The continuous generation of mature blood cells from hematopoietic progenitor cells requires a highly complex series of molecular events. To examine lineage-specific gene expression during the differentiation process, we developed a novel method combiningLacZ reporter gene analysis with in vitro hematopoietic differentiation induction from mouse embryonic stem cells. For a model system using this method, we chose the erythroid and megakaryocytic differentiation pathways. Although erythroid and megakaryocytic cells possess distinct functional and morphologic features, these 2 lineages originate from bipotential erythro-megakaryocytic progenitors and share common lineage-restricted transcription factors. A portion of the 5′ flanking region of the human glycoprotein IIb (IIb) integrin gene extending from base −598 to base +33 was examined in detail. As reported previously, this region is sufficient for megakaryocyte-specific gene expression. However, previous reports that used human erythro-megakaryocytic cell lines suggested that one or more negative regulatory regions were necessary for megakaryocyte-specific gene expression. Our data clearly showed that an approximately 200-base enhancer region extending from −598 to −400 was sufficient for megakaryocyte-specific gene expression. This experimental system has advantages over those using erythro-megakaryocytic cell lines because it recapitulates normal hematopoietic cell development and differentiation. Furthermore, this system is more efficient than transgenic analysis and can easily examine gene expression with null mutations of specific genes.
The process of cell differentiation is regulated through the complex coordination of cell-type–specific gene activation brought about by the interactions of cis- andtrans-acting regulatory elements. The hematopoietic system, one of the stem cell systems, is a useful experimental model for analyzing this coordination because more than 8 distinct blood cell lineages arise from single multipotential hematopoietic stem cells within a relatively short period.1 Recent gene-targeting experiments have demonstrated that various lineage-specific or differentiation-stage–specific transcription factors play crucial roles during this process.2,3 However, our understanding of how lineage commitment is orchestrated by many transcription factors and how lineage-specific gene expression is tightly controlled within individual hematopoietic lineages remains limited.4
In the hematopoietic system, the erythroid and megakaryocytic lineages are considered closely related for 2 reasons. First, both erythrocytes and megakaryocytes differentiate through common committed erythro-megakaryocytic progenitors.5-7 Second, these 2 lineages share several features in common. For example, GATA-1, a principal transcriptional regulator of erythropoiesis, also has an important role in the production of platelets.8-10 Other important transcription factors and cofactors, such as NF-E2 and FOG, which were first discovered in the erythroid lineage, play important roles in platelet production.11-14 In addition, some leukemic cells possess both erythroid and megakaryocytic cell-surface markers.15-17 Although these findings illustrate similarities in the control of the gene expression programs in these 2 lineages, the morphologic and functional features of these 2 lineages are quite different. Therefore, these 2 lineages must possess discrete, specific gene expression programs for commitment to individual cell lineages. Analyzing the regulation of differential gene expression within these 2 closely related lineages should provide clues to our understanding of the mechanism of hematopoietic commitment.
The glycoprotein IIb (GPIIb) gene is an excellent lineage-restricted marker for examining megakaryocyte-specific gene expression because the gene is an early and specific marker of megakaryocytes.18,19 Several groups have analyzed the promoter region of the human GPIIb gene.10,16,17,20,21 All of these studies used the transient expression of reporter genes in human erythro-megakaryocytic leukemic cell lines such as HEL (human erythroleukemia) and K562. These studies showed that 2 kinds of transcription factor binding sites, GATA and Ets binding sites, play important roles in the positive regulation of GPIIb promoter activity.10,17,20 However, some evidence suggests that these 2 sites are not sufficient for megakaryocyte-specific expression and that negative cis-regulatory elements, such as those located at −198/−178 and −124/−99 bp15 or −120/−116 and −102/−93 bp,21 are important for suppressing GPIIb expression in cell lineages other than the megakaryocyte lineage.
The use of “erythro-megakaryocytic” leukemic cell lines may be unsuitable for studying the mechanisms governing the discrimination between erythroid and megakaryocytic lineages because most megakaryocytic and erythroid leukemic cell lines concomitantly express erythroid and megakaryocytic markers, respectively. Most of the data on megakaryocyte-specific gene expression have been obtained by comparing gene expression in erythro-megakaryocytic cell lines with that in irrelevant cell lines such as HeLa.20-22 In addition, several groups using different erythro-megakaryocytic cell lines have reported conflicting results.21 23 To circumvent these dubious situations, it seems preferable to use a different experimental system and to compare the results.
One of the best experimental systems to analyze promoter activity during development and cell differentiation is the transgenic mouse, and this has been validated by the numerous studies of human β-globin locus gene expression.24 However, the analysis of many promoters and their mutants requires many transgenic lines and is tremendously time and labor intensive. We have developed an alternative strategy, which combines promoter-reporter gene analysis and in vitro hematopoietic differentiation from mouse embryonic stem cells (ES cells).25,26 ES cells can give rise to hematopoietic cells, including erythroid and megakaryocyte lineages, when cultured with the macrophage colony-stimulating factor (M-CSF)-deficient OP9 stromal cell line (OP9 system).27-29 The process of hematopoiesis in this system is considered to reflect accurately the early phase of in vivo hematopoietic development.
Here, we show that a 200-bp enhancer element containing a GATA site and an adjacent Ets binding site is necessary and sufficient for megakaryocyte lineage-specific expression. The experimental system developed in this study is a useful, rapid, and reliable method to examine gene-regulatory mechanisms during hematopoiesis.
Materials and methods
Construction of reporter plasmid
The pBS-LacZ vector was constructed by inserting a 4.4-kbHindIII-ApaI fragment of the bacterial LacZgene with a nuclear localization signal (kindly provided by Dr Takeuchi30) into the HindIII and ApaI sites of pBluescript (StrataGene, La Jolla, CA). The human GPIIb promoter from bases −1121 to +516 (transcriptional initiation site is designated as +1) was amplified by polymerase chain reaction (PCR) using primers 5′-TTCCGCTTACCGAGAGAAAA-3′ and 5′-GGGTGGAGTGGTTCATACAA-3′, as described previously.31 The PCR product was cloned into pBluescript and the sequence was confirmed. The sequences of all the PCR products produced in this study were confirmed by DNA sequencing using the Thermosequenase kit (Amersham, Uppsala, Sweden) and a LI-CORauto sequencer (Lincoln, NE).32 The GPIIb fragment was digested with HindIII and MboII to generate a HindIII-MboII segment containing the genomic sequence from nucleotide −598 to +33. The pBS-GPIIb-LacZ parental vector was constructed by inserting this GPIIbHindIII-MboII fragment into the HindIII site of the pBS-LacZ vector. Vector pMC1NEO EF-1 LacZ was constructed by inserting the LacZ gene into the XbaI site of vector pMC1NEO EF-1α.
Various mutants containing deletions or point mutations were produced by PCR.33 To produce a mutant of the region from −118 to −93, all the A's in this region were replaced by C's, the G's by T's, and vice versa. The GATA at site −463 was converted from 5′-TTTATCGG-3′ (the GATA site is underlined) to 5′-TTTAGAGG-3′ using the oligonucleotide 5′-TACAGAAGCCTCAGGTTTTAGAGGGGGCAGCAGCTTCCTTCT-3′. The Ets at site −515 was converted from 5′-AAGGAGG-3′ to 5′-AAGGATT-3′ using the oligonucleotide 5′-CGCCCAAGGTCCTAGAAGGATTAAGTGGGTAAATGCCATATC-3′. The vector pBS-TK-LacZ was constructed by ligating the LacZ gene and the minimal TK promoter, which was obtained by PCR from the pTK vector (Nippon Gene, Toyama, Japan) using primers 5′-CCCGGATCCGGCCCCGCCCAGCGTC-3′ and 5′-CCCAAGCTTAGATCTGCGGCACGCTGTT-3′, into the ApaI and BamHI site of pBluescript, respectively. Four tandem repeats of oligonucleotides including the GATA site (−463) flanked with 13 bp on each end and 4 base pairs ofXhoI-cohesive ends (5′-CGAGAAGCCTCAGGTTTTATCGGGGGCAGCAGCT-3′ and 5′-CTCGAGCTGCTGCCCCCGATAAAACCTGAGGCTT-3′) were cloned into the BamHI site of the pBS-TK-LacZ vector by blunt end ligation. Two tandem repeats of oligonucleotides containing 2 tandem repeats of the distal Ets binding site flanked with 13 bp on both ends and overhanging BamHI-cohesive ends (5′-GATCCAAGGTCCTAGAAGGAGGAAGTGGGTAAATGAAGGTCCTAGAAGGAGGAAGTGGGTAAATGG-3′ and 5′-GATCCCATTTACCCACTTCCTCCTTCTAGGACCTTCATTTACCCACTTCCTCCTTCTAGGACCTTG-3′) were cloned into the BamHI site of the pBS-TK-LacZ vector.
Cell culture and electroporation
D3 ES cells were maintained on embryonic fibroblasts using standard procedures.34 OP9 cells were maintained in α-minimal essential medium supplemented with 20% fetal calf serum and standard antibiotics.27 For differentiation induction, ES cells were seeded onto confluent OP9 cell layers on 6-well plates at a density of 104 cells per well. On day 5 of the differentiation induction, these cells were treated with 0.25% trypsin and seeded onto a fresh confluent OP9 cell layer in the presence of erythropoietin (EPO), thrombopoietin (TPO), or M-CSF to support the differentiation and proliferation of erythroid cells, megakaryocytes, and macrophages, respectively. Erythroid cells and megakaryocytes were analyzed on days 8 and 12 of differentiation induction. Macrophages were analyzed on day 10 or 11.
Reporter plasmids and plasmid pMC1NEO EF-1 containing the neomycin resistance gene were cotransfected at a 20:1 molar ratio into ES cells by electroporation using a gene pulser (Bio-Rad, Hercules, CA) set at 500 μF, 230 V. Transfected ES cells were seeded on a neomycin-resistant layer of embryonic fibroblasts in 10-cm dishes or 6-well plates with G418. The puromycin resistance gene was similarly used to examine gene expression inc-ets-1−/−ES cells, which were established as described previously.35 Dr Yagi (National Institute for Physiological Science, Okazaki, Japan) kindly provided the puromycin-resistant gene and puromycin-resistant STO cells.36 In most experiments, ES cells were harvested from individual wells of 6-well plates when confluent, and the total cells harvested from individual wells were induced to differentiate as described earlier.
Staining and histologic analysis of hematopoietic cells
The hematopoietic cells generated from the transfected ES cells were collected from the supernatants of the induction cultures, and cytospins were produced. These cells were fixed in 1% formaldehyde for 8 minutes and washed 3 times with phosphate-buffered saline (PBS) at room temperature. X-gal staining was carried out overnight at 37°C in PBS containing 1 mg/mL X-gal, 5 mmol/L K3Fe(CN)6, 5 mmol/L K4Fe(CN)6, and 2 mmol/L MgCl2, pH 7.3. Stained cells were washed twice with deionized water. To verify the lineages of the cells, we used acetylcholine esterase (AChE) staining, which is specific for mouse megakaryocytes, and dianisidine staining for hemoglobinized cells. After drying the slides completely, we performed AChE staining at room temperature for 3 hours in 0.1 mol/L PBS containing 0.05% acetylthiocholine iodide, 0.1 mol/L sodium citrate, 30 mmol/L copper sulfate, and 5 mmol/L potassium ferricyanide (pH 6.0). Dianisidine staining for erythroid lineage cells was carried out as described previously.37 Macrophages were not counterstained because more than 95% of the cells harvested from cultures containing M-CSF were macrophages. The percentages ofLacZ-positive megakaryocytes, erythroid cells, and macrophages harvested from the cultures containing EPO, TPO, and M-CSF, respectively, were counted under a microscope. We counted 250 AChE-positive cells, 1000 erythroid cells, and 1000 macrophages to calculate the percentage of LacZ-positive cells. The percentages of LacZ-positive cells were compared statistically with the t test using StatView statistical analysis software.
Results
Selective proliferation of megakaryocytes, erythroid cells, and macrophages by the OP9 differentiation induction system with the addition of growth factors
Our previous studies demonstrated that multipotential hematopoietic progenitors developed from ES cells in the OP9 system.27 38TPO, EPO, or M-CSF was added from day 5 of differentiation induction for the efficient propagation of megakaryocytes, erythroid lineage cells, and macrophages, respectively (Figure1). Preferential proliferation of megakaryocytes was observed in the presence of TPO, and more than 90% of the cells at day 8 expressed AChE, a cytologic marker of murine megakaryocytes. As in our previous studies, primitive embryonic and definitive adult erythroid cells appeared between days 6 and 8 and between days 12 and 15 of the differentiation induction, respectively. In the presence of EPO, more than 90% of the cells were dianisidine-positive hemoglobinized cells at days 8 and 12 of induction. On days 10 and 11, the vast majority (> 95%) of the cells induced in the presence of M-CSF were macrophages, which was confirmed by staining with anti–Mac-1 antibody.
Differentiation induction into megakaryocytes, erythroid cells, and macrophages and LacZ expression driven by the GPIIb promoter and EF1- promoter in these cells.
Hematopoietic cells were generated from ES cells by the addition of TPO (A, E, I), EPO (B, F, J for primitive embryonic erythrocytes and C, G, K for definitive adult erythrocytes), and M-CSF (D, H, L) for megakaryocytes, erythroid cells, and macrophages, respectively. May-Grunwald Giemsa staining was used to detect morphologic features (A-D). The LacZ gene driven by the GPIIb promoter from −598 to +33 (E-H) and the elongation factor-1α promoter (I-L) were transfected into ES cells with a plasmid carrying the neomycin-resistant gene. Differentiation induction was carried out after G418 selection, and the cells were stained with X-gal. Double staining with AChE was performed to confirm LacZ expression in megakaryocytes.
Differentiation induction into megakaryocytes, erythroid cells, and macrophages and LacZ expression driven by the GPIIb promoter and EF1- promoter in these cells.
Hematopoietic cells were generated from ES cells by the addition of TPO (A, E, I), EPO (B, F, J for primitive embryonic erythrocytes and C, G, K for definitive adult erythrocytes), and M-CSF (D, H, L) for megakaryocytes, erythroid cells, and macrophages, respectively. May-Grunwald Giemsa staining was used to detect morphologic features (A-D). The LacZ gene driven by the GPIIb promoter from −598 to +33 (E-H) and the elongation factor-1α promoter (I-L) were transfected into ES cells with a plasmid carrying the neomycin-resistant gene. Differentiation induction was carried out after G418 selection, and the cells were stained with X-gal. Double staining with AChE was performed to confirm LacZ expression in megakaryocytes.
Megakaryocyte-specific gene expression of the GPIIb promoter in the OP9 system
To ascertain the feasibility of the experimental system, we analyzed the GPIIb promoter regions encompassing bases −598 to +33 (−598/+33), −400 to +33 (−400/+33), −100 to +33 (−100/+33), and −598 to −400 plus −100 to + 33 (−598/−400:−100/+33). Constructs consisting of theLacZ reporter gene ligated downstream of the GPIIb promoter and its derivatives were introduced into ES cells by cotransfection with the neomycin resistance gene. For a clonal analysis of the LacZgene, individual stable transformants were cloned after G418 selection. Differentiation induction of individual ES cell clones was performed in the presence of TPO, and the clones that gave rise toLacZ-positive megakaryocytes were selected. The expression ofLacZ in erythroid lineage cells and macrophages was not examined in this preliminary selection. Differentiation induction of these preselected clones was carried out in the presence of TPO, EPO, or M-CSF (Figure 1). The percentages of LacZ-positive cells in megakaryocytes, erythroid cells, and macrophages are shown in Table1. These results show that the shorter the GPIIb promoter regions were, the lower the proportion of positive cells. There were no differences in the intensity of the LacZstaining of the positive cells (data not shown), and thereforeLacZ expression in individual positive cells was all or none. In other words, the percentage of LacZ-positive cells, but not the intensity of LacZ staining, could be used reliably as an index of promoter activity. This all-or-none type of expression will be discussed later in more detail (see Discussion). The expression of theLacZ gene driven by the various GPIIb promoters was completely restricted to megakaryocytes, although the data obtained with the −100/+33 promoter derivative were difficult to interpret because the percentage of LacZ-positive cells was low, even in megakaryocytes. In addition, lineage-restricted LacZ expression was dependent on the promoter activity and not the site of integration because LacZ gene expression driven by the elongation factor-1α (EF-1α) promoter was observed in all 3 hematopoietic lineage cells (Table 1, Figure 1). The percentage ofLacZ-positive megakaryocytes tended to decrease during differentiation when the GPIIb promoter was used. In contrast, no decrease was observed when the EF-1α promoter was used. The reasons for the decrease in GPIIb promoter activity and the difference between these promoters remain unknown.
Clonal analysis of LacZ-positive cells after differentiation induction
| Promoter* . | Percentage of LacZ+ Cells . | ||||
|---|---|---|---|---|---|
| Megakaryocytes . | Erythrocytes . | Macrophages . | |||
| Day 8 . | Day 12 . | Day 8 . | Day 12 . | Day 10 . | |
| GPIIb (−598/+33) | 97.7 62.0 | 42.8 18.2 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 69.5 | 23.6 | <0.1 | <0.1 | <0.1 | |
| 85.8 | 37.4 | <0.1 | <0.1 | <0.1 | |
| 73.4 | 11.8 | <0.1 | <0.1 | <0.1 | |
| 78.0 | 63.5 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−400/+33) | 23.8 <0.1 | 53.4 0.5 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 5.8 | 9.6 | <0.1 | <0.1 | <0.1 | |
| <0.1 | 1.0 | <0.1 | <0.1 | <0.1 | |
| 0.5 | <0.1 | <0.1 | <0.1 | <0.1 | |
| 0.5 | <0.1 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−100/+33) | 0.9 4.7 | <0.1 6.3 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 2.0 | 0.9 | <0.1 | <0.1 | <0.1 | |
| 2.9 | <0.1 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−598/−400: −100/+33) | 48.5 72.2 58.3 | 31.1 18.2 14.1 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 |
| 27.2 | 3.7 | <0.1 | <0.1 | <0.1 | |
| 20.0 | 1.0 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−118/−93 m) | 98.6 3.8 | 96.5 1.9 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 33.5 | 8.2 | <0.1 | <0.1 | <0.1 | |
| 5.8 | 2.5 | <0.1 | <0.1 | <0.1 | |
| 11.4 | 2.1 | <0.1 | <0.1 | <0.1 | |
| 1.8 | 0.5 | <0.1 | <0.1 | <0.1 | |
| 26.8 | 3.0 | <0.1 | <0.1 | <0.1 | |
| 44.6 | 10.2 | <0.1 | <0.1 | <0.1 | |
| 6.5 | 0.9 | <0.1 | <0.1 | <0.1 | |
| EF-1α | ND | 3.5 | 3.4 | 0.2 | 7.0 |
| ND | 28.0 | 22.2 | 2.6 | 19.5 | |
| ND | 49.0 | 52.4 | 5.8 | 55.0 | |
| 6.3 | 9.7 | 5.5 | 1.1 | 2.9 | |
| 6.0 | 6.2 | 0.9 | 0.2 | 4.1 | |
| 25.1 | 32.1 | 10.3 | 6.5 | 20.4 | |
| 3.2 | 2.7 | 1.9 | 0.4 | 0.9 | |
| 32.5 | 32.4 | 33.1 | 9.3 | 20.2 | |
| Promoter* . | Percentage of LacZ+ Cells . | ||||
|---|---|---|---|---|---|
| Megakaryocytes . | Erythrocytes . | Macrophages . | |||
| Day 8 . | Day 12 . | Day 8 . | Day 12 . | Day 10 . | |
| GPIIb (−598/+33) | 97.7 62.0 | 42.8 18.2 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 69.5 | 23.6 | <0.1 | <0.1 | <0.1 | |
| 85.8 | 37.4 | <0.1 | <0.1 | <0.1 | |
| 73.4 | 11.8 | <0.1 | <0.1 | <0.1 | |
| 78.0 | 63.5 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−400/+33) | 23.8 <0.1 | 53.4 0.5 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 5.8 | 9.6 | <0.1 | <0.1 | <0.1 | |
| <0.1 | 1.0 | <0.1 | <0.1 | <0.1 | |
| 0.5 | <0.1 | <0.1 | <0.1 | <0.1 | |
| 0.5 | <0.1 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−100/+33) | 0.9 4.7 | <0.1 6.3 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 2.0 | 0.9 | <0.1 | <0.1 | <0.1 | |
| 2.9 | <0.1 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−598/−400: −100/+33) | 48.5 72.2 58.3 | 31.1 18.2 14.1 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 |
| 27.2 | 3.7 | <0.1 | <0.1 | <0.1 | |
| 20.0 | 1.0 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−118/−93 m) | 98.6 3.8 | 96.5 1.9 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 33.5 | 8.2 | <0.1 | <0.1 | <0.1 | |
| 5.8 | 2.5 | <0.1 | <0.1 | <0.1 | |
| 11.4 | 2.1 | <0.1 | <0.1 | <0.1 | |
| 1.8 | 0.5 | <0.1 | <0.1 | <0.1 | |
| 26.8 | 3.0 | <0.1 | <0.1 | <0.1 | |
| 44.6 | 10.2 | <0.1 | <0.1 | <0.1 | |
| 6.5 | 0.9 | <0.1 | <0.1 | <0.1 | |
| EF-1α | ND | 3.5 | 3.4 | 0.2 | 7.0 |
| ND | 28.0 | 22.2 | 2.6 | 19.5 | |
| ND | 49.0 | 52.4 | 5.8 | 55.0 | |
| 6.3 | 9.7 | 5.5 | 1.1 | 2.9 | |
| 6.0 | 6.2 | 0.9 | 0.2 | 4.1 | |
| 25.1 | 32.1 | 10.3 | 6.5 | 20.4 | |
| 3.2 | 2.7 | 1.9 | 0.4 | 0.9 | |
| 32.5 | 32.4 | 33.1 | 9.3 | 20.2 | |
Hematopoietic cells were generated from ES cells by the addition of TPO, EPO, or M-CSF for megakaryocytes, erythroid cells, and macrophages, respectively. ES cell clones that gave rise to LacZ-positive megakaryocytes were chosen randomly and the percentages of LacZ-positive cells relative to the total cells belonging to each lineage were calculated. Various mutated GPIIb promoters and the control EF-1α promoter were analyzed. The names of the GPIIb promoters are described in the text. ND indicates not determined.
The numbers in parentheses indicate the region of the GPIIb promoter. For example, −598/−400:−100/+33 indicates that a reporter construct consisting of nucleotides −598 to −400 and −100 to +33 of the GPIIb promoter was analyzed.
Although promoter analysis in hematopoietic cells derived from ES cell clones in the OP9 system is more efficient than promoter analysis in a transgenic mouse system, clonal analysis after preselection is still labor intensive. To streamline the process, we pooled ES clones after drug selection, induced differentiation, and then counted theLacZ-positive cells. When 5 to 10 preselected clones were pooled, the percentages of LacZ-positive cells correlated well with those observed in the clonal analysis described earlier (data not shown). Next we induced the differentiation of G418-resistant ES cells in individual wells of 6-well plates without preselection and analyzed the percentages of LacZ-positive cells. To validate the pool analysis, we analyzed pools of ES cells with introduced LacZgenes under the control of various GPIIb promoter derivatives (−598/+33, −100/+33, and −598/−400: −100/+33). Eight million ES cells were electroporated as described in Materials and methods and were cultured in 6-well plates. Individual wells typically contained approximately 50 to 100 independent ES cell clones. The results obtained from pools of individual wells (Table 2) agreed well with those of the clonal analysis (Table 1). Pools from individual wells were used in most subsequent experiments.
Analysis of LacZ-positive cells as pools after differentiation induction
| Promoter* . | Percentages of LacZ+ Cells . | ||||
|---|---|---|---|---|---|
| Megakaryocytes . | Erythrocytes . | Macrophages . | |||
| Day 8 . | Day 12 . | Day 8 . | Day 12 . | Day 10 . | |
| GPIIb (−598/+33) | 32.3 33.2 | 15.4 21.4 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 32.9 | 16.8 | <0.1 | <0.1 | <0.1 | |
| 27.7 | 12.9 | <0.1 | <0.1 | <0.1 | |
| 35.4 | 18.8 | <0.1 | <0.1 | <0.1 | |
| 28.8 | 16.3 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−100/+33) | <0.1 0.8 | <0.1 0.5 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 3.9 | 1.0 | <0.1 | <0.1 | <0.1 | |
| <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | |
| <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | |
| <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−598/−400: −100/+33) | 41.2 21.0 25.0 | 23.1 12.8 16.2 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 |
| 25.0 | 10.3 | <0.1 | <0.1 | <0.1 | |
| 19.1 | 12.3 | <0.1 | <0.1 | <0.1 | |
| 33.3 | 25.0 | <0.1 | <0.1 | <0.1 | |
| Promoter* . | Percentages of LacZ+ Cells . | ||||
|---|---|---|---|---|---|
| Megakaryocytes . | Erythrocytes . | Macrophages . | |||
| Day 8 . | Day 12 . | Day 8 . | Day 12 . | Day 10 . | |
| GPIIb (−598/+33) | 32.3 33.2 | 15.4 21.4 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 32.9 | 16.8 | <0.1 | <0.1 | <0.1 | |
| 27.7 | 12.9 | <0.1 | <0.1 | <0.1 | |
| 35.4 | 18.8 | <0.1 | <0.1 | <0.1 | |
| 28.8 | 16.3 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−100/+33) | <0.1 0.8 | <0.1 0.5 | <0.1 <0.1 | <0.1 <0.1 | <0.1 <0.1 |
| 3.9 | 1.0 | <0.1 | <0.1 | <0.1 | |
| <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | |
| <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | |
| <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | |
| GPIIb (−598/−400: −100/+33) | 41.2 21.0 25.0 | 23.1 12.8 16.2 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 | <0.1 <0.1 <0.1 |
| 25.0 | 10.3 | <0.1 | <0.1 | <0.1 | |
| 19.1 | 12.3 | <0.1 | <0.1 | <0.1 | |
| 33.3 | 25.0 | <0.1 | <0.1 | <0.1 | |
LacZ-positive cells from individual wells as pools were analyzed after differentiation induction. The experiment was similar to that in Table 1.
A lineage-specifying negative regulatory element for megakaryocytes does not exist
To examine which portion(s) of the GPIIb promoter contains the important regulatory element(s), we created serial 30-bp deletion mutants of the −598/+33 GPIIb promoter and analyzed their promoter activities (Figure 2). Only 1 mutant promoter induced a significantly higher percentage of positive cells than the wild-type promoter; this mutant lacked nucleotides between −118 and −89 (Δ−118/−89). As discussed later in more detail, recent studies have suggested that the megakaryocyte-specific expression of GPIIb is regulated by putative negative regulatory elements, which suppress the expression of GPIIb in hematopoietic lineages other than megakaryocytes.15,21,22One of the putative negative regulatory elements was localized in region −118/−93.21 Although the mutant promoter Δ−118/−89 significantly increased the level of expression by up to 150% of the wild-type promoter, the expression ofLacZ was restricted to megakaryocytes (data not shown). To further analyze this putative negative regulatory element, other mutated promoters, which were examined in a previous article,21 were analyzed in the OP9 system. One mutant promoter harboring a deletion between −148 and −59 and another with an altered nucleotide between −118 and −93 (GPIIb −118/−93m) showed megakaryocyte-specific expression (Table 1, Figure 3). Combined with the data obtained from the analysis of the serial 30-bp–deletion mutants, it is clear that the cell lineage specificity of the GPIIb promoter is not controlled by a negative regulatory mechanism in our experimental system.
LacZ expression of serial 30-bp deletion mutants in megakaryocytes.
The names of the mutants indicate the nucleotides deleted from the −598/+33 wild-type GPIIb promoter. For example, Δ−538/−509 shows the data from the promoter lacking the nucleotides between −538 and −509 bp. These constructs were cotransfected to undifferentiated ES cells with a plasmid carrying the neomycin-resistant gene. After G418 selection, ES cells pooled from individual wells of 6-well plates (see text) were differentiated into megakaryocytes with OP9 stromal cells in the presence of TPO. The percentages of LacZ-expressing megakaryocytes were calculated on day 8 of differentiation induction using double staining with AChE and X-gal. The results are shown as the relative percentage of LacZexpression compared with the value for the −598/+33 wild-type promoter. The data are shown as the mean ± standard deviation. The data marked with * show significant differences (P < .05 by t test) compared with the wild-type (−598/+33) promoter. The experiment was performed 2 or 3 times, and representative data are shown.
LacZ expression of serial 30-bp deletion mutants in megakaryocytes.
The names of the mutants indicate the nucleotides deleted from the −598/+33 wild-type GPIIb promoter. For example, Δ−538/−509 shows the data from the promoter lacking the nucleotides between −538 and −509 bp. These constructs were cotransfected to undifferentiated ES cells with a plasmid carrying the neomycin-resistant gene. After G418 selection, ES cells pooled from individual wells of 6-well plates (see text) were differentiated into megakaryocytes with OP9 stromal cells in the presence of TPO. The percentages of LacZ-expressing megakaryocytes were calculated on day 8 of differentiation induction using double staining with AChE and X-gal. The results are shown as the relative percentage of LacZexpression compared with the value for the −598/+33 wild-type promoter. The data are shown as the mean ± standard deviation. The data marked with * show significant differences (P < .05 by t test) compared with the wild-type (−598/+33) promoter. The experiment was performed 2 or 3 times, and representative data are shown.
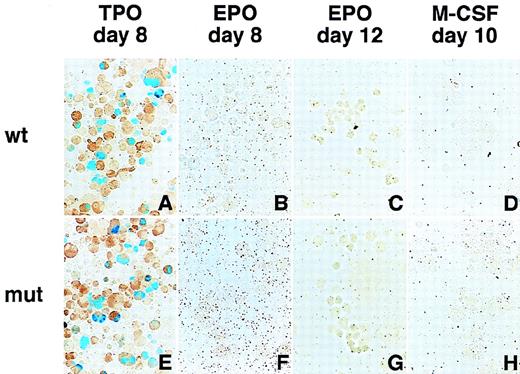
Megakaryocyte-specific expression of the GPIIb promoter containing mutations in the putative negative regulatory element.
Hematopoietic cells were generated from ES cells by the addition of TPO (A, E), EPO (B, F for primitive embryonic erythrocytes and C, G for definitive adult erythrocytes), and M-CSF (D, H) for megakaryocytes, erythroid cells, and macrophages, respectively. The wild-type and mutant GPIIb promoter constructs were transfected and analyzed as described in Figure 2. The data from the wild-type (−598/+33) promoter (A-D) and the mutated promoter in which the nucleotides between −148 and −89 (E-H) were deleted are shown. Promoters lacking −118 to −89 and −148 to −59 show similar results.
Megakaryocyte-specific expression of the GPIIb promoter containing mutations in the putative negative regulatory element.
Hematopoietic cells were generated from ES cells by the addition of TPO (A, E), EPO (B, F for primitive embryonic erythrocytes and C, G for definitive adult erythrocytes), and M-CSF (D, H) for megakaryocytes, erythroid cells, and macrophages, respectively. The wild-type and mutant GPIIb promoter constructs were transfected and analyzed as described in Figure 2. The data from the wild-type (−598/+33) promoter (A-D) and the mutated promoter in which the nucleotides between −148 and −89 (E-H) were deleted are shown. Promoters lacking −118 to −89 and −148 to −59 show similar results.
A positive regulatory element is sufficient for megakaryocyte-specific gene expression
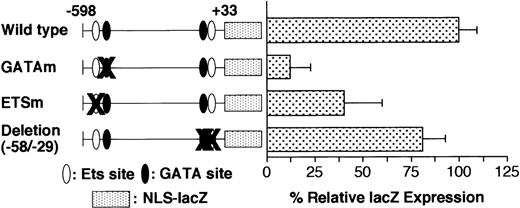
Serial 30-bp deletion analysis revealed a cluster of positive regulatory elements between −538 and −449. The low activity (LacZ expression) of the −400/+33 promoter (Table 1) might be attributable to the positive regulatory elements present in 3 successive 30-bp–deleted regions (Δ−538/−509, Δ−508/−479, and Δ−478/−449). This 90-bp element contains an Ets binding motif located at −515 (Ets−515) and a GATA motif located at −463 (GATA−463), both of which are reported to be critical for full activity of the promoter.20 Presumably, the elimination of Ets−515, GATA−463, or their adjacent elements is responsible for the low activity of these 3 deletion mutants. To study the contribution of these motifs to promoter activity, we performed site-directed mutagenesis of the GATA−463 and Ets−515 sites in the wild-type −598/+33 GPIIb promoter. The expression in the promoter harboring a mutation at the GATA−463 site (GATAm) was only 10% that of the wild-type promoter, whereas the promoter with a mutated Ets−515 binding site (ETSm) exhibited 40% of the activity of the wild-type GPIIb promoter (Figure4). The decrease in LacZ expression driven by the GATAm and ETSm promoters was similar to that of the corresponding serial 30-bp deletion mutants, Δ−478/−449 and Δ−538/−509, respectively. In contrast, eliminating the proximal GATA and Ets binding sites, which are located at −54 and −40, respectively, did not reduce promoter activity (Figure4). These results indicate that the GATA−463 and Ets−515 sites, but not the GATA−54 and Ets−40 sites, are essential components of the regulatory mechanisms governing high-level expression of GPIIb in megakaryocytes. However, the lineage-specific expression of the GPIIb promoter in megakaryocytes is completely maintained even in the absence of both the GATA−463 and Ets−515 sites (Table3).
Effect of deletion and mutation of the GATA and ETS binding sites on GPIIb promoter activity in megakaryocytes.
Wild-type promoter, promoters with mutations in the GATA (GATAm) or Ets binding site (ETSm), and promoters with deletions of the proximal GATA and Ets binding sites were analyzed. The percentages ofLacZ-positive cells per AChE-positive cells were calculated on day 8 of differentiation induction in the presence of TPO. The results are shown as the relative percentage of LacZexpression compared with the value for the −598/+33 wild-type promoter. The data are shown as the mean ± standard deviation. GATAm and ETSm show significant differences from the wild type (P < .05 by t test). Each experiment was performed 2 or 3 times, and representative data are shown.
Effect of deletion and mutation of the GATA and ETS binding sites on GPIIb promoter activity in megakaryocytes.
Wild-type promoter, promoters with mutations in the GATA (GATAm) or Ets binding site (ETSm), and promoters with deletions of the proximal GATA and Ets binding sites were analyzed. The percentages ofLacZ-positive cells per AChE-positive cells were calculated on day 8 of differentiation induction in the presence of TPO. The results are shown as the relative percentage of LacZexpression compared with the value for the −598/+33 wild-type promoter. The data are shown as the mean ± standard deviation. GATAm and ETSm show significant differences from the wild type (P < .05 by t test). Each experiment was performed 2 or 3 times, and representative data are shown.
Lineage-specific expression of LacZ driven by various promoters
| Promoters3-150 . | Megakaryocytes . | Erythrocytes . | Macrophages . | |
|---|---|---|---|---|
| Primitive . | Definitive . | |||
| GPIIb (−598/+33) | ++ | − | − | − |
| GPIIb deletion mutants | + ∼ +++ | − | − | − |
| GATAm | + | − | − | − |
| ETSm | + | − | − | − |
| GPIIb −598/−400:TK | ++ | − | − | − |
| GPIIb −539/−420:TK | + | − | − | − |
| GPIIb −529/−450:TK | + | − | − | − |
| GPIIb −400/−598:TK | + | − | − | − |
| GPIIb TK:−598/−400 | + | − | − | − |
| 4 × GATA:TK | + | + | + | − |
| 4 × ETS:TK | ± | − | − | − |
| EF-1α | + ∼ ++ | + ∼ ++ | + ∼ ++ | + ∼ ++ |
| Promoters3-150 . | Megakaryocytes . | Erythrocytes . | Macrophages . | |
|---|---|---|---|---|
| Primitive . | Definitive . | |||
| GPIIb (−598/+33) | ++ | − | − | − |
| GPIIb deletion mutants | + ∼ +++ | − | − | − |
| GATAm | + | − | − | − |
| ETSm | + | − | − | − |
| GPIIb −598/−400:TK | ++ | − | − | − |
| GPIIb −539/−420:TK | + | − | − | − |
| GPIIb −529/−450:TK | + | − | − | − |
| GPIIb −400/−598:TK | + | − | − | − |
| GPIIb TK:−598/−400 | + | − | − | − |
| 4 × GATA:TK | + | + | + | − |
| 4 × ETS:TK | ± | − | − | − |
| EF-1α | + ∼ ++ | + ∼ ++ | + ∼ ++ | + ∼ ++ |
The absolute percentages of LacZ-positive cells produced with different promoters in megakaryocytes, erythroid cells, and macrophages are shown. − indicates negative; ± indicates very low (<0.5%); + indicates low (0.5% ∼ 15%); ++ indicates medium (15% ∼ 25%); +++ indicates high (>25%).
The names of the promoters are explained in the text.
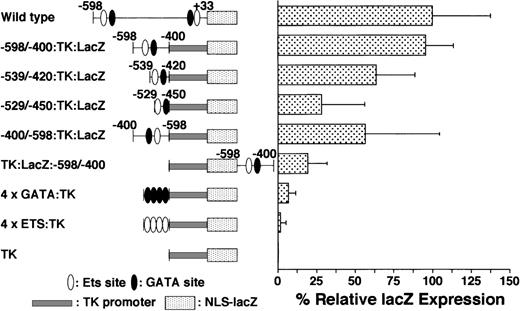
On the basis of the data shown in Tables 1 and 2 and Figure 2, we believe that the −598/−400 promoter region plays an important role in expression of the GPIIb gene during normal megakaryocyte differentiation. To determine whether this region can confer megakaryocyte-specific activity, we constructed and examined a fusion of the GPIIb promoter region −598/−400 to the thymidine kinase minimal promoter of herpes simplex virus (TK promoter). This fusion promoter (−598/−400:TK) demonstrated megakaryocyte-specific expression, and the percentage ofLacZ-positive cells with this promoter was similar to that of the wild-type −598/+33 GPIIb promoter. To delineate the essential region in the −598/−400 promoter, we analyzed the activity of shorter elements within this region. The activities of the promoter regions between −539 and −420 (−539/−420:TK) and between −529 and −450 (−529/−450:TK) were approximately 60% and 30% of that exhibited by −598/−400:TK (Figure 5). Although the level of LacZ expression decreased in these constructs, megakaryocyte-specific expression was maintained even in the −529/−450:TK promoter (Table 3). These results suggest that the GATA−463 and Ets−515 elements may be sufficient to confer the cell specificity of GPIIb expression, but insufficient for full promoter activity.
LacZ gene expression driven by different promoters in megakaryocytes.
The percentages of LacZ-positive cells in megakaryocytes driven by various promoters plus the TK-LacZ gene were examined as described in Figure 2. The details of individual constructs are explained in the text. Data are shown as mean ± standard deviation. All of the results except −598/−400:TK:LacZ are significantly different from the data for the wild type (P < .05 by t test). All of the results except 4 × ETS are significantly different from the results for TK alone (P < .05 by t test). Each experiment was performed 2 or 3 times, and representative data are shown.
LacZ gene expression driven by different promoters in megakaryocytes.
The percentages of LacZ-positive cells in megakaryocytes driven by various promoters plus the TK-LacZ gene were examined as described in Figure 2. The details of individual constructs are explained in the text. Data are shown as mean ± standard deviation. All of the results except −598/−400:TK:LacZ are significantly different from the data for the wild type (P < .05 by t test). All of the results except 4 × ETS are significantly different from the results for TK alone (P < .05 by t test). Each experiment was performed 2 or 3 times, and representative data are shown.
We demonstrated that the region between −598 and −400 of the GPIIb promoter plays an essential role in megakaryocyte-specific expression. Next we examined whether this promoter element can function as an enhancer in the context of stable integration and investigated the positional effects on its expression. Two expression constructs were created to examine whether the element meets the criteria of an enhancer, an element that activates gene expression with a reverse orientation or from the 3′ side of the gene. In 1 construct, the −598/−400 fragment was inserted in the opposite orientation upstream of the TK promoter and LacZ(−400/−598:TK:LacZ). The other construct contained the −598/−400 promoter fragment downstream from the TK promoter and the LacZ gene in the correct orientation (TK:LacZ:−598/−400). These 2 constructs exhibited megakaryocyte-specific activity, although the percentages of positive cells were lower than those observed with the wild-type −598/+33 promoter (Figure 5, Table 3). These results indicate that the −598/−400 promoter fragment containing the GATA−463 and Ets−515 elements governs the lineage-specific expression of GPIIb in megakaryocytes in a position- and orientation-independent manner.
The GATA−463 and Ets−515 elements are not sufficient for megakaryocyte-specific expression
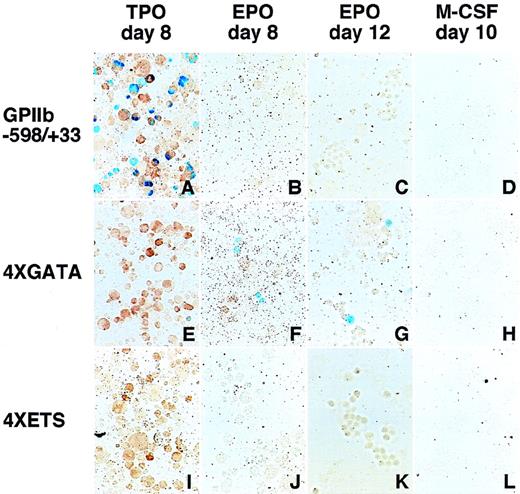
To examine whether the sequences flanking the GATA−463 and Ets−515 sites are sufficient to mediate megakaryocyte-specific gene expression, we tested the double-stranded oligonucleotides encompassing each site and the 13 bp flanking each end of the sites for their ability to direct the expression of LacZ in hematopoietic cells (Figure 5). These oligonucleotides were linked upstream from aLacZ reporter gene driven by the TK promoter. Four tandem repeats of the GATA-site oligonucleotide (4 × GATA:TK) droveLacZ expression not only in megakaryocytes, but also in erythroid cells (Table 3, Figure 6). The percentage of LacZ-positive erythroid cells was slightly lower than that of LacZ-positive megakaryocytes. However, noLacZ-positive macrophages could be detected. In contrast to the 4 × GATA-TK promoter, the 4 × ETS-TK promoter did not drive significant expression of LacZ in megakaryocytes, erythroid cells, or macrophages. (Table 3, Figure 6).
LacZ expression in erythroid and megakaryocytic lineages driven by the 4 × GATA promoter.
Hematopoietic cells were generated from ES cells by the addition of TPO (A, E, I), EPO (B, F, J for primitive embryonic erythrocytes and C, G, K for definitive adult erythrocytes), and M-CSF (D, H, L) for megakaryocytes, erythroid cells, and macrophages, respectively. The wild-type (−598/+33) (A-D), the 4 × GATA (E-H), and the 4 × ETS promoters (I-L) were transfected into ES cells; differentiation induction was carried out; and the cells were stained with X-gal. Double staining with AChE was performed to confirmLacZ expression in megakaryocytes (A, E, I). The 4 × GATA promoter was active in megakaryocytes (E), primitive embryonic erythrocytes (F), and definitive adult erythrocytes (G), but was inactive in macrophages (H).
LacZ expression in erythroid and megakaryocytic lineages driven by the 4 × GATA promoter.
Hematopoietic cells were generated from ES cells by the addition of TPO (A, E, I), EPO (B, F, J for primitive embryonic erythrocytes and C, G, K for definitive adult erythrocytes), and M-CSF (D, H, L) for megakaryocytes, erythroid cells, and macrophages, respectively. The wild-type (−598/+33) (A-D), the 4 × GATA (E-H), and the 4 × ETS promoters (I-L) were transfected into ES cells; differentiation induction was carried out; and the cells were stained with X-gal. Double staining with AChE was performed to confirmLacZ expression in megakaryocytes (A, E, I). The 4 × GATA promoter was active in megakaryocytes (E), primitive embryonic erythrocytes (F), and definitive adult erythrocytes (G), but was inactive in macrophages (H).
c-Ets-1 is not involved in the regulation of the GPIIb promoter
As shown in Figure 4, we demonstrated that the Ets−515 site plays an important role in the transcriptional activity of the GPIIb gene. One previous study reported that c-Ets-1, which is a member of the Ets family of transcription factors, can bind to this element and regulate expression. It was suggested that c-Ets-1 is important for megakaryocytic differentiation.20 To determine whether c-Ets-1 is involved in regulating GPIIb promoter activity, we investigated wild-type −598/+33 GPIIb promoter activity in c-ets-1 null ES cells (Table4).35 Megakaryocytic differentiation from c-ets-1 null ES cells was similar to that from control ES cells. Moreover, the level of LacZ expression did not differ between megakaryocytes generated fromc-ets-1+/+ andc-ets-1−/−ES cells, suggesting that at least in our system, c-Ets-1 does not play a role in megakaryocytic differentiation, or if it does, then its role is redundant.
Clonal analysis of LacZ-positive cells after differentiation induction from c-ets-1+/+ and c-ets-1−/− ES cells
| Genotype of ES Cells . | Percentages of LacZ+ Cells . | ||||
|---|---|---|---|---|---|
| Megakaryocytes . | Erythrocytes . | Macrophages . | |||
| Day 8 . | Day 12 . | Day 8 . | Day 12 . | Day 10 . | |
| +/+ | 84.8 | 93.1 | <0.1 | <0.1 | <0.1 |
| +/+ | 5.0 | 2.9 | <0.1 | <0.1 | <0.1 |
| +/+ | 89.8 | 45.2 | <0.1 | <0.1 | <0.1 |
| +/+ | 66.5 | 8.3 | <0.1 | <0.1 | <0.1 |
| +/+ | 65.1 | 32.8 | <0.1 | <0.1 | <0.1 |
| +/+ | 91.0 | 96.7 | <0.1 | <0.1 | <0.1 |
| +/+ | 65.2 | 50.2 | <0.1 | <0.1 | <0.1 |
| +/+ | 67.0 | 86.3 | <0.1 | <0.1 | <0.1 |
| −/− | 85.3 | 91.0 | <0.1 | <0.1 | <0.1 |
| −/− | 89.0 | 91.0 | <0.1 | <0.1 | <0.1 |
| −/− | 95.9 | 77.1 | <0.1 | <0.1 | <0.1 |
| −/− | 51.6 | 27.6 | <0.1 | <0.1 | <0.1 |
| −/− | 80.5 | 85.5 | <0.1 | <0.1 | <0.1 |
| −/− | 80.9 | 22.5 | <0.1 | <0.1 | <0.1 |
| −/− | 67.8 | 70.7 | <0.1 | <0.1 | <0.1 |
| Genotype of ES Cells . | Percentages of LacZ+ Cells . | ||||
|---|---|---|---|---|---|
| Megakaryocytes . | Erythrocytes . | Macrophages . | |||
| Day 8 . | Day 12 . | Day 8 . | Day 12 . | Day 10 . | |
| +/+ | 84.8 | 93.1 | <0.1 | <0.1 | <0.1 |
| +/+ | 5.0 | 2.9 | <0.1 | <0.1 | <0.1 |
| +/+ | 89.8 | 45.2 | <0.1 | <0.1 | <0.1 |
| +/+ | 66.5 | 8.3 | <0.1 | <0.1 | <0.1 |
| +/+ | 65.1 | 32.8 | <0.1 | <0.1 | <0.1 |
| +/+ | 91.0 | 96.7 | <0.1 | <0.1 | <0.1 |
| +/+ | 65.2 | 50.2 | <0.1 | <0.1 | <0.1 |
| +/+ | 67.0 | 86.3 | <0.1 | <0.1 | <0.1 |
| −/− | 85.3 | 91.0 | <0.1 | <0.1 | <0.1 |
| −/− | 89.0 | 91.0 | <0.1 | <0.1 | <0.1 |
| −/− | 95.9 | 77.1 | <0.1 | <0.1 | <0.1 |
| −/− | 51.6 | 27.6 | <0.1 | <0.1 | <0.1 |
| −/− | 80.5 | 85.5 | <0.1 | <0.1 | <0.1 |
| −/− | 80.9 | 22.5 | <0.1 | <0.1 | <0.1 |
| −/− | 67.8 | 70.7 | <0.1 | <0.1 | <0.1 |
Hematopoietic cells were generated from c-ets-1+/+ and c-ets-1−/− ES cells after transfection of the GPIIb wild-type promoter-LacZ construct. The format of the table is the same as in Table 1.
Discussion
We have established a new experimental system to analyze promoter activity with a combination of reporter gene analysis and in vitro hematopoietic differentiation from ES cells. This experimental system can be used as an alternative to transgenic mice. We analyzed megakaryocyte-specific expression of the GPIIb promoter region between nucleotides −598 and −400 using LacZ reporter gene constructs. Our data show that this −598 to −400 region functions as an enhancer that is necessary and sufficient for megakaryocyte-specific expression and that no lineage-specific negative regulatory element exists from −598 to +33.
The OP9 system is useful for promoter analysis in hematopoietic cells
Recently, some in vitro ES cell–based hematopoietic differentiation systems have been developed. Most systems use the formation of embryoid bodies for the initial step of differentiation induction.25,26 Although embryoid body formation allows analysis of the frequency of hematopoietic precursors, it is insufficient to obtain reasonable numbers of mature hematopoietic cells. We have demonstrated that large numbers of erythrocytes (both primitive embryonic and definitive adult erythrocytes), megakaryocytes, and macrophages are propagated by the addition of the appropriate growth factors. More than 4 × 105 cells of individual lineages were induced from 104 ES cells. The combination of the differentiation induction system and LacZpromoter analysis enabled us to analyze hematopoietic lineage-specific gene expression more easily and efficiently than in a transgenic animal study.39 The LacZ expression pattern in individual positive cells was all or none, and the percentage ofLacZ-positive cells could be used reliably as an index of promoter activity. Patterns consistent with all-or-none responses have been observed in in vivo studies in which reporter gene expression has been analyzed at the single-cell level.40 41 One reason for the lack of larger differences in LacZ intensity might be the relatively small numbers ofLacZ gene copies integrated into ES cells. Although we did not examine all of the clones, only a few copies were integrated under the electroporation conditions used in this study.
The other advantage of this experimental system is the easy genetic manipulation of ES cells. Our analysis ofc-ets-1−/− ES cells suggests that the role of different genes in regulating different promoter regions can be examined easily using double knockout ES cells. Lien et al42 examined regulation of the human gp91-phox gene using a combination of transfer of YAC clones into ES cells and differentiation induction into myeloid cells by the embryoid body method. Gene expression of gp91-phox was monitored by staining with a monoclonal antibody against gp91-phox, but lineage-specific expression was barely examined. The complexity of this system may have precluded examination of lineage-specific expression. Our data show that not only large DNA fragments, such as YAC, but also small promoter fragments, such as those used in a transient expression system, can be well characterized using our experimental system. Therefore, this new method should facilitate the study of hematopoietic lineage-specific expression.
A negative regulatory element in the GPIIb promoter
So far, 3 studies of GPIIb gene regulation have defined “silencer domains” in its promoter region.15,21,22 Two studies analyzed the human GPIIb gene and the other examined the rat GPIIb gene. The location of the silencer element differed in these 3 studies. Fong and Santoro15 determined that 2 sites (−198 to −178 bp and −124 to −99 bp) were involved in the silencer effect in the human GPIIb promoter (−598 to +32 bp) by examining PMA-induced differentiation of K562 cells into megakaryocytes. Prandini et al21 also detected silencer elements in the human GPIIb promoter (−120 to −116 bp and −102 to −93 bp) that partially overlapped the element detected in the study of Fong and Santoro.15 This element was also described in another erythro-megakaryocytic human leukemic cell line, HEL. These studies of the human GPIIb promoter suggest a common silencer site around −120 to −116 bp with the sequence 5′-ATGAG-3′. Prandini et al21suggested that the sequence 5′-ATGAG-3′ is found in the 5′-flanking regions of several megakaryocyte-specific promoters and may be a common silencer of these genes. However, this region is not conserved in the rat and mouse GPIIb promoter regions. In our study, the human GPIIb promoter deletion mutant Δ−118/−89, which removes part of the putative negative regulatory element, had higher transcriptional activity than the control promoter in megakaryocytes. But lineage-specific expression of the promoter was not affected by the deletion at all. In contrast to the previous study,21 a promoter mutated at the negative regulatory element, GPIIb −118/−93m, did not show lineage abrogation. Our study suggests that a negative regulatory element might exist in the region −118 to −89. However, the activity of this element is restricted to megakaryocytes and does not function to determine lineage-specific gene expression. Unexpectedly, the deletion mutant (Δ−208/−179) lacking the other putative negative regulatory element at −198 to −178 bp showed significantly reduced expression. This result suggests that the region from −198 to −178 bp may control expression positively rather than negatively. The other putative negative regulatory element reported by Shou et al22 is a nonconsensus SP1-binding site between −145 and −125 of the rat GPIIb promoter. Deletion mutants lacking the human homologous region, Δ−178/−149 and Δ−148/−119, did not demonstrate significantly different transcriptional activity compared with the control promoter. Here again, lineage-specific expression of these deletion mutants was observed. In summary, our study does not provide any evidence for a lineage-specifying negative regulatory element in the GPIIb promoter, suggesting that only positive regulation is necessary and sufficient for megakaryocyte-specific gene expression.
There are 2 major differences between our experimental system and other systems. First, the types of cells are different. Our system used ES cells and their differentiating progeny within 3 hematopoietic lineages. The differentiation process and developmental characteristics of this in vitro differentiation system are considered very similar to those seen in ontogeny. Meanwhile, all the studies reporting negative regulatory elements in the GPIIb promoter were performed using transient expression in human erythro-megakaryocytic leukemic cell lines. However, leukemic cell lines such as HEL are not necessarily representative of normal megakaryocytes because they express the gene products of other hematopoietic lineages. We believe that using a totipotent ES cell line may avoid the limitations inherent in such leukemic cell lines.
The second issue is the species of cells examined. The discrepancies between our observations and those of others may be attributed to species differences of the cells used. It seems unlikely, however, that the discrepant report of an Sp-1–binding putative negative regulatory element by Shou et al22 is due to species differences because this promoter region is conserved in the human, rat, and mouse, and the homology of Sp-1 among these species is very high. It is difficult to evaluate the other 2 putative negative regulatory elements reported because those promoter elements are not conserved in the human and mouse. However, tissue-specific expression elements are generally well conserved among species, allowing expression analysis of human genes in transgenic mouse systems. It also may be possible that the difference between transient transfection and stable transformation caused some of the discrepancy.
Combinatorial regulation of a cis-regulatory element containing GATA and Ets binding sites
The promoter region between bases −598 and −400 of the human GPIIb promoter showed full and lineage-specific expression. In contrast, the 4 × GATA:TK promoter exhibited activity (LacZ expression) not only in megakaryocytes but also in erythroid cells. However, this promoter did not produce activity in macrophages. Thus, GATA-1, which is the only member of the GATA transcription family expressed in erythroid and megakaryocytic lineages, can drive the expression of this promoter in both lineages. The promoter containing both GATA−463 and Ets−515 (−529/−450:TK) has megakaryocyte-specific expression. Taken together, our data suggest that some member(s) of the etsgene family, rather than GATA factors, function to restrict expression to the megakaryocyte lineage. It is not yet known which member of theets gene family is critical for megakaryocyte-specific gene expression. Candidates are c-Ets-1, Spi-1, Fli-1, and PU.1, all of which are expressed in megakaryocytes.23,43 44 The data using c-ets-1double knockout ES cells demonstrated that c-Ets-1 is not essential for the lineage-specific expression.
The observation that the promoter regions of many megakaryocyte-specific genes contain both GATA and Ets binding sites underlines the importance of these 2 binding sites.21However, there is no general rule concerning the orientation, the surrounding nucleotide sequence, and the length separating these 2 sites. Our study shows that adjacent regions around the binding sites and the region flanked by these 2 sites influence transcriptional activity. The regulation of GPIIb gene expression may be more complex than the many studies to date have suggested.
Acknowledgments
We thank Dr Shigekazu Nagata, Kirin Brewery Co Ltd, and Snow Brand Co Ltd for the kind gift of EF1-α promoter, human recombinant EPO and TPO, and human recombinant M-CSF; and Drs Janusz Kabarowski and Anne B. Satterthwaite for critical reading of the manuscript.
Supported in part by grants from the Ministry of Education, Science, Sports and Culture; Japanese Society for Promotion of Sciences (JSPS-RFTF98L01101); the Cell Science Research Foundation; and the Osaka Foundation for Promotion of Clinical Immunology.
Reprints:Toru Nakano, Department of Molecular Cell Biology, Research Institute for Microbial Diseases, Osaka University, 3-1 Yamadaoka, Suita 565-0871, Osaka, Japan; e-mail:tnakano@biken.osaka-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal