Chronic myelogenous leukemia (CML) begins with an indolent chronic phase but inevitably progresses to a fatal blast crisis. Although the Philadelphia chromosome, which generates p210bcr/abl, is a unique chromosomal abnormality in the chronic phase, additional chromosomal abnormalities are frequently detected in the blast crisis, suggesting that superimposed genetic events are responsible for disease progression. To investigate whether loss of p53 plays a role in the evolution of CML, we crossmated p210bcr/abl-transgenic (BCR/ABLtg/−) mice with p53-heterozygous (p53+/−) mice and generated p210bcr/abl-transgenic, p53-heterozygous (BCR/ABLtg/−p53+/−) mice, in which a somatic alteration in the residual normal p53 allele directly abrogates p53 function. TheBCR/ABLtg/−p53+/− mice died in a short period compared with their wild-type (BCR/ABL−/−p53+/+), p53 heterozygous (BCR/ABL−/−p53+/−), and p210bcr/abl transgenic (BCR/ABLtg/−p53+/+) litter mates. They had rapid proliferation of blast cells, which was preceded by subclinical or clinical signs of a myeloproliferative disorder resembling human CML. The blast cells were clonal in origin and expressed p210bcr/abl with an increased kinase activity. Interestingly, the residual normal p53 allele was frequently and preferentially lost in the tumor tissues, implying that a certain mechanism facilitating the loss of p53 allele exists in p210bcr/abl-expressing hematopoietic cells. Our study presents in vivo evidence that acquired loss of p53 contributes to the blastic transformation of p210bcr/abl-expressing hematopoietic cells and provides insights into the molecular mechanism for blast crisis of human CML.
In chronic myelogenous leukemia (CML), a clonal disorder of multipotential hematopoietic stem cells, excessive proliferation of immature and mature myeloid cells occurs.1The cytogenetic hallmark of CML is the Philadelphia chromosome,2 created by t(9;22)(q34;q11), in which the amino-terminal bcr gene on chromosome 22 is fused to most of the c-abl proto-oncogene on chromosome 9, thereby creating an 8.5-kilobase (kb) bcr/abl chimeric messenger RNA encoding a 210-kd hybrid protein (p210bcr/abl).3-5The p210bcr/abl has much greater kinase activity than the normal 145-kd c-abl gene product, and this is believed to play a critical role in the pathogenesis of CML.6
The clinical course of CML is characterized by hematologically and temporally distinct stages.1 In the initial stage, called chronic phase, the disease is indolent and the leukemic cells retain an ability to differentiate into mature granulocytes. But after several years of the chronic phase, the disease inevitably accelerates and ultimately progresses to the terminal fatal stage, called blast crisis, which involves aggressive proliferation of immature hematopoietic cells arrested at an early stage of differentiation. Although the molecular mechanisms responsible for the transition from the chronic phase to the blast crisis are not fully understood, the appearance of additional and nonrandom chromosomal abnormalities in the blast phase strongly suggests that superimposed genetic events account for the disease progression.7 One of the most frequently observed chromosomal abnormalities is isochromosome 17q, or i(17q),7where p53 tumor-suppressor gene is mapped.8 Studies have shown that structural alterations (ie, rearrangement, deletion, and mutation) of the p53 gene are extremely rare in the chronic phase but frequently appear in the blast crisis, strongly suggesting that the functional loss of p53 is essentially involved in the disease evolution.9-11
To understand the complex processes involved in the pathogenesis of human CML, it is necessary to develop animal models that recapitulate the clinical features of the disease. Several different approaches have been used to create a murine model for human CML. Earlier attempts focused on bone marrow transplantation (BMT) experiments. Mice that were lethally irradiated and given transplants of BM (BM) cells infected with p210bcr/abl-expressing retroviruses had CML-like granulocyte hyperplasia and other hematopoietic malignancies, such as myelomonocytic leukemias, macrophage tumors, pre-B- and T-cell lymphomas, reticulum-cell sarcomas, and erythroid tumors.12-14 Another approach recently used is transplantation of peripheral blood (PB) or BM cells from patients with CML into sublethally irradiated nonobese diabetic (NOD) or NOD/severe combined immunodeficient (SCID) mice. A population of the implanted CML cells was successfully engrafted and proliferated in the recipient mice, which eventually showed a CML-like hematologic disorder.15 16
On the other hand, generation of transgenic mice is an attractive approach. However, transgenic models for human CML have been unsuccessful until recently, probably owing to the lack of appropriate promoters.17 To overcome this problem, we cloned the promoter of mouse tec gene,18,19 which is preferentially expressed in hematopoietic progenitor cells,20 and generated transgenic mice expressing p210bcr/abl under the control of the tecpromoter.21 Although the founder mice showed massive proliferation of lymphoblasts shortly after birth and were diagnosed as having acute lymphoblastic leukemia (ALL), the transgenic offspring, after a long latency period, reproducibly exhibited a myeloproliferative disorder closely resembling human CML.21The PB smear showed remarkable granulocyte hyperplasia, and the BM was hypercellular, with a predominance of myeloid cells at various stages of differentiation.21 Because these pictures represent cardinal features of human CML, our transgenic mice can be regarded as a model for human CML.
Compared with the BMT and NOD or NOD/SCID mice approach, our transgenic approach has several advantages. First, the disease spectrum and disease frequency in the BMT approach are markedly influenced by the infection conditions22,23 and the implantation efficiency and survival of the engrafted CML cells in NOD or NOD/SCID mice vary significantly, depending on the donor patients and recipient mice.15 16 In contrast, our transgenic mice reproducibly exhibit CML-like granulocyte hyperplasia, with disease penetrance of about 100% (among the 87 transgenic mice thus far generated, 84 [about 97%] developed CML and the other 3 [about 3%] developed ALL [Honda et al, unpublished data]). Second, although the mice in the other 2 models have p210bcr/abl only in the hematopoietic compartment, our transgenic mice have p210bcr/abl transgene in every type of cell and can be crossmated with other transgenic or knockout mice to investigate the possible promoting or suppressing effect of a specific gene on p210bcr/abl in vivo. Thus, our transgenic mice can be regarded as a stable and inheritable model for human CML. Because of these advantages, we used this model to examine whether loss of p53 contributes to the evolution to blast crisis of CML in vivo. With this aim, we crossmated p210bcr/abl-transgenic mice with p53-heterozygous mice and generated p210bcr/abl-transgenic, p53-heterozygous mice, in which a somatic alteration in the residual p53 gene directly abrogates p53 function.
Materials and methods
Mice
Mice that were p210bcr/abl transgenic (BCR/ABLtg/−) were generated by using the mouse tec promoter (−1948 to +22) and the bcr/abl(p210) complementary DNA (cDNA; b3a2 type) as previously described.21 Because the founder mice were generated by using eggs derived from C57Bl/6 × DBA F2 (BDF2) mice and the transgenic progeny were generated by mating the transgenic mice with BDF1 mice, the genetic background of theBCR/ABLtg/− mice was a mixture of C57Bl/6 and DBA. The p53-heterozygous (p53+/−) mice were generated by using TT2 ES cells inserted with a neomycin-resistance cassette into the second exon of the p53 gene as previously described.24 Because the TT2 ES cells were established from an F1 embryo between C57Bl/6 and CBA25 and the p53-heterozygous mice were generated by mating the chimeric mice with C57Bl/6 mice, the genetic background of the p53+/− mice was a mixture of C57Bl/6 and CBA. Therefore, the background of the mice used in this experiment was a mixture of C57Bl/6, DBA, and CBA. Identification of genotypes forBCR/ABL and p53 was carried out by hybridizingBamHI-digested tail DNA with bcr/abl cDNA andSacI-digested tail DNA with the 5′ flanking region of the mouse p53 gene, as previously described.21 24
Pathological analysis
Autopsies were performed on dead or moribund animals. Smears and stamp specimens of leukemic tissues were stained with Wright-Giemsa stain. Tissues were also fixed in 10% neutral-buffered formaldehyde and subjected to routine light microscopical examination. All the organs were examined grossly and representative slices were prepared for hematoxylin-eosin staining.
Western blot, immunoprecipitation, and in vitro kinase assay
For detection of p210bcr/abl-transgene product, proteins were extracted by homogenizing leukemic tissues in RIPA lysis buffer (150 mmol/L sodium chloride, 50 mmol/L TRIS chloride [pH 7.4], 1% Triton X-100, 0.05% sodium dodecyl sulfate, and 1% sodium deoxycholate) with 50 U/mL of aprotinin and were probed with the anti-Abl monoclonal antibody AB3 (Oncogene Science, Manhasset, NY) as previously described.21 For detection of the kinase activity of the p210bcr/abl-transgene product, protein aliquots were incubated with AB3 and immunoprecipitated proteins were subjected to in vitro kinase assay as previously described.21 For detection of p53 protein, nuclear fractions of the tissues were subjected to immunoprecipitation/Western blot analysis using anti-p53 antibodies (Oncogene Science) as previously described.24
Flow cytometric analysis
Cells were stained with fluorescein isothiocyanate-conjugated or phycoerythrin-conjugated commercial monoclonal antibodies, including anti-Thy-1.2, anti-B220, anti-CD4, and anti-CD8 (Pharmingen, San Diego, CA), according to the manufacturer's instructions. The stained cells were washed 3 times with phosphate-buffered saline and analyzed on a FACScan (Becton Dickinson, Sunnyvale, CA).
DNA extraction and Southern blot analysis
Genomic DNAs were extracted from tumor tissues and digested with restriction enzymes. Southern blot analysis was performed by using phosphorus (P) 32-dCTP–labeled mouse TCRγ as a probe as previously described.21
Polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) analysis
PCR-SSCP analysis was performed essentially as previously described.26 In brief, genomic DNAs were amplified by using several different sets of primers with α32P-dCTP. The PCR products were mixed with formamide dye, electrophoresed on acrylamide gels with or without glycerol, and autoradiographed.
Results
Early death and blast-cell proliferation in p210bcr/abl-transgenic, p53-heterozygous mice
Crossmating of p210bcr/abl-transgenic (BCR/ABLtg/−) mice with p53-heterozygous (p53+/−) mice produced mice with 4 different genotypes, which wereBCR/ABL−/−p53+/+(wild-type),BCR/ ABL−/−p53+/−(p53 heterozygous),BCR/ABLtg/−p53+/+(p210bcr/abl transgenic), andBCR/ABLtg/−p53+/−(p210bcr/abl transgenic, p53 heterogeneous). The genotypes of the 58 total offspring were determined by Southern blot assessment using tail DNAs (Figure 1A). The numbers of the mice with each genotype were 16 forBCR/ABL−/−p53+/+(27.5%), 13 forBCR/ABL−/−p53+/−(22.4%), 15 forBCR/ABLtg/−p53+/+(25.9%), and 14 forBCR/ABLtg/−p53+/−(24.1%). These results were in good agreement with the expected ratio based on the Mendelian rule (25% each), indicating that the crossmating did not affect the embryonic development of the mice.
Studies of offspring generated by crossmatingBCR/ABLtg/− mice with p53+/−mice.
(A) Genotyping of the mice. The upper lanes show the results of Southern blot testing. The arrow indicates the position of thebcr/abl transgene; G, the position of the germline; and T, the targeted alleles of the p53 gene. The lower lanes show the genotypes of the mice. (B) Survival curves for the mice, according to genotype. Thin dotted lines indicate the percentage of surviving mice with theBCR/ABL−/−p53+/+genotype; thick dotted lines, theBCR/ABL−/−p53+/−genotype; thin solid lines, theBCR/ABLtg/−p53+/+genotype; and thick solid lines, theBCR/ ABLtg/−p53+/−genotype. (C) Changes in the white blood cell (WBC) count and mortality of the mice. The white circle indicates the WBC counts ofBCR/ ABL−/−p53+/+mice; black circle,BCR/ABL−/−p53+/−mice; white square,BCR/ ABLtg/−p53+/+mice; and black square,BCR/ABLtg/−p53+/−mice. The bar indicates SD; the arrow shows the point at which a mouse died.
Studies of offspring generated by crossmatingBCR/ABLtg/− mice with p53+/−mice.
(A) Genotyping of the mice. The upper lanes show the results of Southern blot testing. The arrow indicates the position of thebcr/abl transgene; G, the position of the germline; and T, the targeted alleles of the p53 gene. The lower lanes show the genotypes of the mice. (B) Survival curves for the mice, according to genotype. Thin dotted lines indicate the percentage of surviving mice with theBCR/ABL−/−p53+/+genotype; thick dotted lines, theBCR/ABL−/−p53+/−genotype; thin solid lines, theBCR/ABLtg/−p53+/+genotype; and thick solid lines, theBCR/ ABLtg/−p53+/−genotype. (C) Changes in the white blood cell (WBC) count and mortality of the mice. The white circle indicates the WBC counts ofBCR/ ABL−/−p53+/+mice; black circle,BCR/ABL−/−p53+/−mice; white square,BCR/ ABLtg/−p53+/+mice; and black square,BCR/ABLtg/−p53+/−mice. The bar indicates SD; the arrow shows the point at which a mouse died.
All mice were kept under the same maintenance conditions and were continually observed. The hematopoietic variables of the PB were routinely examined. SixteenBCR/ABL−/−p53+/+mice were all healthy during the observation period (Figure 1B, thin dotted line). Among the 13BCR/ABL−/−p53+/−mice, 1 mouse died with a gross subcutaneous tumor at 11 months of age (data not shown), but the other 12 mice survived in a healthy state for more than 1 year with no symptoms of illness (Figure B, thick dotted line). No obvious hematologic changes were observed in these mice (Figure 1C, upper panels). Among the 15BCR/ABLtg/−p53+/+ mice, 1 mouse died at 3 months of age from ALL and the remaining 14 gradually exhibited CML after 6 months of age, as previously reported21 (Figure 1C, left bottom panel). About half of the mice died within 1 year of age (Figure 1B, thin solid line). In contrast to the mice with these genotypes,BCR/ABLtg/−p53+/−mice showed high morbidity and mortality rates (Figure 1B, thick solid line). Eleven of 14BCR/ABLtg/−p53+/−mice died within 1 year. Although detailed analysis was not possible in 2 mice because of advanced autolysis, 9 mice were subjected to pathological, biochemical, and molecular analyses. A common macroscopic aspect of these mice was enlargement of the thymus (Table1). Thymoma was frequently associated with pleural effusion, and splenomegaly was observed in some mice (Table 1). Wright-Giemsa staining showed that blast cells with no granules and with morphologic characteristics of lymphoblasts proliferated markedly in the thymus (Figure 2A) and pleural effusion (Figure 2B).
Characterization ofBCR/ABLtg/−p53+/− leukemic mice
| Mouse No. . | Sex/ Age at Diagnosis, mo . | Macroscopic Tumor Sites . | Surface Markers . | DNA Status (TCR probe) . | p210bcr/abl Expression . | p53 Gene Status . |
|---|---|---|---|---|---|---|
| 1 | F/8.0 | Thymus, PE, Spleen | Thy1.2+, CD4−, CD8− | G/R | Yes | G/T |
| 2 | M/2.8 | Thymus, PE | Thy1.2+, CD4+, CD8+ | G/R | Yes | Loss/T |
| 3 | F/11.5 | Thymus, PE, Spleen | ND | ND | Yes | G/T |
| 4 | M/9.7 | Thymus, PE | Thy1.2+, CD4+, CD8+ | G/R | Yes | G/T |
| 5 | F/3.0 | Thymus | Thy1.2+, CD4+, CD8+ | G/R | Yes | Loss/T |
| 6 | F/3.2 | Thymus | Thy1.2+, CD4+, CD8+ | G/R | Yes | Loss/T |
| 7 | M/4.0 | Thymus | Thy1.2+, CD4−, CD8+ | G/R | Yes | Loss/T |
| 8 | M/2.3 | Thymus, PE | Thy1.2+, CD4+, CD8− | G/R | Yes | Loss/T |
| 9 | M/4.1 | Thymus | ND | ND | Yes | Loss/T |
| Mouse No. . | Sex/ Age at Diagnosis, mo . | Macroscopic Tumor Sites . | Surface Markers . | DNA Status (TCR probe) . | p210bcr/abl Expression . | p53 Gene Status . |
|---|---|---|---|---|---|---|
| 1 | F/8.0 | Thymus, PE, Spleen | Thy1.2+, CD4−, CD8− | G/R | Yes | G/T |
| 2 | M/2.8 | Thymus, PE | Thy1.2+, CD4+, CD8+ | G/R | Yes | Loss/T |
| 3 | F/11.5 | Thymus, PE, Spleen | ND | ND | Yes | G/T |
| 4 | M/9.7 | Thymus, PE | Thy1.2+, CD4+, CD8+ | G/R | Yes | G/T |
| 5 | F/3.0 | Thymus | Thy1.2+, CD4+, CD8+ | G/R | Yes | Loss/T |
| 6 | F/3.2 | Thymus | Thy1.2+, CD4+, CD8+ | G/R | Yes | Loss/T |
| 7 | M/4.0 | Thymus | Thy1.2+, CD4−, CD8+ | G/R | Yes | Loss/T |
| 8 | M/2.3 | Thymus, PE | Thy1.2+, CD4+, CD8− | G/R | Yes | Loss/T |
| 9 | M/4.1 | Thymus | ND | ND | Yes | Loss/T |
PE indicates pleural effusion; G/R, germline/rearranged; G/T, germline/targeted; and ND, not done.
Pathological analysis ofBCR/ABLtg/−p53+/− leukemic mice.
Figures 2A to 2D show Wright-Giemsa staining of stamped or smeared specimens of hematopoietic tissues. Massive proliferation of lymphoblasts is observed in the thymus (A) and in the pleural effusion (B). In mice with late onset of acute transformation, proliferation of granulocytes is evident in the peripheral blood (C) and the bone marrow shows marked hyperplasia of myeloid cells (D). Figures 2E to 2J show hematoxylin-eosin staining of tissue from the following: the lung (2E and 2F), liver (2G and 2H), and kidney (2I and 2J). Infiltration of leukemic cells was detected around the blood vessels. The boxed areas in Figures 2E, 2G, and 2I are magnified in Figures 2F, 2H, and 2J, respectively.
Pathological analysis ofBCR/ABLtg/−p53+/− leukemic mice.
Figures 2A to 2D show Wright-Giemsa staining of stamped or smeared specimens of hematopoietic tissues. Massive proliferation of lymphoblasts is observed in the thymus (A) and in the pleural effusion (B). In mice with late onset of acute transformation, proliferation of granulocytes is evident in the peripheral blood (C) and the bone marrow shows marked hyperplasia of myeloid cells (D). Figures 2E to 2J show hematoxylin-eosin staining of tissue from the following: the lung (2E and 2F), liver (2G and 2H), and kidney (2I and 2J). Infiltration of leukemic cells was detected around the blood vessels. The boxed areas in Figures 2E, 2G, and 2I are magnified in Figures 2F, 2H, and 2J, respectively.
The age at disease onset ranged from 2 to 11 months (Figure 1B, thick solid line, and Table 1). In mice with disease onset at a young age, no obvious hematologic changes were observed in PB (Figure 1C, right bottom panel), although the BM showed predominance of myeloid cells (data not shown). On the other hand, mice with disease onset in old age had typical symptoms of CML before thymomas developed; they showed leukocytosis with granulocyte proliferation in the PB (Figure 1B, right bottom panel, and Figure 2C) and marked myeloid hyperplasia in the BM (Figure 2D). Proliferation of blast cells in the PB was observed in some mice (data not shown). Pathological analysis showed that the blast cells infiltrated several major organs, such as the lung, liver, and kidney, in all mice examined (Figure 2E-2J).
Expression and enhanced kinase activity of the p210bcr/abl-transgene product in the tumor tissues
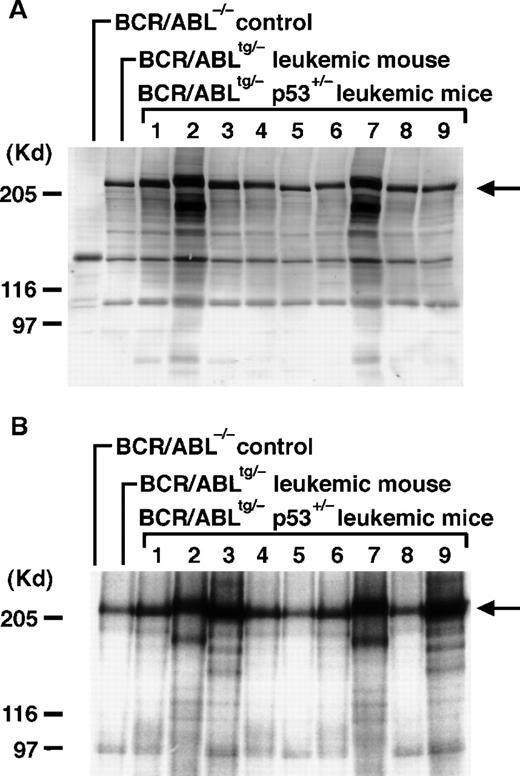
To examine whether the blast cells inBCR/ABLtg/−p53+/− leukemic mice expressed the p210bcr/abl-transgene product, proteins extracted from the enlarged thymus or pleural effusion were blotted with the anti-Abl monoclonal antibody AB3. As shown in Figure3A (arrow), the 210-kd band was detected in all the leukemic tissues, indicating that blast cells expressed the p210bcr/abl-transgene product. Blotting the same samples with antiphosphotyrosine antibody showed that the expressed p210bcr/abl was tyrosine phosphorylated (data not shown). To examine whether the expressed p210bcr/abl had an enzymatically active kinase activity, proteins extracted from the tumor tissues were subjected to in vitro kinase assays. As shown in Figure 3B, phosphorylated 210-kd band was observed in all the leukemic tissues (arrow), indicating that p210bcr/abl contained an increased kinase activity.
Expression, kinase activity, and TCR rearrangements in p210bcr/abl in the leukemic tissues ofBCR/ABLtg/−p53+/− mice.
For the expression assessment (A), 50 μg of protein aliquots extracted from the enlarged thymus or pleural effusion of 9BCR/ABLtg/−p53+/−leukemic mice (no. 1-9 on Table 1) was separated by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane, and probed with the anti-Abl monoclonal antibody AB3 (1:500). To determine kinase activity (B), 1 mg of protein aliquot extracted from the same tissues shown in Figure 3A was incubated with AB3 (1:200) and the immunoprecipitated proteins were subjected to in vitro kinase assays. Phosphorylated proteins were separated by 6% SDS-PAGE, dried, and autoradiographed. For both the expression and kinase activity assessments, the thymus of a normalBCR/ABL−/− control mouse was the negative control, and the thymus of aBCR/ABLtg/− leukemic mouse21was the positive control. The arrow indicates the position of p210bcr/abl; the positions of the molecular markers are shown on the left. To describe TCR rearrangements (C), 5 μg of DNAs digested with EcoRI was separated by 0.7% agarose gel, blotted to a nylon membrane, and probed with a phosphorus 32-dCTP–labeled TCRγ probe. DNA extracted from normal thymus tissue was used as a control (denoted by C). Molecular markers are shown on the left.
Expression, kinase activity, and TCR rearrangements in p210bcr/abl in the leukemic tissues ofBCR/ABLtg/−p53+/− mice.
For the expression assessment (A), 50 μg of protein aliquots extracted from the enlarged thymus or pleural effusion of 9BCR/ABLtg/−p53+/−leukemic mice (no. 1-9 on Table 1) was separated by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane, and probed with the anti-Abl monoclonal antibody AB3 (1:500). To determine kinase activity (B), 1 mg of protein aliquot extracted from the same tissues shown in Figure 3A was incubated with AB3 (1:200) and the immunoprecipitated proteins were subjected to in vitro kinase assays. Phosphorylated proteins were separated by 6% SDS-PAGE, dried, and autoradiographed. For both the expression and kinase activity assessments, the thymus of a normalBCR/ABL−/− control mouse was the negative control, and the thymus of aBCR/ABLtg/− leukemic mouse21was the positive control. The arrow indicates the position of p210bcr/abl; the positions of the molecular markers are shown on the left. To describe TCR rearrangements (C), 5 μg of DNAs digested with EcoRI was separated by 0.7% agarose gel, blotted to a nylon membrane, and probed with a phosphorus 32-dCTP–labeled TCRγ probe. DNA extracted from normal thymus tissue was used as a control (denoted by C). Molecular markers are shown on the left.
Expression of T-cell antigen by blast cells and their clonal origin
To determine the cell lineage of the leukemias, blast cells in the enlarged thymus or pleural effusion from 7BCR/ABLtg/−p53+/−leukemic mice were subjected to flow cytometric analysis. Staining the cells with anti-Thy1.2 and anti-B220 antibodies showed that all the tumor cells were positive for Thy1.2 antigen but negative for B220 (Table 1). Staining the cells with anti-CD4 and anti-CD8 antibodies revealed that 1 tumor was double negative (CD4−CD8−), 4 tumors were double positive (CD4+ CD8+), and the other 2 were single positive for CD4 (CD4+ CD8−) and CD8 (CD4− CD8+), respectively (Table 1). To investigate the clonality of the blast cells, DNAs extracted from the tumor tissues were subjected to Southern blot analysis using TCR as a probe. The results showed that all the tumor tissues carried rearrangements in the TCR locus, indicating that the blast cells were clonal in origin (Figure 3C and Table 1).
Frequent and preferential loss of the wild-type p53 allele and absence of p53 protein in the leukemic cells in BCR/ABLtg/−p53+/− mice
The rapid proliferation of blast cells in theBCR/ABLtg/−p53+/−mice strongly suggested that a somatic mutation or mutations occurred in the residual wild-type p53 gene. To address this issue, PCR-SSCP analysis was performed. This assessment can detect mutations in almost all of the coding sequence (codon 5 to 356) and the splice donor and acceptor sites in introns 2 through 9 of the mouse p53 gene.26 No band shifts were observed in any regions of the p53 gene under 2 different conditions, indicating that the leukemias did not carry point mutations (data not shown). On the other hand, we found a polymorphic site in exon 5 that allowed us to distinguish the PCR product of the wild-type allele from that of the null allele. Sequencing revealed that codon 134 of the wild-type allele was GCG (coding Ala), whereas that of the null allele was GTG (coding Val).
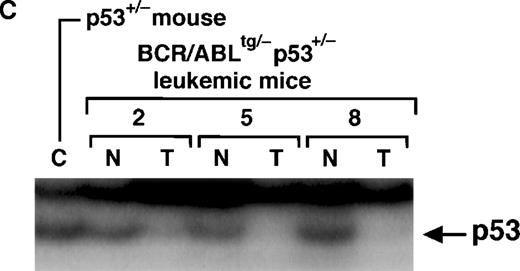
A schematic model of the region encompassing the polymorphic site and the results of PCR-SSCP are shown in Figure4A and Figure 4B, respectively. As shown in the left 2 lanes in Figure 4B, the p53+/− mouse was a heterozygote of alleles A and B, whereas theBCR/ABLtg/− mouse was a homozygote of allele B. Because all the tissues examined in theBCR/ABLtg/−p53+/−leukemic mice harbored allele A (numbers 1-9 in Figure 4B),BCR/ABLtg/−p53+/−mice were considered to be heterozygotes of allele A from the p53+/− mouse (null) and allele B from theBCR/ABLtg/− mouse (wild-type). Interestingly, among 9 tumors that developed inBCR/ ABLtg/−p53+/−mice (T lanes in Figure 4B), 6 tumors (no. 2 and no. 5-9) contained only allele A (null) and clearly lost allele B (wild-type). This allelic loss was obviously a somatic event, since the normal tissues (N lanes of no. 2, 5, and 8 in Figure 4B) retained allele B. The absence of the p53 protein in the tumor tissues was confirmed by immunoprecipitation/Western blot analysis using anti-p53 antibodies (Figure 4C). These data demonstrated that the residual wild-type p53 allele was frequently and preferentially lost in the tumor tissues in theBCR/ABLtg/−p53+/−leukemic mice and that this led to the absence of the p53 protein. The characteristics of the leukemias that developed inBCR/ABLtg/−p53+/−mice are summarized in Table 1.
Polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) analysis of the p53 gene and expression of p53 protein in the leukemic tissues ofBCR/ABLtg/−p53+/− mice.
(A) Schematic model of the region of the mouse p53 gene that showed a polymorphic pattern between the p53 wild-type allele and the p53 null allele. The nucleotide sequences and amino acids of the wild-type allele and the null allele, which showed a polymorphism, are also shown. The box denotes exon 5; black bars, adjacent introns; white triangles, locations of the primers used for amplification (p5-1 and p5-2); and the diamond, the polymorphic site. (B) Results of PCR-SSCP. DNAs extracted from the tumor tissues (T) of 9BCR/ABLtg/−p53+/−leukemic mice (no. 1-9) were subjected to PCR-SSCP analysis. In mice 2, 5, and 8, DNAs extracted from the normal tissues (N) were also analyzed to provide an internal control. DNAs extracted from the normal tissues of a p53+/− mouse and aBCR/ABLtg/− mouse were used as controls for detecting the migration patterns of the parental alleles. Black triangles denote the position of allele A (null); and white triangles, the position of allele B (wild-type). (C) Absence of p53 protein. Proteins extracted from tumor tissues (T) and normal tissues (N) of mice 2, 5, and 8 were subjected to immunoprecipitation/Western blot analysis using anti-p53 antibodies. Normal tissue from a p53+/− mouse was used as a control (denoted by C). The arrow indicates the position of p53.
Polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) analysis of the p53 gene and expression of p53 protein in the leukemic tissues ofBCR/ABLtg/−p53+/− mice.
(A) Schematic model of the region of the mouse p53 gene that showed a polymorphic pattern between the p53 wild-type allele and the p53 null allele. The nucleotide sequences and amino acids of the wild-type allele and the null allele, which showed a polymorphism, are also shown. The box denotes exon 5; black bars, adjacent introns; white triangles, locations of the primers used for amplification (p5-1 and p5-2); and the diamond, the polymorphic site. (B) Results of PCR-SSCP. DNAs extracted from the tumor tissues (T) of 9BCR/ABLtg/−p53+/−leukemic mice (no. 1-9) were subjected to PCR-SSCP analysis. In mice 2, 5, and 8, DNAs extracted from the normal tissues (N) were also analyzed to provide an internal control. DNAs extracted from the normal tissues of a p53+/− mouse and aBCR/ABLtg/− mouse were used as controls for detecting the migration patterns of the parental alleles. Black triangles denote the position of allele A (null); and white triangles, the position of allele B (wild-type). (C) Absence of p53 protein. Proteins extracted from tumor tissues (T) and normal tissues (N) of mice 2, 5, and 8 were subjected to immunoprecipitation/Western blot analysis using anti-p53 antibodies. Normal tissue from a p53+/− mouse was used as a control (denoted by C). The arrow indicates the position of p53.
Discussion
CML provides an appropriate model for a disease in which the gain of an acquired genetic abnormality contributes directly to the disease progression. The p53 is one of the genes whose alterations are frequently detected in the blast crisis of human CML. Therefore, the functional defect of p53 is believed to play a key role in the evolution of the disease, but no direct evidence has been shown.9-11 Skorski et al27 transfected p210bcr/abl-expressing retroviruses into p53-deficient and wild-type BM cells. They showed that the p210bcr/abl-positive, p53-deficient BM cells had blast-like morphologic characteristics and an increased colony-forming ability compared with p210bcr/abl-expressing, wild-type BM cells. In addition, when injected into SCID mice, p210bcr/abl-positive, p53-deficient cells showed more rapid disease progression than p210bcr/abl-expressing, wild-type cells. These results demonstrated that the disruption of p53 function conferred a more aggressive leukemogenic potential on p210bcr/abl-positive hematopoietic cells and induced a more malignant phenotype. However, because the experimental procedures, such as culture of BM cells and retrovirus transfection, were performed in vitro, the synergistic effect of p210bcr/abl and loss of p53 in vivo could not be fully understood. We addressed this issue by generating p210bcr/abl-transgenic, p53-heterozygous mice, in which a somatic event in the residual p53 gene directly abrogates p53 function.
The crossmating of p210bcr/abl-transgenic mice with p53-heterozygous mice resulted in generation of mice with 4 different genotypes:BCR/ABL−/−p53+/+,BCR/ABL−/−p53+/−,BCR/ ABLtg/−p53+/+, andBCR/ABLtg/−p53+/−. TheBCR/ABL−/−p53+/+mice were reasonably healthy (Figure 1B and 1C). Of 13BCR/ ABL−/−p53+/−mice, 1 mouse died of a skin tumor (data not shown). This result is not consistent with those of previous p53+/− studies, in which malignancies, including T-cell lymphoma, occurred at a higher ratio.28,29 Although the reason for this difference is not clear, one possibility is the genetic background of the mice, as previously suggested.30 The genetic background of the mice in our study was a mixture of C57Bl/6, DBA, and CBA, whereas other p53+/− studies used mice containing the 129 strain28,29 and a study showed that an increased ratio of 129 strain accelerates disease development.30 BCR/ABLtg/−p53+/+ mice had CML-like disease and ALL (Figure 1B and 1C), as expected.21 In contrast to these mice,BCR/ABLtg/−p53+/−mice died in a short period (Figure 1B and 1C) and with rapid proliferation of blast cells (Figure 2A and 2B). The leukemic cells were highly malignant, as indicated by massive infiltration into tissues (Figure 2E-2J). They expressed p210bcr/ablwith an enhanced kinase activity (Figure 3A, 3B). In addition, the residual wild-type p53 allele was frequently and preferentially lost in leukemic tissues (Figure 4B), which led to the absence of p53 expression (Figure 4C). These results demonstrate that the functional loss of p53 essentially contributed to the blastic transformation of the p210bcr/abl-expressing hematopoietic cells in vivo.
InBCR/ABLtg/−p53+/−mice, the blastic transformation was observed in mice with a wide range of ages (Figure 1B and 1C and Table 1). This was possibly because the disease onset depended on when an acquired event, including loss of p53, occurred in the p210bcr/abl-expressing hematopoietic cells. In mice with early disease onset, the BM showed a predominance of myeloid cells (data not shown), although no abnormalities were observed in the PB (Figure 1B and 1C). On the other hand, in mice with late disease onset, the PB showed excessive proliferation of granulocytes (Figure 2C) and the BM showed marked hyperplasia of myeloid cells (Figure 2D). Therefore, subclinical or clinical signs of CML already existed when acute leukemias developed in the mice. These results indicate that CML preceded the acute phase in these mice, in recapitulation of the clinical course of human CML.
It is interesting that the loss of the wild-type p53 allele was observed in the leukemic tissues at a high incidence but that normal tissues retained the wild-type allele (Figure 4B). Thus, the acquired loss of the wild-type p53 allele was not a random event; rather, there is a certain mechanism by which genetic abnormalities preferentially occurred in the hematopoietic cells. It could be supposed that the expression of p210bcr/abl might contribute to the process or processes leading to the allelic loss. The p210bcr/abl might accelerate the mutation rate or promote genetic instability (as reported in p190bcr/abl transgenic mice31), which would facilitate the loss of the wild-type p53 allele and consequently cause the blastic transformation. Further studies will be required to clarify the molecular mechanism or mechanisms underlying the preferential loss of p53 in the p210bcr/abl-expressing hematopoietic cells in the transgenic mice.
It should be noted that all the leukemias were clonal T-cell tumors, as demonstrated by flow cytometry and gene-rearrangement analysis (Figure3C and Table 1). These results indicate that the loss of p53 occurred in hematopoietic cells that had already been committed to the T-cell lineage. This idea was further supported by the finding that hematopoietic cells of other lineages, such as myeloid, contained the wild-type p53 allele (data not shown). The reason for the remarkable susceptibility of the T cells to p53 loss remains elusive. One possibility is that gene rearrangements in the TCR loci triggered gene alterations (possibly in cooperation with p210bcr/abl) and led to loss of the wild-type p53 allele. The finding that p53 loss occurred preferentially in relatively young mice (number 2 and numbers 5-9 in Figure 4C and Table 1) supports this notion, since physiologic TCR rearrangements occur actively in the thymus of young animals. In the 3 mice in which leukemia developed at a relatively old age, no mutations were detected in the p53 gene (numbers 1, 3, and 4 in Figure 4C and Table 1). In these mice, the mechanism that caused blastic transformation remains unknown. We consider it possible that the reduction of p53 protein in p53+/−cells enhanced the oncogenicity of p210bcr/abl and caused malignant transformation.32 Alternatively, the decrease in p53 protein might have facilitated activation of oncogenes or inactivation of tumor-suppressor genes, with which p210bcr/abl exerted its fully oncogenic potential.
Mouse models for clonal expansion of p210bcr/abl-expressing hematopoietic cells have been demonstrated in BMT experiments.33-36 Serial transplantation of murine CML cells into syngeneic recipient mice frequently caused acute leukemias. The leukemic cells contained the same proviral integration site as the primary CML cells, indicating that they originated from the same clone. Although the molecular mechanisms responsible for the disease progression have not clearly been identified in BMT models, it may be that a secondary genetic event or events, such as loss of p53 as observed in our study, might contribute to the blastic transformation. One difference between our transgenic model and the BMT experiments is the disease phenotype. Our transgenic mice had T-cell leukemias exclusively (Table 1), whereas the recipient mice in the BMT experiments had myeloid and B-cell acute leukemias in addition to T-cell leukemias.33-36 This phenotypic difference might be due to the genetic backgrounds of the mice. The genetic background of our transgenic mice is a mixture of C57Bl/6, DBA, and CBA, whereas the mice used in the BMT experiments were of the BALB/C strain.33-36 An expanded study would provide more information about the phenotypic disparity between our transgenic mice and the mice in the BMT experiments.
There is a feature of our model that is dissimilar to human CML. A T-cell phenotype is rarely observed in the blast crisis of human CML.1 One possible reason is that T-cell lineage is rarely involved in human CML, whereas every type of cell in our transgenic mice has the transgene and the thymic cells might be more susceptible to a somatic event or events causing blastic transformation. An animal model for CML in which loss of p53 could be controlled in spacial and temporal ways would clarify this issue.
In this study, we showed thatBCR/ABLtg/−p53+/−mice had marked proliferation of blast cells in a short period, in which the residual wild-type p53 allele was frequently and preferentially lost. These results provide in vivo evidence that an acquired loss of p53 function contributed a proliferative advantage to the p210bcr/abl-expressing hematopoietic cells and caused blastic transformation. Our findings provide insights into the molecular mechanism of blast crisis of human CML. In addition, our transgenic mice are a useful animal model for examining the biologic effect of a specific gene on the malignant transformation of p210bcr/abl-expressing hematopoietic cells in vivo.
Acknowledgments
We thank Yoshikazu Oh-hira for preparing pathological specimens and Tsuyoshi Takahashi, Yoichi Imai, and Koichiro Yuji for technical assistance. We also thank T. W. Mak for providing the mouse TCR probe.
Supported in part by grants-in-aid from the Ministry of Education, Science and Culture of Japan.
Reprints:Hisamaru Hirai, Third Department of Internal Medicine, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; e-mail: hhirai-tky@umin.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal