Chediak-Higashi syndrome (CHS) is a rare autosomal recessive disorder in which an immune deficiency occurs in association with pigmentation abnormalities. Most patients who do not undergo bone marrow transplantation die of a lymphoproliferative syndrome, though some patients with CHS have a relatively milder clinical course of the disease. The large size of the LYST gene, defective in CHS, has made it difficult to screen for mutations in a large number of patients. Only 8 mutations have been identified so far, and all lead to a truncated LYST protein. We conducted protein truncation tests on this gene in 8 patients with CHS. Different LYST mutations were identified in all subjects through this approach, strengthening the observation of a high frequency of truncated LYST proteins as the genetic cause of CHS.
Chediak-Higashi syndrome (CHS; MIM, 214 500) is an autosomal recessive disease characterized by partial oculocutaneous albinism, mild predisposition to pyogenic infections, and abnormally large granules in many different cell types.1-3 A patient's susceptibility to infection may be explained by defects observed in T-cell cytotoxicity4 and natural killer cell activity5,6 as well as in chemotaxis7 and bactericidal capacity8,9 of granulocytes and monocytes. Unless they undergo allogenic bone marrow transplantation (BMT),10 many patients with CHS die young because of the so-called accelerated phase, which is characterized by the infiltration of most organs with activated T lymphocytes and macrophages and by pancytopenia and coagulation disorder.11 In some patients, however, the disease takes a relatively milder clinical course and the patients survive to adulthood with few or even no severe infections.
At the cellular level, enlarged vesicles or granules are easily detected in the cytoplasm of most cell types. These structures are preferentially perinuclear and are associated with impaired compartmentalization or sorting of many different lysosomal protein components, suggesting that CHS may involve defective trafficking of specific proteins to organelles.4 12
A similar disorder is reported in other species,13including mice (beige mouse), minks (Aleutian), rats, cats, cows, and whales. Mutants in all species examined have similar morphologic features, such as altered size and altered distribution of lysosomes.13 Manifestations of the accelerated phase, however, have never been reported in animal models of CHS.13 Homology between that disease in beige mice and CHS was recently supported by the mapping of the CHS locus to chromosome 1q42-q43 in a segment corresponding to the beige locus region on mouse chromosome 13.14,15 It was confirmed by the sequence homology shared between the CHS and the beige coding sequence gene (LYST).16-18 The full-length human LYST gene coding segment encompasses 13.5 kb and encodes a predicted 3801 amino acid polypeptide. Pathologic mutations in this gene have been reported in 9 patients.17,19 20 Interestingly, only nonsense mutations and frameshifts of the LYST gene have been identified so far.
Considering the length of the LYST coding sequence and the unknown genomic structure of this gene, the detection of mutations is a technical challenge. We therefore established a protein truncation test21-23 to look for mutations in LYST mRNA from 8 patients with CHS. All the patients had premature termination codon mutations on both alleles.
Patients, materials, and methods
Patients
All patients analyzed in this study fulfilled the criteria for the Chediak-Higashi syndrome24: partial albinism with typical pigment clumping in the hair shaft, giant granulations easily detected in polymorphonuclear cells, and defective cytotoxic activity of natural killer cells. In 7 patients, the disease entered the accelerated phase (Table 1). Informed consent was obtained from patients or families for this study.
Clinical data of 8 patients with CHS
| Patient . | Age at Diagnosis Current Age (y) . | Accelerated Phases . | BMT . | Consanguinity Family History . | Neurologic Manifestations . | ||
|---|---|---|---|---|---|---|---|
| Age at Onset . | Severity . | Suspected Triggering Factor . | |||||
| 1 | 5.5 | 5.5 | ++ | EBV | + | − | − |
| 6.0* | − | ||||||
| 2 | 2.5 | 2.5 | + | EBV | + | + | Low intellectual capacity (QD = 80) |
| 22 | + | ||||||
| 3 | 14 | 14 | ++ | ? | − | + | Mental retardation |
| 16 | − | ||||||
| 4 | 1 | 1.6 | ++ | EBV | + | + | Poor performance at school |
| 14* | 14† | − | |||||
| 5 | 1.3 | 1.3 | ++ | EBV | − | − | |
| 1.5* | + | − | |||||
| 6 | 7.8 | 0 | − | − | − | − | |
| 10 | + | − | |||||
| 7 | <1 | 2 | ++ | EBV | − | + | Low intellectual capacity (QD = 75) |
| 11 | + | ||||||
| 8 | <1 | 1 | ++ | ? | + | + | |
| 1.4* | + | − | |||||
| Patient . | Age at Diagnosis Current Age (y) . | Accelerated Phases . | BMT . | Consanguinity Family History . | Neurologic Manifestations . | ||
|---|---|---|---|---|---|---|---|
| Age at Onset . | Severity . | Suspected Triggering Factor . | |||||
| 1 | 5.5 | 5.5 | ++ | EBV | + | − | − |
| 6.0* | − | ||||||
| 2 | 2.5 | 2.5 | + | EBV | + | + | Low intellectual capacity (QD = 80) |
| 22 | + | ||||||
| 3 | 14 | 14 | ++ | ? | − | + | Mental retardation |
| 16 | − | ||||||
| 4 | 1 | 1.6 | ++ | EBV | + | + | Poor performance at school |
| 14* | 14† | − | |||||
| 5 | 1.3 | 1.3 | ++ | EBV | − | − | |
| 1.5* | + | − | |||||
| 6 | 7.8 | 0 | − | − | − | − | |
| 10 | + | − | |||||
| 7 | <1 | 2 | ++ | EBV | − | + | Low intellectual capacity (QD = 75) |
| 11 | + | ||||||
| 8 | <1 | 1 | ++ | ? | + | + | |
| 1.4* | + | − | |||||
Died.
Relapse after bone marrow graft rejection.
Patients 5 and 6 are siblings.
BMT, bone marrow transplantation; CHS, Chediak–Higashi syndrome; EBV, Epstein–Barr virus.
RNA extraction and reverse transcription/nested polymerase chain reaction
Using standard techniques, total RNA was extracted as described elsewhere19 from fibroblast cell lines or from Epstein-Barr virus-transformed lymphoblastoid cell lines established from patients with CHS. The entire coding region (13.5 kb) of the LYST gene was reverse-transcribed in 9 overlapping nested reactions (mean size, 1.4 kb), and 1 μg RNA was used as a template for the first-strand cDNA synthesis. Reverse transcription (RT) was performed according to the manufacturer's (Roche Diagnostics, Neylan, France) instruction from a specific reverse primer LYSTXR (X = 1-9) (Table2) in a total volume of 20 μL. A 30-μL polymerase chain reaction (PCR) mix containing primers LYSTXF (forward) and LYSTXR (reverse) was then added to the RT reaction, and the first round of amplification was performed, consisting of 40 cycles of 94°C for 1 minute, specific annealing temperature for each pair of primers (see Table 2) for 1 minute, and 72°C for 5 minutes. A 2-μL aliquot of the RT-PCR was used in a subsequent round of nested PCR (nested RT-PCR) using inner primers (T7-LYSTX'F and LYSTX'R) (Table 2) in a final volume of 50 μL (30 cycles were performed as above). The inner forward primers were designed to include a translation initiation site and a T7 promoter sequence for the initiation of transcription by T7 RNA polymerase. One fifth of the nested RT-PCR reaction was analyzed on 1% agarose gel.
LYST primers
| Name RT-PCR . | Position 5′-3′ cDNA* . | Annealing Temperature (°C) . | Purpose . | PCR Products (bp) . | Name Nested RT-PCR . | Position 5′-3′ cDNA . | Annealing Temperature (°C) . | Purpose . | PCR Products (bp)‡ . |
|---|---|---|---|---|---|---|---|---|---|
| LYST1F LYST1R | 127-146 1740-1755 | 51 | Fragment 1 | 1628 | LYST1′Fa LYST1′R | 148-167 1651-1670 | 54 | Fragment 1′ | 1522 |
| LYST2F LYST2R | 1215-1234 2789-2808 | 56 | Fragment 2 | 1593 | LYST2′Fa LYST2′R | 1529-1547 2669-2688 | 52 | Fragment 2′ | 1157 |
| LYST3F LYST3R | 2437-2456 4185-4201 | 52 | Fragment 3 | 1764 | LYST3′Fa LYST3′R | 2641-2661 4057-4075 | 51 | Fragment 3′ | 1434 |
| LYST4F LYST4R | 3311-3319 4855-4874 | 52 | Fragment 4 | 1563 | LYST4′Fa LYST4′R | 3549-3566 4594-4613 | 50 | Fragment 4′ | 1064 |
| LYST5F LYST5R | 4459-4476 6245-6265 | 50 | Fragment 5 | 1806 | LYST5′Fa LYST5′R | 4523-4539 5957-5977 | 52 | Fragment 5′ | 1454 |
| LYST6F LYST6R | 5800-5820 7619-7636 | 53 | Fragment 6 | 1836 | LYST6′Fa LYST6′R | 5918-5938 7530-7550 | 52 | Fragment 6′ | 1632 |
| LYST7F LYST7R | 7424-7444 9149-9169 | 52 | Fragment 7 | 1745 | LYST7′Fa LYST7′R | 7477-7497 9038-9053 | 52 | Fragment 7′ | 1576 |
| LYST8F LYST8R | 8950-8970 10314-10334 | 53 | Fragment 8 | 1384 | LYST8′Fa LYST8′R | 9008-9027 10257-102761 | 53 | Fragment 8′ | 1268 |
| LYST9F LYST9R | 10120-10137 11696-11716 | 54 | Fragment 9 | 1596 | LYST9′Fa LYST9′R | 0184-10204 11677-11697 | 54 | Fragment 9′ | 1513 |
| Name RT-PCR . | Position 5′-3′ cDNA* . | Annealing Temperature (°C) . | Purpose . | PCR Products (bp) . | Name Nested RT-PCR . | Position 5′-3′ cDNA . | Annealing Temperature (°C) . | Purpose . | PCR Products (bp)‡ . |
|---|---|---|---|---|---|---|---|---|---|
| LYST1F LYST1R | 127-146 1740-1755 | 51 | Fragment 1 | 1628 | LYST1′Fa LYST1′R | 148-167 1651-1670 | 54 | Fragment 1′ | 1522 |
| LYST2F LYST2R | 1215-1234 2789-2808 | 56 | Fragment 2 | 1593 | LYST2′Fa LYST2′R | 1529-1547 2669-2688 | 52 | Fragment 2′ | 1157 |
| LYST3F LYST3R | 2437-2456 4185-4201 | 52 | Fragment 3 | 1764 | LYST3′Fa LYST3′R | 2641-2661 4057-4075 | 51 | Fragment 3′ | 1434 |
| LYST4F LYST4R | 3311-3319 4855-4874 | 52 | Fragment 4 | 1563 | LYST4′Fa LYST4′R | 3549-3566 4594-4613 | 50 | Fragment 4′ | 1064 |
| LYST5F LYST5R | 4459-4476 6245-6265 | 50 | Fragment 5 | 1806 | LYST5′Fa LYST5′R | 4523-4539 5957-5977 | 52 | Fragment 5′ | 1454 |
| LYST6F LYST6R | 5800-5820 7619-7636 | 53 | Fragment 6 | 1836 | LYST6′Fa LYST6′R | 5918-5938 7530-7550 | 52 | Fragment 6′ | 1632 |
| LYST7F LYST7R | 7424-7444 9149-9169 | 52 | Fragment 7 | 1745 | LYST7′Fa LYST7′R | 7477-7497 9038-9053 | 52 | Fragment 7′ | 1576 |
| LYST8F LYST8R | 8950-8970 10314-10334 | 53 | Fragment 8 | 1384 | LYST8′Fa LYST8′R | 9008-9027 10257-102761 | 53 | Fragment 8′ | 1268 |
| LYST9F LYST9R | 10120-10137 11696-11716 | 54 | Fragment 9 | 1596 | LYST9′Fa LYST9′R | 0184-10204 11677-11697 | 54 | Fragment 9′ | 1513 |
T7-primer with promoter and sequence for initiation of translation LYST primer sequence at 3′-position. The respective open reading frames start with the codon.
Length without T7 primer sequence, as described above.
RT-PCR, reverse transcription–polymerase chain reaction.
Protein truncation test.
Full-length RT-PCR products were subjected to the protein truncation test (PTT) to search for truncated mutations. Peptides were produced in the TNT-T7 Coupled Reticulocyte Lysate System (Promega, Lyon, France) according to the manufacturer's instructions in the presence of35S cysteine (Amersham, France) in a reaction volume of 12.5 μL. The derived protein products were separated on 12% sodium dodecyl sulphate-polyacrylamide gels and detected by autoradiography.
Direct sequencing.
PCR products showing an abnormal pattern in the PTT analysis were directly sequenced using dye terminator cycle sequencing kits on an ABI 377 automatic sequencer (PE Applied Biosystems, Foster City, CA). They were amplified using a forward primer identical to that used in the PTT-PCR without the presence of the T7 polymerase binding site and the start codon. Several mutations were sequenced in 2 different cDNA preparations, and all the mutations were confirmed on the opposite strand in at least 1 PCR. When blood samples from the patients' parents were available, heterozygous mutations were determined by direct sequencing of each specific LYST region that encompassed the mutation(s) previously identified in the probands.
Results
Clinical phenotype
The 8 patients analyzed in this study belong to 7 different families. Five patients are offspring of genetically related parents (Table 1). An accelerated phase occurred in 7 of these patients; in all but 1, it occurred before they were 6 years of age (Table 1). The accelerated phase was characterized in all by fever, hepatosplenomegaly, pancytopenia, hypertriglyceridemia, and fibrinopenia. In 5 of them (patients 1, 2, 3, 4, 5), it was characterized by the presence of mononuclear cells in the cerebrospinal fluid. In patient 2 the course of the accelerated phase was less severe. Essentially it consisted of hepatosplenomegaly and mild leucopenia that remained stable without treatment during the 6 months preceding successful BMT. In 5 of these patients, occurrence of the accelerated phase clearly correlated with Epstein-Barr virus (EBV) infection. Either a high EBV-specific antibody response or the EBV genome was detected at the time of onset; sometimes both were detected. In the 2 remaining patients, there was no assessment of EBV infection. Three patients died before (patient 5) or during (patients 1 and 8) BMT, and 1 patient (patient 4) was successfully treated by BMT. However, the total disappearance of donor cells was observed 11 years later after transplantation, and the patient died of a new accelerated phase before a second BMT could be performed. Three patients had milder forms of the disease. Patients 7 and 3 had a single accelerated phase at the age of 2 years and 14 years, respectively, that resolved rapidly under specific treatment (corticoids in addition to etoposide or cyclosporin A respectively) without further relapse to date (Table 1). The accelerated phase never occurred in patient 6, who is now 10 years old. This milder expression of CHS was not found in the other affected siblings from the same families. Indeed, the brother of patient 6 (patient 5) died of a severe form of the disease before he was 2 years of age. Patient 7 had 1 younger sister and 1 cousin who died before they were 2 years of age and 3 other affected cousins aged 11, 16, and 40 years, who are still alive.
In 4 patients older than 10 years of age (patients 2, 3, 4, and 7) at the time of investigation, mild to severe mental retardation, including slow ideation in the first 3 patients, was noted. School performance of these patients is poor (Table 1). In addition to pronounced mental deficiency and poorly developed speech, patient 3 has loss of muscle stretch reflexes, weakness, and distal atrophy.
Mutations
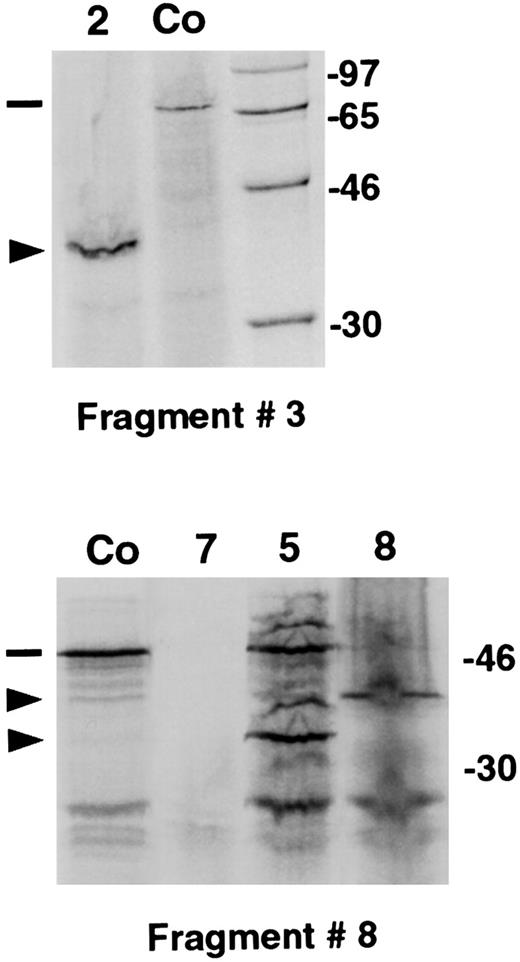
Full-length RT-PCR products for the 9 LYST overlapping fragments were obtained from the cDNA of each CHS patient studied. The products were then analyzed by PTT. All the patients exhibited an abnormal electrophoresis pattern for 1 or 2 of the 9 RT-PCR fragments studied. Truncated proteins were generated from cDNA fragment 3 from patients 1 and 2 (Figure1), from cDNA fragment 5 of patient 5 (for half the product), from cDNA fragment 6 of patient 4, from cDNA fragment 7 of patient 8, and from cDNA fragment 8 in the remaining patients (Table 3). No peptide could be detected for patient 7 after in vitro transcription and translation of fragment 8 (Figure 1). Slightly reduced band sizes of fragment 8 were observed for the samples of patients 5 and 8 (Figure1). For all patients except patient 5, the presence of a truncated protein was associated with the total disappearance of the corresponding normal product. In each case, the size of the band made it possible to localize the site of the mutation and to focus the sequencing step. The absence of a peptide suggested a mutation located at the 5′ end of the RT-PCR fragment analyzed. Direct cDNA sequencing using appropriate primers led, in each case, to the identification of the disease-causing mutation. They consisted of a nonsense mutation in patient 2, a 1-bp deletion in patients 1, 3, 5, and 8, or a deletion of several base pairs in most of the remaining patients, each of which led to a sequence frameshift (Table 3 and Figure 2). On 1 allele, a 10-bp insertion was observed in patient 5. Both heterozygous mutations identified in patient 5 were also detected in his sister (patient 6), whereas LYST cDNA of his father and mother displayed the 5317 delA and the 10-bp insertion at nucleotide 9228, respectively. In the family of patient 1, further analysis enabled us to demonstrate that the homozygous mutation identified in the proband was the result of a LYST mutation inherited from his mother, associated with a maternal, uniparental isodisomy of chromosome 1.25
Mutation screening by protein truncation test.
PTT analysis shows abnormal fragment 3 pattern in patient 2 and abnormal fragment 8 pattern in patients 5, 7, and 8 in comparison with the control (Co). (dashes) Wild-type, full-length products. (arrows) Abnormal bands. Nonspecific additional bands can be seen in some samples. Marker and molecular weights of the marker proteins (in kd) are shown on the right-hand side of the gels.
Mutation screening by protein truncation test.
PTT analysis shows abnormal fragment 3 pattern in patient 2 and abnormal fragment 8 pattern in patients 5, 7, and 8 in comparison with the control (Co). (dashes) Wild-type, full-length products. (arrows) Abnormal bands. Nonspecific additional bands can be seen in some samples. Marker and molecular weights of the marker proteins (in kd) are shown on the right-hand side of the gels.
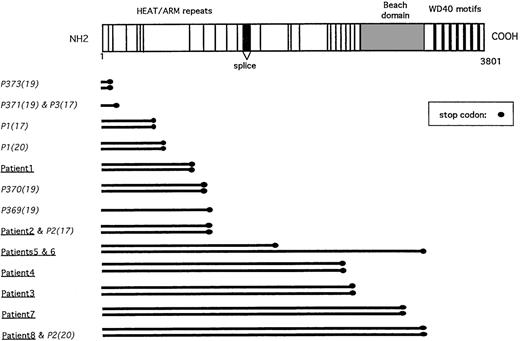
Representation of the LYST gene mutations.
Schematic representation of the LYST protein with its known motifs (HEAT and ARM repeats, BEACH domain, WD40 repeats), as previously described.17 LYST gene mutations described herein (underlined), as well as previously reported (italics), with their respective references in parentheses, are listed.
Representation of the LYST gene mutations.
Schematic representation of the LYST protein with its known motifs (HEAT and ARM repeats, BEACH domain, WD40 repeats), as previously described.17 LYST gene mutations described herein (underlined), as well as previously reported (italics), with their respective references in parentheses, are listed.
LYST mutations detected by protein truncation test
| Patient . | Zygosity . | DNA Sequence Change . | Effect on LYST mRNA . | Effect on LYST Protein . |
|---|---|---|---|---|
| 1 | Homozygous | 2620delT | Frameshift | Truncated at position 874 |
| 2 | Homozygous | C3310T | Nonsense | Truncated at position 1103 |
| 5 | Heterozygous | 5317delA | Frameshift | Truncated at position 1773 |
| 6 | 9228 + 10bpins | Frameshift | Truncated at position 3210 | |
| 4 | Homozygous | del7060-7066 | Frameshift | Truncated at position 2354 |
| 3 | Homozygous | 7555delT | Frameshift | Truncated at position 2519 |
| 7 | Homozygous | del9106-9161 | Frameshift | Truncated at position 3036 |
| 8 | Homozygous | 9590delA | Frameshift | Truncated at position 3197 |
| Patient . | Zygosity . | DNA Sequence Change . | Effect on LYST mRNA . | Effect on LYST Protein . |
|---|---|---|---|---|
| 1 | Homozygous | 2620delT | Frameshift | Truncated at position 874 |
| 2 | Homozygous | C3310T | Nonsense | Truncated at position 1103 |
| 5 | Heterozygous | 5317delA | Frameshift | Truncated at position 1773 |
| 6 | 9228 + 10bpins | Frameshift | Truncated at position 3210 | |
| 4 | Homozygous | del7060-7066 | Frameshift | Truncated at position 2354 |
| 3 | Homozygous | 7555delT | Frameshift | Truncated at position 2519 |
| 7 | Homozygous | del9106-9161 | Frameshift | Truncated at position 3036 |
| 8 | Homozygous | 9590delA | Frameshift | Truncated at position 3197 |
In addition to the normal-sized product expected from the position of the primers, shorter PCR products were also amplified in some patients after the second PCR round, though at a less abundant level (data not shown). After gel purification of the extra bands, direct cDNA sequencing was performed. In each case, amplification started at the 5′ or the 3′ part of the expected product with unspecific annealing of 1 primer. Sequence anomalies were still detectable when corresponding shorter products were amplified. These additional PCR bands probably accounted for the shorter PTT extra bands detected in some of the samples. Thus any biologic relevance of these alternative transcripts is unlikely.
Discussion
The lysosomal trafficking regulator (LYST) gene was recently identified and was found to have mutated in patients with Chediak-Higashi syndrome16,17 and in beige mice.18 In each species, all the LYST gene alterations published so far result in a truncated protein because of nonsense or frameshift mutations.17,19,20 This observation—in addition to the large size of the LYST cDNA (13.5 kb), the complexity of the gene, and the scattering of mutations across the coding sequence with no apparent clustering detected—makes PTT suitable for mutation screening of the LYST gene.21-23 26 Furthermore, mutations identified by PTT have immediate clinical relevance, whereas amino acid substitutions might still turn out to be false positive as a result of rare polymorphisms with no causal relationship to the disease phenotype. We thus established the technical conditions necessary to study LYST gene mutations by using this approach, and we analyzed 8 patients with CHS in 7 different families.
PTT analysis enabled us to identify mutations in all the patients studied. Six mutations were homozygous at the protein and at the nucleic acid sequence levels (Table 3). Four mutations occurred in patients from consanguineous families. In the remaining patient, the homozygous mutation was the result of a maternal chromosome 1 isodisomy inherited from a heterozygous carrier mother.25 In sibling patients 5 and 6, 2 heterozygous mutations inherited from each of their unrelated parents led to a stop codon. Thus, in addition to the 9 patients with CHS previously reported in the literature,16,17,19 20 the LYST gene mutations identified in the 7 families reported herein also exhibited truncations of the LYST protein (Figure 2). These results justify the use of PTT for rapid analysis of this huge gene. They also suggest that missense substitutions in the LYST gene may result in a milder phenotype, a different clinical phenotype, or the absence of any phenotype. These should be determined by studying a larger series of patients. Alternatively, these results may indicate that the presence of the terminal part of the LYST protein, which contains the WD40 motifs suspected to play a role in protein–protein interaction, may be a determining factor in the function of the LYST protein. Analysis of additional patients would help to answer these intriguing questions.
Although the immunologic defect in CHS implicates defective chemotaxis of granulocytes, delayed intracellular microorganism killing, and lack of natural killer cell cytotoxic activity, some of these features are inconstantly observed in the patients and are probably not directly responsible for the severity of this disorder. The primary immunologic symptom that determines the prognosis of CHS is the accelerated phase. It has been reported that the onset of the accelerated phase and the patient's age at onset are key prognostic factors of CHS.2 Our results strengthen this observation. Among the 6 patients in whom the accelerated phase occurred before the age of 6, 5 died after the initial onset (4 patients) or a relapse (1 patient). Only 1 patient had early onset and no relapse 9 years later. In contrast, each patient with no or late (14 years of age) development of the accelerated phase is still alive without having undergone BMT.
A clear correlation between genotype and accelerated phase occurrence was not observed among this limited number of patients. Indeed, within a given family, variation in the severity of the phenotypic expression of CHS can be observed. Patient 5 died at the age of 18 months during the course of an accelerated phase, whereas his older sister is still immunologically asymptomatic at 10 years of age. In her, the diagnosis was made only because of the diagnosis of CHS in her younger brother. A similar observation was made in the family of patient 7. Two affected siblings died after onset of the accelerated phase before they were 2 years of age, whereas 3 other relatives who did not undergo BMT and have not had symptoms of the accelerated phase are still alive at 11, 16, and 40 years of age. In addition, variations in the phenotypic expression of the disease can be observed among unrelated patients with identical LYST mutations. Patient 2, in whom the accelerated phase occurred at the age of 2 years, shares a mutation with a previously reported patient who had late-onset CHS expression.17Finally, it is striking to note that the phenotypic expression of this disease does not seem to correlate with the length of the residual LYST truncated protein. In both patients with terminal truncation of the LYST protein, classical severe expression of the disease was observed, whereas less severe forms were observed in patients with more proximal truncations (patients 3 and 7). However, studying additional patients and better determining the functional domains of the LYST protein are necessary to draw definitive conclusions about the phenotypic consequences of these mutations.
These data indicate that neither the clinical expression in another family nor the determination of that family's mutation can easily help in therapeutic decisions; the overall outcome of CHS is generally poor. These data also indicate that other factors of genetic or environmental origin could modify the clinical expression of CHS. In this respect, it is worth noting the role of EBV infection as a trigger of the accelerated phase. To some extent, variability in the accelerated phase is reminiscent of the most variable outcome of X-linked lymphoproliferation, a genetic condition in which the SAP/SH2D1A/DHSP27-29 gene defect often leads to a condition phenotypically close to the accelerated phase of CHS once patients have been infected by EBV. Additional studies of patients with CHS are required to assess better the roles of LYST mutations on the function of this protein and of other intervening factors in the outcome of the disease.
Acknowledgments
We thank the families involved in the study for their cooperation. We also thank Drs M. Duval, F. Bertey, W. Friedrich, and the physicians of the Department of Pediatric Immunology and Hematology, Hôpital Necker-Enfants Malades, for their part in the recruitment and follow-up of patients. We thank S. Tuffery for helpful technical discussions regarding PTT.
Supported in part by grants from l'Institut National de la Santéet de la Recherche Médicale (INSERM), l'Association Vaincre les Maladies Lysosomiales, l'Association Française contre les Myopathies, l'Assistance Publique Hôpitaux de Paris, and the CDCHT-ULAM-383-91 (Merida-Venezuela).
Reprints:Geneviève de Saint Basile, INSERM U429, Hôpital Necker-Enfants Malades, 149 rue de Sèvres, 75743 Paris, Cedex 15, France; e-mail: sbasile@necker.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal