblastomas.3,4 Among myeloid leukemias, acute promyelocytic leukemia (APL) was found to be particularly sensitive to retinoic acid5 and more than a decade has passed since the demonstration that all-trans retinoic acid (tRA) could induce remission in patients with APL by stimulating differentiation of the leukemic cells.6 7 Understanding the pathogenesis and retinoid responsiveness of APL is important for expanding the application of retinoids in cancer treatment and for developing additional differentiation therapies.
In 1977 Rowley and colleagues8 described a specific association of APL with a t(15;17) chromosomal translocation. Subsequent to the demonstration of the therapeutic benefit of tRA in APL, the breakpoint on chromosome 17 was identified to be within a gene encoding a retinoic acid receptor, RARα.9-12 The breakpoint on chromosome 15 was identified to be within a novel gene,PML.13-17 Expression of the PMLRARαfusion is a consistent feature of the disease in the vast majority of APL patients.18 Since these discoveries, efforts have been directed at understanding the role of PMLRARα in leukemogenesis and response to therapy.
In addition to the common t(15;17) translocation, other chromosomal translocations have been identified in rare cases of APL. These translocations also result in fusions to RARα and include fusions with PLZF in t(11;17)(q23;q21),19NPM in t(5;17)(q32;q21),20 and NuMA in t(11;17)(q13;q21).21 The partners of RARαin the APL fusions are all nuclear but otherwise have limited commonality. This fact raises the possibility that all the translocations contribute to APL pathogenesis by generating abnormal retinoic acid receptors that share common transcriptional properties.
RARα is a ligand inducible transcription factor. In the absence of its ligand, tRA, RARα generally acts as a transcriptional repressor by recruiting corepressor molecules, including SMRT and N-CoR, which in turn recruit histone deacetylases. In the presence of ligand, RARα generally acts to induce transcription by releasing corepressor molecules and recruiting coactivators.22 When compared with RARα,PMLRARα has context dependent effects on transcription.13,15,17 23-25 For example, depending on cell type and the transcriptional element assayed, PMLRARα can decrease or increase basal transcription in the absence of ligand. Similarly, PMLRARα can exhibit both dominant negative activity and superactivation in the presence of tRA. Whether transcriptional activation and/or transcriptional repression byPMLRARα is necessary or sufficient for leukemogenesis has not been experimentally addressed.
Although almost all APL patients respond to tRA therapy, resistance to this agent often develops in patients so treated.26,27 It has been suggested that enhanced metabolism of tRA, increased expression of the cellular retinoic acid binding protein II, and increased expression of the multidrug resistance gene product may contribute to clinical tRA resistance (reviewed in Ding et al28 and Imaizumi et al29). However, alterations of the PMLRARα protein itself have been described in some retinoic acid resistant subclones of the NB4 APL cell line, as well as in some patients with a disease that was clinically resistant to tRA.28-31 The observed mutations in PMLRARαincluded amino acid changes that impair the ability of the protein to bind retinoic acid and to activate transcription. These findings provided evidence that loss of ligand responsiveness byPMLRARα can play a role in clinical tRA resistance.
We previously developed a murine myeloid leukemia model that recapitulates many of the features of APL.32 We have now generated additional transgenic mice to assess the role of hormone responsiveness by PMLRARα in both leukemogenesis and tRA response, as well as the sufficiency of transcriptional repression at retinoic acid response elements (RAREs) in initiation of leukemia.
Materials and methods
Preparation of plasmid constructs
The m4 mutation was introduced into the PMLRARα andRARα open-reading frames by a 2-step polymerase chain reaction (PCR) mutagenesis protocol, using 2 mutagenic primers, 5′-GATCACGCCGAAGATGGAGATCCC-3′ and 5′-ATCTTCGGC GTGATCACCCGCTC-3′. The resulting PCR-generated fragment was digested with Bcl I and Xba I and was transferred into the pSG5-RARα or pSG5-PMLRARα background. The results of mutagenesis were verified by sequencing. A pGEX-KG construct was used to express either glutathione S-transferase (GST) or a GST-SMRT fusion (encoding codons 751-1495 of human SMRT) inEscherichia coli.33
Protease resistance assay
35S-radiolabeled PMLRARα andPMLRARαm4 mutant proteins were synthesized in vitro by the TnT procedure. For the trypsin assay, 1 μL aliquots of in vitro translation product were diluted to a final volume of 20 μL each with 50 mmol/L Tris-Cl (pH 7.4) containing either tRA or an equivalent volume of ethanol carrier. Proteolysis was initiated by adding from 0 to 8 μgs of trypsin-TPCK per sample (trypsin pretreated with tosyl-L-phenylalanine chloromethyl ketone). The samples were then incubated at room temperature for 10 minutes. The proteolysis was terminated by addition of 14 μL of 5 × denaturing polyacrylamide gel electrophoresis (PAGE) sample buffer and the samples were rapidly frozen on dry ice. The samples were subsequently thawed, boiled for 10 minutes, resolved by denaturing PAGE, and visualized by autoradiography.
Transient transfections
CV-1 cell transfections were performed by a lipofection method as recommended by the manufacturer (Lipofectin, Gibco-BRL). Approximately 7 × 104 cells were transfected with 25 ng of the pSG5-RARα or pSG5-PMLRARα plasmids (representing “wild-type” or the m4 mutant), 100 ng of pCMV-lacZ (used as an internal normalization control for the efficiency of the transfection procedure) and 100 ng of the ptk-luciferase-βRARE reporter. Five hours after transfection, the cells were transferred into media either lacking or containing 1 μmol tRA. Cells were harvested 48 hours after transfection and the levels of luciferase and β-galactosidase were determined.34 35
In vitro receptor/corepressor binding assays
GST-fusion proteins were expressed in E coli and were purified and immobilized by binding to glutathione agarose as previously described.3435S-methionine–labeled full-length RARα, RARαm4, PMLRARα, andPMLRARαm4 proteins were synthesized by a coupled in vitro transcription and translation system (Promega TnT kit, Promega, Madison, WI). The radiolabeled proteins were subsequently incubated with the immobilized GST fusion proteins in HEMG buffer in the presence or absence of tRA, the agarose matrix was extensively washed and bound proteins were eluted with free glutathione and analyzed by denaturing PAGE.33 The electrophoretograms were visualized and quantified by phosphorimager analysis (Molecular Dynamics STORM system, Molecular Dynamics, Sunnyvale, CA).
Generation of transgenic mice
Western blotting and immunofluorescence
Western blotting was performed as previously described with a rabbit polyclonal antiserum raised against a GST-fusion protein, encompassing amino acids 420-462 of the human RARα protein (anti-RARαF).32,39 Whole-cell lysates of bone marrow from control and transgenic mice were subjected to denaturing PAGE on 8% or 12% SDS-polyacrylamide gels and were transferred to nitrocellulose. Immunofluorescence analysis of bone marrow cells was performed essentially as described40 but using the anti-RARαF antiserum at a 1:150 dilution.
Isolation of cells from tissues, cell staining, and fluorescence-activated cell sorting
Peripheral blood counts
Blood was analyzed on a Hemavet veterinary hematology analyzer to assess white blood cell counts, hemoglobin, and platelet counts. White blood cell differential counts were performed on peripheral blood smears.
Methylcellulose cultures
Bone marrow cells were cultured in duplicate in 35 mm petri dishes in Methocult M3230 methylcellulose medium (StemCell Technologies, Vancouver, BC) supplemented with either 50 units/mL G-CSF (Boehringer Mannheim), or 2.5 ng/mL GM-CSF (StemCell Technologies) plus 2% Xg63Ag8-653-IL341 conditioned medium. One milliliter cultures contained 5 × 104 viable bone marrow cells. Analysis was as previously described.40
Transplantations
Cells isolated from bone marrow and spleens of leukemic animals were resuspended in buffered saline and injected into the tail veins of 6- to 12-week-old FVB/N mice, 5 × 106 viable cells/recipient. Nonleukemic bone marrow isolated from PMLRARαm4transgenic founder #4048 was transplanted into lethally irradiated FVB/N mice as previously described.32
Treatment with all-trans retinoic acid
Leukemic mice were treated by subcutaneous implantation of 21-day release pellets containing 5 mg tRA or placebo (Innovative Research of America). Morphologic differentiation by tRA was assessed on days 4 and 11 of therapy.
Results
Generation of transgenic mice
We generated transgenic mice expressing a PMLRARα unable to activate transcription as well as transgenic mice expressing anRARα with dominant negative activity. For this purpose, we introduced a Leu to Pro mutation at amino acid 398 of RARαinto cDNAs encoding PMLRARα andRARα (Figure 1A). This mutation was originally identified by Shao and colleagues31in a retinoic acid resistant subclone of human APL cells (NB4-R4) and was designated the m4 mutation. The m4 mutation impairs ligand binding, abrogates ligand-induced transcriptional activation, and blocks ligand-induced release of SMRT corepressor.31,42Furthermore, PMLRARαm4 and RARαm4 act as dominant negative inhibitors of tRA-induced transcription.31
PMLRARm4 and RARm4.
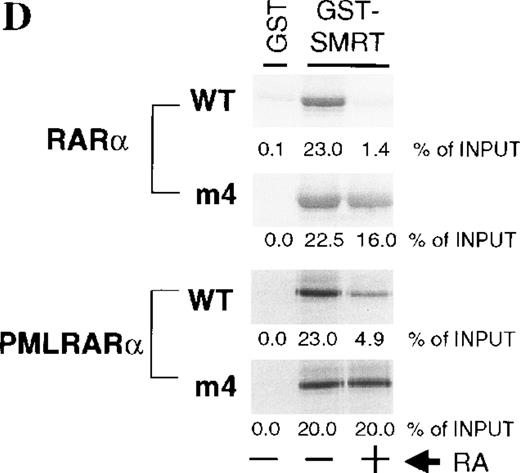
A, PMLRARαm4: the Leu to Pro point mutation at codon 398 ofRARα was introduced into a human PMLRARα cDNA whose chromosome 15 breakpoint lies in breakpoint cluster 1.16The PML portion of the fusion is shown with hatching with selected structural domains labeled and shown in white. The B-F domains that encompass the RARα portion of the fusion are labeled and functional regions are noted. RARαm4: the Leu to Pro point mutation was introduced into a human RARα1 cDNA. B, Hormone binding by PMLRARα (“Wild-type,” WT) andPMLRARαm4 (m4 mutant). Radiolabeled proteins synthesized by in vitro transcription and translation were incubated without or with increasing amounts of trypsin (indicated above the panels) in the absence or presence of 1 μmol tRA. The protein products were resolved by denaturing PAGE and visualized by autoradiography. The arrows show the position of the full-length undigested proteins. Smaller bands represent partially degraded products. C, Dominant negative activity of the m4 mutant proteins. CV-1 cells were transiently transfected with pSG5 constructs containing no exogenous receptor, RARα (WT),RARαm4, PMLRARα (WT), and PMLRARαm4. Luciferase activity expressed from a cotransfected βRARE-luciferase reporter gene was normalized to β-galactosidase activity from a cotransfected pCH210-LacZ plasmid. D, Decreased hormone-induced dissociation of SMRT corepressor by the m4 mutant proteins. GST-SMRT fusion protein was synthesized in E coli and was immobilized on glutathione agarose. The different receptor proteins were synthesized by in vitro transcription and translation and were incubated with the immobilized GST-SMRT in the absence or presence of 1 μmol tRA, as indicated below the panels. Equivalent amounts of GST-SMRT and radiolabeled receptor protein were used for each panel. Nonrecombinant GST, immobilized on glutathione agarose, was used in parallel as a negative control. The radiolabeled receptors remaining bound to the GST or GST-SMRT matrix after washing were eluted, were resolved by denaturing PAGE, and were visualized and quantified by phosphorimager analysis. The amount of radiolabeled receptor bound to the GST or GST-SMRT matrix, relative to the amount of receptor input in each binding reaction, is displayed beneath each panel.
PMLRARm4 and RARm4.
A, PMLRARαm4: the Leu to Pro point mutation at codon 398 ofRARα was introduced into a human PMLRARα cDNA whose chromosome 15 breakpoint lies in breakpoint cluster 1.16The PML portion of the fusion is shown with hatching with selected structural domains labeled and shown in white. The B-F domains that encompass the RARα portion of the fusion are labeled and functional regions are noted. RARαm4: the Leu to Pro point mutation was introduced into a human RARα1 cDNA. B, Hormone binding by PMLRARα (“Wild-type,” WT) andPMLRARαm4 (m4 mutant). Radiolabeled proteins synthesized by in vitro transcription and translation were incubated without or with increasing amounts of trypsin (indicated above the panels) in the absence or presence of 1 μmol tRA. The protein products were resolved by denaturing PAGE and visualized by autoradiography. The arrows show the position of the full-length undigested proteins. Smaller bands represent partially degraded products. C, Dominant negative activity of the m4 mutant proteins. CV-1 cells were transiently transfected with pSG5 constructs containing no exogenous receptor, RARα (WT),RARαm4, PMLRARα (WT), and PMLRARαm4. Luciferase activity expressed from a cotransfected βRARE-luciferase reporter gene was normalized to β-galactosidase activity from a cotransfected pCH210-LacZ plasmid. D, Decreased hormone-induced dissociation of SMRT corepressor by the m4 mutant proteins. GST-SMRT fusion protein was synthesized in E coli and was immobilized on glutathione agarose. The different receptor proteins were synthesized by in vitro transcription and translation and were incubated with the immobilized GST-SMRT in the absence or presence of 1 μmol tRA, as indicated below the panels. Equivalent amounts of GST-SMRT and radiolabeled receptor protein were used for each panel. Nonrecombinant GST, immobilized on glutathione agarose, was used in parallel as a negative control. The radiolabeled receptors remaining bound to the GST or GST-SMRT matrix after washing were eluted, were resolved by denaturing PAGE, and were visualized and quantified by phosphorimager analysis. The amount of radiolabeled receptor bound to the GST or GST-SMRT matrix, relative to the amount of receptor input in each binding reaction, is displayed beneath each panel.
Before producing transgenic animals, we validated the characteristics of our PMLRARαm4 and RARαm4 cDNA constructs, comparing our results with those previously reported. The effects of the m4 mutation on ligand binding had been assessed by evaluating the binding of 3H-tRA to PMLRARαm4 orRARαm4 in nuclear extracts of transiently transfected Cos-1 cells.31 In these assays, no ligand binding was observed. We used a sensitive protease-resistance assay to determine whether the m4 mutation fully abolished ligand binding. Condensation of a hormone-binding domain of a nuclear receptor around the hormone ligand can result in a protease-resistant core. Gain of protease resistance has been seen with many nuclear hormone receptors and has been used as a measure of ligand occupancy.43 44 RadiolabeledPMLRARα and PMLRARαm4 protein were incubated with increasing amounts of trypsin in the absence or presence of 1 μmol tRA. The products were then resolved by denaturing PAGE and visualized by phosphor imaging (Figure 1B). PMLRARα was readily degraded by proteolytic treatment in the absence of hormone, but produced a protease-resistant polypeptide in the presence of hormone.PMLRARαm4 also exhibited some protease resistance in the presence of 1 μmol tRA, although it was less resistant thanPMLRARα. Ten- to 20-fold higher concentrations of hormone were required to produce a resistant polypeptide fromPMLRARαm4 than from PMLRARα (data not shown).
Although tRA was able to bind weakly to PMLRARαm4, when we examined the effects of the m4 mutation on transcriptional activity and association with SMRT corepressor, our results were similar to published analyses. PMLRARα, PMLRARαm4,RARα, and RARαm4 were transiently expressed in CV-1 cells and transcription from a βRARE response element in the absence or presence of tRA was assessed. In contrast to PMLRARα andRARα, the m4 mutant proteins strongly inhibited transcriptional activation (Figure 1C). In cell-free protein assays to measure receptor interaction with SMRT, the m4 mutants, in contrast toRARα, were unable to release the corepressor on addition of 1 μmol tRA (Figure 1D). As has been previously reported,42 45PMLRARα was itself somewhat less efficient at releasing corepressor than was RARα.
Expression of the transgenes
The MRP8 promoter element can drive transgene expression in myeloid cells, including myeloblasts, neutrophils, and monocytes.32,36,40 Western blotting of bone marrow was performed using a rabbit polyclonal antiserum raised to humanRARαF.32 The results are summarized in Tables1 and 2, and representative data are shown (Figure 2A and B). Although the murine peptide differs from the human by only 4 of 43 amino acids, the antiserum recognizes murine RARα poorly and as a result endogenous murine RARα is not seen on these blots. PMLRARαm4 protein was present in 5 of 7 lines ofMRP8-PMLRARαm4 transgenic mice analyzed. Levels of expression in 2 of the lines appeared comparable to levels of PMLRARα in our highest expressing MRP8-PMLRARα mice (Figure 2A).RARαm4 protein was present in 5 of 7 lines ofMRP8-RARαm4 transgenic mice analyzed. Levels of expression in 2 of the lines appeared to exceed the levels of PMLRARα in our highest expressing MRP8-PMLRARα mice (Figure 2B).
PMLRARm4 transgenic mice
| Mouse . | Transgene Expression in Bone Marrow . | Age at Death (days) . | Reason for Death . |
|---|---|---|---|
| 4042 | + | 93 | Leukemia |
| 4048* | ++ | 37 | Abnormal Skin Growth† |
| 4048.2 | 189 | Leukemia | |
| 4048.3 | 215 | Leukemia | |
| 4048.4 | 259 | Leukemia | |
| 4048.5 | 328 | Leukemia | |
| 4048.6 | 341 | Abdominal Sarcoma‡ | |
| 4099 | ++ | 156 | Leukemia |
| 4100 | − | 548 | Study1-153 |
| 4104 | + | 235 | Leukemia |
| 4105 | + | 235 | Abnormal Skin Growth |
| 4114 | − | 235 | Abnormal Skin Growth |
| Mouse . | Transgene Expression in Bone Marrow . | Age at Death (days) . | Reason for Death . |
|---|---|---|---|
| 4042 | + | 93 | Leukemia |
| 4048* | ++ | 37 | Abnormal Skin Growth† |
| 4048.2 | 189 | Leukemia | |
| 4048.3 | 215 | Leukemia | |
| 4048.4 | 259 | Leukemia | |
| 4048.5 | 328 | Leukemia | |
| 4048.6 | 341 | Abdominal Sarcoma‡ | |
| 4099 | ++ | 156 | Leukemia |
| 4100 | − | 548 | Study1-153 |
| 4104 | + | 235 | Leukemia |
| 4105 | + | 235 | Abnormal Skin Growth |
| 4114 | − | 235 | Abnormal Skin Growth |
Relative levels based on visual comparison of western blots. ++: expression equal to or greater than PMLRARα expression in the highest expressing line of MRP8-PMLRARα transgenic mice (line 556). +: expression present but less than PMLRARα expression in line 556. +/−: minimal expression. −: no signal on western blot.
When nonleukemic founder 4048 was euthanized at 37 days of age, its bone marrow was transplanted into lethally irradiated nontransgenic FVB/N mice. These mice were designated as 4048.1-4048.6. Mouse 4048.1 was euthanized for assessment of transgene expression and the other 5 recipient mice were monitored for illness.
Founders 4042, 4048, 4099, 4105, and 4114 exhibited abnormal skin growth similar to that seen in our MRP8-PMLRARα transgenic mice (data not shown).
Radiation can induce sarcomas in FVB/N recipients of transgenic marrow.
This founder was euthanized at 18 months of age for examination and assessment of transgene expression.
RARm4 transgenic mice
| Founder . | Transgene Expression in Bone Marrow . | Cohort . |
|---|---|---|
| 4142 | ++ | 26 mice. 10-18 mo |
| 4151 | + | 10 mice. 10-18 mo |
| 4155 | ND | Founder, 13 mo |
| No offspring. | ||
| 4157 | +/− | Founder, 9 months* |
| 4159 | + | 2 mice, 11 & 18 mo |
| 4160 | − | Founder, 9 mo* |
| 4192 | ++ | 5 mice, 10-18 mo |
| 4195 | − | Founder, 9 mo* |
| Founder . | Transgene Expression in Bone Marrow . | Cohort . |
|---|---|---|
| 4142 | ++ | 26 mice. 10-18 mo |
| 4151 | + | 10 mice. 10-18 mo |
| 4155 | ND | Founder, 13 mo |
| No offspring. | ||
| 4157 | +/− | Founder, 9 months* |
| 4159 | + | 2 mice, 11 & 18 mo |
| 4160 | − | Founder, 9 mo* |
| 4192 | ++ | 5 mice, 10-18 mo |
| 4195 | − | Founder, 9 mo* |
See note to Table 1.
Founders 4157, 4160, and 4195 (and their offspring) were monitored for illness for up to 9 months. Monitoring was then discontinued given very low to absent transgene expression.
ND: not determined.
Expression of the transgenes.
A, B, Whole cell lysates of bone marrow were subjected to denaturing PAGE and Western blotting using a rabbit polyclonal antihumanRARαF domain antibody. Signals corresponding to transgenically expressed PMLRARα and RARα proteins are indicated by arrows. Locations of size markers are indicated by lines. (A) Protein expression in nonleukemic bone marrow of the highest expressing MRP8-PMLRARα transgenic line (Tg556-PR) and in 2 of the MRP8-PMLRARαm4 mice, Tg4048-PRm4 (nonleukemic bone marrow) and Tg4099-PRm4 (leukemic bone marrow). 8% SDS-polyacrylamide. (B) Protein expression in nonleukemic bone marrow Tg556-PR and in the marrows of healthy MRP8-RARαm4 transgenic mice from 3 lines, Tg4142-Rm4, Tg4151-Rm4, and Tg4192-Rm4. 12% SDS-polyacrylamide. C, Immunofluorescence analysis of bone marrow neutrophilic cells, anti-RARαF antiserum and Hoechst 33258, 1300 × .
Expression of the transgenes.
A, B, Whole cell lysates of bone marrow were subjected to denaturing PAGE and Western blotting using a rabbit polyclonal antihumanRARαF domain antibody. Signals corresponding to transgenically expressed PMLRARα and RARα proteins are indicated by arrows. Locations of size markers are indicated by lines. (A) Protein expression in nonleukemic bone marrow of the highest expressing MRP8-PMLRARα transgenic line (Tg556-PR) and in 2 of the MRP8-PMLRARαm4 mice, Tg4048-PRm4 (nonleukemic bone marrow) and Tg4099-PRm4 (leukemic bone marrow). 8% SDS-polyacrylamide. (B) Protein expression in nonleukemic bone marrow Tg556-PR and in the marrows of healthy MRP8-RARαm4 transgenic mice from 3 lines, Tg4142-Rm4, Tg4151-Rm4, and Tg4192-Rm4. 12% SDS-polyacrylamide. C, Immunofluorescence analysis of bone marrow neutrophilic cells, anti-RARαF antiserum and Hoechst 33258, 1300 × .
To further substantiate that the transgenes were expressed in myeloid cells, bone marrow was also analyzed by immunofluorescence using the anti-RARαF antiserum. Cells with ring-shaped nuclei, as revealed by a fluorescent DNA-binding dye, are primarily neutrophilic. Transgene expression in such neutrophilic cells was observed in bothPMLRARαm4 and RARαm4 transgenic animals (Figure2C). A range of staining intensity was seen and neutrophilic cells without visibly detectable protein were also present. The speckled nuclear staining present in PMLRARαm4 transgenic mice was similar to that seen in PMLRARα transgenic mice. The nuclear staining observed in RARαm4 mice lacked the distinct speckles present in the other transgenics.
PMLRARm4 transgenic mice develop leukemia
Leukemias developed in 4 of 7 founders/lines ofMRP8-PMLRARαm4 transgenic mice (Table 1), a frequency similar to that encountered previously in MRP8-PMLRARα transgenic mice.32 The latency until leukemia onset, 3 to 11 months, was also comparable to that seen in our MRP8-PMLRARα mice (Table 1; see also Brown et al32). Assessment of leukemia penetrance was hampered by the fact that because of early illness, poor reproduction, or lack of transgene transmission, we did not obtain transgenic offspring for any of the lines in which leukemias developed. Nevertheless, it was apparent that PMLRARαm4 could readily initiate leukemia: 3 of 7 independent founder mice developed leukemia and 4 of 5 mice that were reconstituted with the nonleukemic bone marrow of a fourth independent founder also developed leukemia (Table1).
The leukemias arising in the PMLRARαm4 transgenic mice were acute leukemias with promyelocytic features. The peripheral blood of the leukemic animals was characterized by anemia, thrombocytopenia, and the presence of leukemic cells at the blast/promyelocyte stage of neutrophilic differentiation (Table 3). The leukemic bone marrows had large numbers of early myeloid cells, many of which had numerous azurophilic primary granules (Figure3B). These early cells also stained strongly with Sudan Black B (Figure 3D). The morphology and strong Sudan Black B staining were indicative of the promyelocytic character of the leukemic cells. The leukemias caused hepatomegaly and splenomegaly and were invasive, being present not only in the periportal areas of the liver but also invading the liver parenchyma (Figure 3F).
PMLRARm4 leukemias blood
| Mice . | WBC . | HGB . | PLT . | Lymph . | Blast + Pro . | Imm Neut . | Mat Neut . | Mono . | Eosin . |
|---|---|---|---|---|---|---|---|---|---|
| Control | 3.0 ± 2.1 | 13.6 ± 0.3 | 1064 ± 69 | 89.9 ± 5.4 | 0 | 0 | 7.9 ± 3.9 | 1.7 ± 2.0 | 0.4 ± 0.2 |
| PR | 4.5 ± 5.0 | 7.8 ± 1.8 | 334 ± 69 | 87.2 ± 7.2 | 6.8 ± 4.7 | 3.2 ± 2.2 | 2.0 ± 1.5 | 0.5 ± 0.8 | 0.3 ± 0.8 |
| PRm4 | 67.7 ± 50.7 | 7.8 ± 1.2 | 483 ± 192 | 48.8 ± 25.3 | 16.9 ± 14.0 | 13.9 ± 10.5 | 17.6 ± 19.1 | 2.4 ± 3.5 | 0.3 ± 0.5 |
| Leukemia | |||||||||
| 4042 | 101.33-150 | 8.73-150 | 6313-150 | 52 | 11 | 11 | 17 | 8 | 1 |
| 4048.2 | 3.73-150 | 93-150 | 3173-150 | 88 | 8 | 3 | 1 | 0 | 0 |
| 4048.3 | Not increased | NA | NA | 23 | 9 | 20 | 47 | 0 | 1 |
| 4048.5 | 6.6 | 7.7 | 389 | 61 | 13.5 | 16.5 | 3.5 | 5.5 | 0 |
| 4099 | 1413-150 | 7.43-150 | 7423-150 | 49 | 45 | 3 | 3 | 0 | 0 |
| 4104 | 86.1 | 6.1 | 335 | 20 | 15 | 30 | 34 | 1 | 0 |
| Mice . | WBC . | HGB . | PLT . | Lymph . | Blast + Pro . | Imm Neut . | Mat Neut . | Mono . | Eosin . |
|---|---|---|---|---|---|---|---|---|---|
| Control | 3.0 ± 2.1 | 13.6 ± 0.3 | 1064 ± 69 | 89.9 ± 5.4 | 0 | 0 | 7.9 ± 3.9 | 1.7 ± 2.0 | 0.4 ± 0.2 |
| PR | 4.5 ± 5.0 | 7.8 ± 1.8 | 334 ± 69 | 87.2 ± 7.2 | 6.8 ± 4.7 | 3.2 ± 2.2 | 2.0 ± 1.5 | 0.5 ± 0.8 | 0.3 ± 0.8 |
| PRm4 | 67.7 ± 50.7 | 7.8 ± 1.2 | 483 ± 192 | 48.8 ± 25.3 | 16.9 ± 14.0 | 13.9 ± 10.5 | 17.6 ± 19.1 | 2.4 ± 3.5 | 0.3 ± 0.5 |
| Leukemia | |||||||||
| 4042 | 101.33-150 | 8.73-150 | 6313-150 | 52 | 11 | 11 | 17 | 8 | 1 |
| 4048.2 | 3.73-150 | 93-150 | 3173-150 | 88 | 8 | 3 | 1 | 0 | 0 |
| 4048.3 | Not increased | NA | NA | 23 | 9 | 20 | 47 | 0 | 1 |
| 4048.5 | 6.6 | 7.7 | 389 | 61 | 13.5 | 16.5 | 3.5 | 5.5 | 0 |
| 4099 | 1413-150 | 7.43-150 | 7423-150 | 49 | 45 | 3 | 3 | 0 | 0 |
| 4104 | 86.1 | 6.1 | 335 | 20 | 15 | 30 | 34 | 1 | 0 |
Data are shown as arithmetic means ± SD. WBC: White blood cell count in 1000/μL. HGB: Hemoglobin in gms/dL. PLT: Platelet Count in 1000/μL. Percentages of nucleated leukocytes were derived from 200 cell differential counts of Wright's Giemsa stained blood smears. Lymph: lymphocytes. Blast + Pro: blasts & promyelocytes. Imm Neut: neutrophilic myelocytes & metamyelocytes. Mat Neut: neutrophilic band, mature ring, & polymorphonuclear forms. Mono: monocytes. Eosin: eosinophils. WBC, HGB, and PLT: Control n = 7, PR (MRP8-PMLRARα) primary leukemias n = 10, PRm4 (MRP8-PMLRARαm4) n = 5 (leukemic mice 4104 & 4048.5 and transplant recipients of leukemias 4042, 4048.2, and 4099). Differential counts: Control n = 7, PR primary leukemias n = 6, PRm4 primary leukemias n = 6.
WBC, HGB, PLT obtained on initial transplant recipients. Peripheral blood was not obtained from leukemic mouse 4048.4. NA: not available.
Acute leukemia in MRP8-PMLRARm4 transgenic mice.
(A, C, E) Samples from control mice. (B, D, F) Samples from leukemicPMLRARαm4 mice. (B) Founder 4099. (D) Transplanted leukemia 4048.2. (F) founder 4042. (A, B) Bone marrow, Wright's Giemsa stain, 500 × . (C, D) Bone marrow, Sudan Black B stain, 965 × . (E, F) Liver, Hematoxylin and eosin stain, 200 × .
Acute leukemia in MRP8-PMLRARm4 transgenic mice.
(A, C, E) Samples from control mice. (B, D, F) Samples from leukemicPMLRARαm4 mice. (B) Founder 4099. (D) Transplanted leukemia 4048.2. (F) founder 4042. (A, B) Bone marrow, Wright's Giemsa stain, 500 × . (C, D) Bone marrow, Sudan Black B stain, 965 × . (E, F) Liver, Hematoxylin and eosin stain, 200 × .
The leukemias were readily transplantable to histocompatible normal mice. Four of the leukemias were each transplanted by intravenous injection into 6 healthy unirradiated nontransgenic FVB/N animals. The cells engrafted and leukemias developed in all 24 recipient mice. The leukemias were subsequently maintained by serial transplantation in vivo and by cryopreservation.
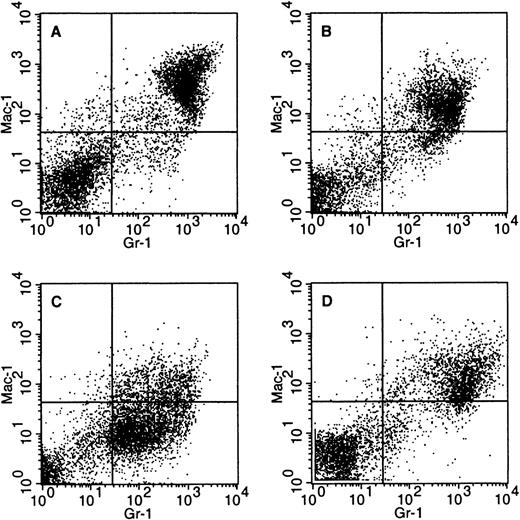
The PMLRARαm4 leukemias exhibited variability. White blood cell counts ranged from normal to markedly elevated (Table 3). Peripheral blood leukocytes included significant numbers of maturing neutrophilic cells in some but not all cases. Similarly, although the bone marrows of some mice were effaced with promyelocytes, neutrophilic cells maturing beyond the promyelocyte stage were present in other animals (Table 4). Flow cytometric analysis with Gr-1 and Mac-1 markers revealed that leukemia 4099 had the low-level expression pattern typical of leukemias in ourPMLRARα transgenic mice, but that the 4042 and 4048.2 leukemias expressed these markers at moderately high levels (Figure4). The observation that some of thePMLRARαm4 leukemias were associated with increased white blood cell counts and significant numbers of maturing neutrophilic cells in the blood and bone marrow contrasts with the leukemic phenotype previously observed in our PMLRARα transgenic mice (Tables 3 and 4).
PMLRARm4 leukemias bone marrow
| Mice . | Blast + Pro . | Imm Neut . | Mat Neut . | Erythroid . | Lymph . | Eosin . |
|---|---|---|---|---|---|---|
| Control | 2.0 ± 0.7 | 16.4 ± 2.4 | 27.0 ± 5.0 | 40.3 ± 6.5 | 12.1 ± 1.9 | 2.3 ± 0.9 |
| PR | 81.3 ± 8.1 | 4.5 ± 2.6 | 0.7 ± 0.7 | 8.7 ± 7.7 | 4.5 ± 2.2 | 0.3 ± 0.3 |
| PRm4 | 65.7 ± 27.1 | 13.1 ± 14.4 | 12.7 ± 17.5 | 3.5 ± 2.4 | 4.4 ± 2.4 | 0.5 ± 1.1 |
| Leukemia | ||||||
| 4042 | 44.5 | 37.5 | 5.5 | 4 | 6 | 2.5 |
| 4048.3 | 66.5 | 11 | 15.5 | 0.5 | 6.5 | 0 |
| 4048.5 | 94.5 | 0.5 | 0 | 3.5 | 1.5 | 0 |
| 4099 | 90 | 4.5 | 0.5 | 2.5 | 2 | 0 |
| 4104 | 33 | 12 | 42 | 7 | 6 | 0 |
| Mice . | Blast + Pro . | Imm Neut . | Mat Neut . | Erythroid . | Lymph . | Eosin . |
|---|---|---|---|---|---|---|
| Control | 2.0 ± 0.7 | 16.4 ± 2.4 | 27.0 ± 5.0 | 40.3 ± 6.5 | 12.1 ± 1.9 | 2.3 ± 0.9 |
| PR | 81.3 ± 8.1 | 4.5 ± 2.6 | 0.7 ± 0.7 | 8.7 ± 7.7 | 4.5 ± 2.2 | 0.3 ± 0.3 |
| PRm4 | 65.7 ± 27.1 | 13.1 ± 14.4 | 12.7 ± 17.5 | 3.5 ± 2.4 | 4.4 ± 2.4 | 0.5 ± 1.1 |
| Leukemia | ||||||
| 4042 | 44.5 | 37.5 | 5.5 | 4 | 6 | 2.5 |
| 4048.3 | 66.5 | 11 | 15.5 | 0.5 | 6.5 | 0 |
| 4048.5 | 94.5 | 0.5 | 0 | 3.5 | 1.5 | 0 |
| 4099 | 90 | 4.5 | 0.5 | 2.5 | 2 | 0 |
| 4104 | 33 | 12 | 42 | 7 | 6 | 0 |
Data are shown as arithmetic means ± SD. Percentages of nucleated cells were derived from 400 cell differential counts of Wright's Giemsa stained bone marrow smears. Abbreviations: see legend to Table 3. Control n = 6, PR (MRP8-PMLRARα) primary leukemias n = 6, PRm4 (MRP8-PMLRARαm4) primary leukemias n = 5. Bone marrow smears were not available for leukemic mice 4048.2 and 4048.4.
Variable surface marker expression in PMLRARm4leukemias.
Bone marrow cells were stained with Gr-1 and Mac-1 antibodies that recognize myeloid surface antigens. Dead cells were eliminated from the analysis on the basis of staining with propidium iodide. (A) Control. (B) Leukemic mouse 4042 (C) Leukemic mouse 4099 (D) Transplanted leukemia 4048.2.
Variable surface marker expression in PMLRARm4leukemias.
Bone marrow cells were stained with Gr-1 and Mac-1 antibodies that recognize myeloid surface antigens. Dead cells were eliminated from the analysis on the basis of staining with propidium iodide. (A) Control. (B) Leukemic mouse 4042 (C) Leukemic mouse 4099 (D) Transplanted leukemia 4048.2.
Although PMLRARαm4 leukemias were invasive transplantable diseases, they were not always associated with an aggressive clinical course. In our experience with PMLRARα leukemias, the interval between when a leukemic mouse appears ill and when it has progressed to a moribund condition is usually very short, on the order of 1 to 7 days. We were therefore surprised when we noted that mice ill with PMLRARαm4 leukemia did not necessarily exhibit rapid deterioration. Leukemic mice 4042 and 4099 were euthanized at the time their leukemias were initially apparent as determined by visible signs of illness. The primary transplant recipients of these 2 leukemias did not rapidly deteriorate after their leukemias became clinically apparent, living with their disease for weeks to months. Leukemic mice 4048.2 and 4104 were not euthanized at the time their leukemias were initially apparent, but were killed more than 2 weeks later. On transplantation, these leukemias were more rapidly fatal (Table5). Serial transplantation of the leukemias was, in some instances, accompanied by changes in features of the disease that may reflect the accumulation of additional genetic abnormalities. For example, although the first recipients of leukemia 4042 survived with the leukemia for an extended period and exhibited high peripheral white blood cell counts, mice that received the third serial transplant of this leukemia died rapidly without developing a peripheral blood leukocytosis.
Survival of primary recipients of PMLRAR and PMLRARm4 leukemias
| . | Survival of Primary Transplant Recipients (days) . | ||
|---|---|---|---|
| Mean . | Median . | Range . | |
| PMLRARα Leukemias | 38 | 34 | 31-60 |
| PMLRARαm4 Leukemias | |||
| 4042 | 135 | 136 | 119-154 |
| 4048.2 | 44 | 42 | 42-50 |
| 4099 | 71 | 73 | 60-85 |
| 4104 | 31 | 31 | 28-34 |
| . | Survival of Primary Transplant Recipients (days) . | ||
|---|---|---|---|
| Mean . | Median . | Range . | |
| PMLRARα Leukemias | 38 | 34 | 31-60 |
| PMLRARαm4 Leukemias | |||
| 4042 | 135 | 136 | 119-154 |
| 4048.2 | 44 | 42 | 42-50 |
| 4099 | 71 | 73 | 60-85 |
| 4104 | 31 | 31 | 28-34 |
Results obtained with 3 different PMLRARα leukemias and 4 different PMLRARαm4 leukemias that were transplanted into healthy FVB/N mice are shown (5-6 recipients for each leukemia). ThePMLRARα leukemias were similar to each other and the combined data are presented.
All-trans retinoic acid does not cause differentiation ofPMLRARm4 leukemias
Mice that were recipients of 3 different PMLRARαm4leukemias (4042, 4048.2, 4099) were treated with placebo or tRA to ascertain the effects of the Leu to Pro mutation on tRA responsiveness. For our PMLRARα leukemias, tRA generally causes a rapid rise in the peripheral leukocyte count as leukemic promyelocytes differentiate to mature neutrophils (25-fold average increase in leukocyte count on day 4 of therapy). Morphologic differentiation is readily apparent in the bone marrow of tRA-treated animals (Figure5A and B; see also Brown et al32) and regression of the leukemia is seen in histologic sections of liver (Figure 5C and D). In contrast, examination of the bone marrow and liver of tRA-treated PMLRARαm4 leukemic mice revealed that tRA did not induce morphologic differentiation and disease regression (Figure 5E through L). In addition, unlikePMLRARα leukemias, retinoic acid therapy did not cause a rapid rise in the peripheral white blood cell count (1.5-fold average increase in leukocyte count on day 4 of therapy).
Retinoic acid response of PMLRARm4 leukemias.
Leukemic mice were treated with placebo (A, C, E, G, I, K) or tRA (B, D, F, H, J, L). (A-D) PMLRARα expressing leukemia. (E-H)PMLRARαm4 leukemia 4099. (I-J) PMLRARαm4 leukemia 4042. (K-L) PMLRARαm4 leukemia 4048.2. (A, B, E, F, I-L) Bone marrow, Wright's Giemsa stain, 350 × . (C, D, G, H) Liver, Hematoxylin and eosin stain, 140 × . Effects of 11 days of tRA therapy are shown.
Retinoic acid response of PMLRARm4 leukemias.
Leukemic mice were treated with placebo (A, C, E, G, I, K) or tRA (B, D, F, H, J, L). (A-D) PMLRARα expressing leukemia. (E-H)PMLRARαm4 leukemia 4099. (I-J) PMLRARαm4 leukemia 4042. (K-L) PMLRARαm4 leukemia 4048.2. (A, B, E, F, I-L) Bone marrow, Wright's Giemsa stain, 350 × . (C, D, G, H) Liver, Hematoxylin and eosin stain, 140 × . Effects of 11 days of tRA therapy are shown.
We also assessed the clinical effectiveness of tRA in the treatment of leukemias that arose in the PMLRARαm4 transgenic mice. To this end, we studied the effect of tRA on survival of mice that were recipients of 2 independent leukemias, leukemia 4048.2 and the aggressive variant of leukemia 4042 that arose on serial passaging. Mice that received leukemic cells by intravenous injection were treated with placebo or tRA when ill. Four recipients of leukemia 4042 treated with placebo died on days 5 and 6 of therapy and 5 mice treated with tRA also died rapidly, on days 5 to 10. Although the difference in survival was statistically significant (P = .04 by log-rank test), the rapid demise of tRA treated animals contrasted with our previous experience with PMLRARαleukemias.32 46 Unexpectedly, although tRA did not cause morphologic differentiation of leukemia 4048.2 (Figure 5K through L), it nevertheless substantially prolonged survival of the leukemic animals: 3 placebo-treated mice died on days 8 and 9 of therapy, whereas 3 tRA-treated mice were still alive at the end of 21 days of tRA treatment (P = .015 by log-rank test).
RARm4 transgenic mice are healthy
None of the MRP8-RARαm4 transgenic mice developed leukemia in up to 18 months of observation (Table 2). This result contrasts with our observations in the MRP8-PMLRARα andMRP8-PMLRARαm4 transgenic mice: in these mice, leukemias developed in more than half of the independently derived founders/lines beginning at 3 months of age, and by 10 months of age leukemia had appeared in one third of the mice of the highest expressingPMLRARα line32 and, as noted previously, in 4 of 5 mice derived from a high-expressing PMLRARαm4 founder.
Although leukemias did not develop in the RARαm4 transgenic mice, we investigated whether the RARαm4 protein altered neutrophil development in 1 of the highest expressing lines, line 4142. Expression of RARαm4 did not alter peripheral white blood cell counts. In the bone marrow, there was a trend toward increased immature neutrophilic cells, but this trend did not reach statistical significance (controls n = 9, line 4142 mice n = 6,P > .05, data not shown). In our previous work, we had observed that MRP8-PMLRARα and MRP8-PEBP2βMYH11transgenic mice exhibited a modest shift in the bone marrow toward immature neutrophilic cells that was accompanied by increased expression of the Mac-1 cell surface antigen (Kogan et al40and unpublished observations). When stained with Mac-1, the mean fluorescence of the myeloid cells in the bone marrow ofRARαm4 transgenic mice was 2.4-fold greater than that observed in controls (controls n = 5, line 4142 mice n = 5, data not shown). This increase was statistically significant (P = .002 by Student t test) and is consistent with the trend toward increased immature neutrophilic cells observed by morphologic examination. We also compared the colony-forming units present in the bone marrow of control and RARαm4 transgenic mice. Bone marrow cells from groups of 3 healthy untreated mice were grown in methylcellulose cultures in the presence of either G-CSF or a combination of GM-CSF and IL-3. Neither the number of colony-forming units nor the morphology of the cells as assessed on cytospins were significantly different between RARαm4 transgenic and control mice (data not shown).
Discussion
We and others had demonstrated that directing expression of thePMLRARα fusion protein to immature mouse myeloid cells initiated leukemias with promyelocytic features.32,47 48 We have now similarly expressed altered forms of PMLRARα andRARα that are unable to respond to retinoic acid. The results show that retinoic acid responsiveness of PMLRARα, including transcriptional activation by tRA, is dispensable for leukemogenesis. In addition, we found that the ability of the PMLRARα protein to respond to tRA plays an important though perhaps not exclusive role in the therapeutic effects of retinoic acid. Furthermore, our finding that a RARα with dominant negative activity did not readily initiate leukemia suggests that the PML portion of PMLRARαplays an essential role in leukemic transformation.
Leukemogenesis cannot be explained by inappropriate transcriptional activation by PMLRAR
Alterations in transcription factors have been shown to play a central role in the pathogenesis of leukemias, lymphomas, and other malignancies.49,50 Changes in both transcriptional activation and repression can be important for the pathogenic effects of these alterations. PMLRARα retains the abilities ofRARα to repress and to activate transcription of retinoic acid receptor target genes. Although in some settings PMLRARαcan act to inappropriately repress transcription, when compared withRARα, it can also increase transcription in the absence or presence of ligand.13,15,17 23-25 The possible role of transcriptional activation in the pathogenesis of APL has not been previously tested.
By expressing PMLRARαm4 in the myeloid cells of transgenic mice, we directly assessed whether ligand-induced transcriptional activation by PMLRARα is required for leukemic transformation. The ability of PMLRARα variants to initiate leukemia has not been heretofore assessed. Grignani and colleagues45,51,52 examined the ability of altered forms ofPMLRARα to inhibit differentiation of the U937 promonocytic cell line. In this cell line, mutations that abolished the ability ofPMLRARα to interact with corepressors abrogated the ability of PMLRARα to inhibit differentiation.45 Although studies in U937 cells did not specifically address the role of transcriptional activation in inhibition of differentiation, thePMLRARα variants that were most effective at blocking differentiation retained the ability to act as strong transcriptional activators.52PMLRARαm4 is unable to activate transcription. Our finding that it readily initiates leukemia demonstrates that although PMLRARα can enhance the transcription of RARα target genes, this ability to activate transcription in response to ligand plays no role in leukemogenesis.
Retinoic acid responsiveness may influence leukemic phenotype
Although RA binding, corepressor release, and transcriptional activation are not required for leukemogenesis, these activities of thePMLRARα protein may influence the characteristics of the leukemias initiated by PMLRARα. Leukemias that developed in our original PMLRARα transgenic mice were characterized by normal white blood cell counts, bone marrow effaced by cells at the promyelocyte stage of maturation, and a rapidly fatal course. In contrast, PMLRARαm4 leukemias were more variable: some of these leukemias were characterized by increased white blood cell counts, persistence of neutrophilic maturation beyond the promyelocyte stage, and an indolent clinical course. Because transgenes integrate randomly into the genome, we cannot fully exclude the possibility that the differences between PMLRARα and PMLRARαm4leukemias were due to variation in the level or pattern of protein expression in the independent founders. Furthermore, caution in interpreting such variation is warranted considering the modest number of leukemias that arose in this study. Nevertheless, the differences observed between the PMLRARα and PMLRARαm4leukemias might reflect an effect of endogenous ligands on the behavior of the leukemic cells. Physiologic levels of retinoic acid may bind toPMLRARα and thereby decrease repression or increase activation of target genes.
The possibility that ligand-responsiveness may influence leukemic phenotype is supported by a study of leukemias initiated by a PLZFRARα transgene. PLZFRARα is expressed as a result of a t(11;17)(q13;q21) translocation in rare APL patients who have a disease that is clinically resistant to tRA.53,54 Similar to PMLRARαm4,PLZFRARα does not activate transcription in response to tRA.25,55 Leukemias in Cathepsin G-PLZFRARαtransgenic mice resembled human chronic myeloid leukemia and displayed less of a block in neutrophilic differentiation than leukemias inCathepsin G-PMLRARα transgenic mice.56 The differences between Cathepsin G-PLZFRARα and Cathepsin G-PMLRARα leukemias are strikingly parallel to those we observed between MRP8-PMLRARαm4 and MRP8-PMLRARα leukemias. Given the view that retinoids play a role in fostering neutrophilic maturation, it was unexpected that the fusion proteins that are less responsive to retinoic acid were associated with leukemias with a greater degree of differentiation. Retinoids are not, however, simply differentiative agents. Depending on the experimental system and the maturational state of the cells, retinoids can stimulate or inhibit both proliferation and differentiation of myeloid cells (reviewed in Purton et al57). We speculate that retinoid-responsiveness of PMLRARα facilitates differentiation arrest at the promyelocyte stage of neutrophil maturation.
A dominant negative RAR does not appear sufficient to initiate leukemia
A number of lines of evidence support the hypothesis that dominant negative inhibition by PMLRARα of RARα may underlie the pathogenesis of APL. First, retinoic acid can enhance neutrophilic differentiation.58,59 Second, dominant negativeRARα can inhibit neutrophilic maturation of primary cells.60 Third, comparisons between PMLRARα andPLZFRARα have focused attention on the role of these proteins as transcriptional repressors that can interfere with normal activation of retinoic acid responsive genes.34,42,45,56,61 Fourth, the 4 described translocations involving RARα in APL result in fusions to PML, PLZF, NPM, and NuMA, proteins that do not appear to share common functions. In light of the evidence that transcriptional repression is important in the pathogenesis of APL,34,42,45,56 61 the lack of similarities between the 4RARα partners raises the possibility that the fusion proteins contribute to APL by acting as dominant negative RARαs. The fact that PMLRARα and PMLRARαm4 readily initiated leukemias, whereas RARαm4 did not, strongly suggests that the PML domain does more than simply confer dominant negative activity ontoRARα.
The PML domain of PMLRARα may also contribute to transformation by disrupting the normal function of PML, or, alternatively, by altering the DNA binding characteristics ofPMLRARα relative to RARα. PML can act as a growth suppressor62-65 and loss of PML predisposes mice to the development of a variety of tumors when the mice are treated with the tumor initiator dimethylbenzanthracene.66 Cells lacking PML grow more quickly than normal cells and are resistant to apoptosis induced by a variety of agents.66,67 These findings indicate that decreased PML function may contribute to transformation by allowing for increased proliferation and increased survival. In support of the hypothesis that disruption of PML activity byPMLRARα contributes to APL is a report that loss of PML increases the penetrance of leukemia in Cathepsin G-PMLRARαtransgenic mice.68 An alternative explanation for the differences we have observed in the leukemogenicity ofPMLRARαm4 and RARαm4 is that the PML domain changes binding site specificity69 70: The genes whose transcription is repressed by PMLRARαm4 may differ from those repressed by RARαm4 and such differences could account for the absence of leukemia in RARαm4 transgenic mice.
A recent study of retrovirally infected murine bone marrow demonstrated that transcriptional repression by RARα fusion proteins, dominant negative RARα, or unliganded normal RARαcan inhibit neutrophilic differentiation and immortalize primary hematopoietic cells.71 These observations support a role for transcriptional repression in APL pathogenesis and raised the possibility that dominant negative RARα would act as potent leukemogen. In vitro immortalization does not, however, always correspond with leukemic behavior in vivo. For example, although HRX-ENL both immortalizes cells and causes leukemia in mice,72 immortalized murine myeloid cell lines such as FDC-P cells are nonleukemic when injected into histocompatible animals.73 The fact that MRP8-PMLRARαm4 but not MRP8-RARαm4 mice developed leukemia is not inconsistent with the hypothesis that transcriptional repression is necessary for leukemic transformation. Furthermore, although our results show that the leukemogenic potential of PMLRARα extends beyond its ability to inhibit RARα function, our findings cannot be interpreted to indicate that dominant negative RARα would not contribute to leukemia when expressed in mice under conditions other than those we have used.
The expression of RARαm4 in the bone marrow cells of our transgenic mice had a modest impact on neutrophil differentiation. The lack of a strong effect of the MRP8-RARαm4 transgene compared with prior in vitro studies of retrovirally expressed dominant negativeRARα could reflect a number differences in experimental methods, including protein expression levels in hematopoietic progenitors, in vitro growth conditions, and mouse strain. In addition, expression of the transgene was heterogeneous and its effects on differentiation of total bone marrow may consequently have been masked by the presence of neutrophilic cells with low or absentRARαm4 protein. Our results are, however, concordant with studies of mice that lack RARα1 andRARγ (which comprise the vast majority ofRARs expressed in myeloid cells):RARα1−/−, RARγ−/− mice displayed no apparent hematopoietic defect in vivo and the neutrophilic cells from these mice showed a modest maturation defect in vitro.74
Retinoic acid binding to PMLRAR is essential for retinoic acid-induced differentiation of leukemic promyelocytes
The leukemias that arose in the PMLRARαm4 transgenic mice did not differentiate in response to retinoic acid. The m4 mutation was originally described in a subclone of the NB4 cell line selected to grow in retinoic acid. Our results parallel the previously observed association of the m4 mutation with resistance to differentiation, and further demonstrate that the ability of tRA to cause differentiation of leukemic cells requires direct effects of tRA on the PMLRARαfusion protein.
Interestingly, tRA may have been of some therapeutic benefit despite the fact that differentiation of the leukemic blasts was not observed. Such a therapeutic effect of tRA has been observed in at least 1 case of human APL exhibiting marked in vitro resistance to the differentiative effects of tRA.28 The mechanism by which tRA was of benefit in these individual murine and human leukemias is not known. However, the possibility that tRA might improve survival even when it does not cause morphologic differentiation draws attention to the fact that novel therapies may have unanticipated effects that contribute to their clinical impact. This finding also suggests that tRA therapy may, in combination with other agents, continue to be of some benefit to APL patients identified as having resistant disease.
Acknowledgments
We thank Daphne Haas-Kogan and H. Jeffrey Lawrence for critical reading of the manuscript, and Meijuan Zhou for technical assistance.
Supported by grants CA 4338 and CA 75985 from the National Institutes of Health and by funds from the G.W. Hooper Research Foundation. S.C.K. is a recipient of a Burroughs Wellcome Fund Career Award.
Reprints:Scott C. Kogan, Department of Laboratory Medicine, Room M524, Box 0100, 505 Parnassus Ave, University of California, San Francisco, CA 94143-0100.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal