One of the most common translocations in acute myeloid leukemia (AML) is the t(8;21), which produces the fusion gene AML1-MTG8. We have developed a sensitive competitive reverse transcriptase-polymerase chain reaction (RT-PCR) assay forAML1-MTG8 transcripts, coupled with a competitive RT-PCR for the ABL transcript as a control to accurately estimate the level of amplifiable RNA. We have shown that AML1-MTG8 andABL transcripts have equal degradation rates. Thus, this method is useful for multicenter studies. We studied 25 patients with t(8;21) AML by means of serial analysis done on bone marrow (BM) and peripheral blood (PB) samples from 21 patients. Our analysis showed that, in general, a successful induction chemotherapy produces a reduction of 2 to 3 log in the level of AML1-MTG8, followed by a further 2 to 3 log after consolidation/intensification chemotherapy. Levels up to 1 × 103 and 1 × 102 molecules/μg of RNA in BM and PB, respectively, were compatible with durable remission. On the other hand, 5 patients with levels of 0.71 × 105to 2.27 × 105 molecules/μg of RNA in BM and 2.27 × 103 to 2.27 × 104 molecules/μg of RNA in PB had hematologic relapse within 3 to 6 months. Our data indicate that serial quantitation of AML1-MTG8 transcripts is useful in identifying patients at high risk of relapse and may offer an opportunity for clinical intervention to prevent hematologic relapse. This approach was applied successfully in a patient who had an allogeneic BM transplantation. We also suggest that PB may be used an alternative to BM for quantitating AML1-MTG8 transcripts.
The t(8;21) translocation fuses 2 genes, AML1on chromosome 21 and MTG8 on chromosome 8, to produce the fusion gene AML1-MTG8 on the derivative chromosome 8.1 It is one of the most common chromosomal translocations in acute myeloid leukemia (AML), occurring in approximately 20% of adult and 40% of pediatric cases of AML M2. Although patients with t(8;21) AML have a relatively good prognosis, relapse remains the commonest cause of treatment failure. Most published studies, including ours,2-7 have reported the persistence of AML1-MTG8fusion transcripts in patients in long-term remission. Although some studies8,9 have shown absence of the fusion transcripts in patients in long-term remission, both sets of studies found that qualitative detection of AML1-MTG8 fusion transcripts has a limited value in monitoring minimal residual disease (MRD) and predicting relapse in patients with t(8;21) AML. We recently developed a quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) method to estimate levels ofAML1-MTG8 fusion transcripts and, by inference, levels of MRD, at different phases of the disease.10 We here report our evaluation of this method in assessments of sequential peripheral blood (PB) and bone marrow (BM) samples from a larger group of AML patients with t(8;21). We also report the identification of critical threshold levels for AML1-MTG8 fusion transcripts that enable distinction between patients in durable remission and those at high risk of relapse.Materials and methods
Patients
In all patients, the diagnosis of AML M2 was made by morphologic and cytochemical examination of bone marrow aspirates. The presence of t(8;21) was confirmed by conventional cytogenetic analysis. Twenty-five patients (median age, 36 years; range, 15-67 years) were entered into this study. All patients had complete hematologic remission after one course of induction chemotherapy with standard doses of daunorubicin or mitoxantrone, cytarabine, thioguanine, or etoposide. Eighteen patients received DAT (daunorubicin, cytarabine [cytosine arabinoside], and thioguanine) 3 + 10/3 + 7, 2 patients received ADE (cytarabine, daunorubicin, and etoposide), and 3 patients received MAE (mitoxantrone, cytarabine, and etoposide), according to UK Medical Research Council AML protocols. Between 2 and 4 courses of intensive postremission chemotherapy were given to all patients except one, who refused further treatment. These chemotherapy courses included DAT or ADE or MAE, MACE (amsacrine [m-AMSA], cytarabine, and etoposide), MIDAC (mitoxantrone and intermediate-dose cytarabine), or intermediate-dose cytarabine. Four patients received an allogeneic or autologous BM/PB stem-cell transplantation in the first remission. BM and PB samples were collected from patients at presentation and at different intervals during remission, after BM transplantation, and at relapse. The patients studied had been in remission from 1 to 125 months. Informed consent was obtained from patients according to established procedures in each center, and samples were collected as part of the treatment protocols, which were approved by the local hospital ethics committees.
Samples and RNA preparation
Mononuclear cells from PB and BM samples were obtained by Ficoll-Hypaque density gradient centrifugation and either stored at −80°C or used immediately for RNA isolation. RNA was isolated from the Kasumi-1 cell line11 or patients' mononuclear cells by the guanidinium-phenol-chloroform method of Chomczynski and Sacchi,12 with minor modifications.
Reverse transcription
Approximately 5 μg of total RNA was used for the synthesis of complementary DNA (cDNA) n a 20-μL reaction. The reaction contained 0.25 μg of random hexamers, 200 U of Moloney's murine leukemia virus RT, and 40 U of RNAsin. The reverse transcription reaction was performed at 37°C for 1 hour, then at 45°C for 30 minutes, and at 75°C for 5 minutes.
Control gene (ABL RT-PCR)
To assess the quality and quantity of amplifiable RNA isolated from samples, qualitative and quantitative RT-PCR amplification ofABL gene transcripts was performed. PCR amplification was done as previously described.13ABL transcript was amplified from 2 μL of cDNA in a 25-μL reaction containing primers CA3 and A2 at 97°C for 1 minute 30 seconds, 64°C for 50 seconds, and 72°C for 1 minute, (1 cycle); 97°C for 30 seconds, 64°C for 50 seconds, and 72°C for 1 minute (40 cycles); and 72°C for 5 minutes (1 cycle). PCR products were electrophoresed on a 2% agarose gel.
AML1-MTG8 RT-PCR
Two microliters of cDNA was subjected to 2 rounds of PCR amplification for the AML1-MTG8 transcripts. The first-round PCR was performed in a 50-μL reaction containing primers 11 and 12 at 95°C for 2 minutes (1 cycle); 93°C for 50 seconds, 56°C for 50 seconds, and 72°C for 1 minute (40 cycles); and 72°C for 5 minutes (1 cycle). Two microliters of first-round product was used in a 50-μL second-round PCR reaction containing primers TS and 24 under the same PCR conditions. Second-round products were electrophoresed on a 2% agarose gel. This PCR protocol is a transcript-specific amplification designed to amplify the main (inframe) AML1-MTG8transcript that is detected in all patients with t(8;21).14-16
ABL competitive RT-PCR
For accurate estimation of the level of AML1-MTG8 fusion transcripts, we have developed a quantitative RT-PCR method for theABL transcript as a control for the level of amplifiable RNA in samples. ABL competitor was prepared by using the same principle (splicing by overlap extension technique) employed forAML1-MTG8 transcript's competitor.10 Two microliters of cDNA was mixed with 2 μl of competitor DNA for theABL gene transcript and subjected to a one-round PCR amplification as described above. The expected band sizes forABL transcript and competitor are 276 base pairs (bp) and 235 bp, respectively.
AML1-MTGB competitive RT-PCR
Two microliters of cDNA from samples positive for AML1-MTG8 was mixed with 2 μL of competitor DNA10 and subjected to PCR amplification as described above. Each sample was quantified at every order of magnitude and then at every half order of magnitude. The point of equivalence was assessed by gel densitometry. The level ofAML1-MTG8 present in samples was adjusted according to the level of ABL transcript present to give an accurate number ofAML1-MTG8 molecules per microgram of RNA.
ABL and AML1-MTG8 degradation rates
To evaluate the degradation rates of the transcripts of both theABL and AML1-MTG8 fusion genes, samples from patients and the Kasumi-1 cell line were divided into 3 parts. One part was subjected to Ficoll-Hypaque density gradient centrifugation as described above, then frozen directly at −80°C; the other 2 parts were incubated at room temperature for 24 and 48 hours, respectively, before centrifugation. The levels of ABL andAML1-MTG8 transcripts were estimated in all 3 parts to determine the rates of degradation.
Reproducibility and accuracy of assays
In all tests, negative and positive controls were used. Negative controls included reactions with no RNA or no cDNA or t(8;21)-negative cell lines, such as K562. Kasumi-1 cell line was used as a positive control. All necessary precautions were taken to avoid contamination. These included the use of a specially designed UV-flow cabinet, PCR-designated pipettes, and filtered tips for all PCR preparations. All tests were done twice to confirm the results.
Sequences of PCR primers:
11 5′ AGCCATGAAGAACCAGG 3′
12 5′ AGGCTGTAGGAGAATGG 3′
TS 5′ CCCCGAGAACCTCGAAATCGT 3′
24 5′ GTTGTCGGTGTAAATGAA 3′
A2 5′ TTCAGCGGCCAGTAGCATCTGACTT 3′
CA3 5′ TGTTGACTGGCGTGATGTAGTTGCTTGG 3′
Results
Degradation rates for ABL and AML1-MTG8 transcripts
The degradation rates of the ABL and AML1-MTG8transcripts were equal. After incubation of samples at room temperature, the levels of both transcripts decreased equally, by 0.5 log after 24 hours and by 1 log after 48 hours. These results indicate that ABL is a suitable control gene for quantitation ofAML1-MTG8. RNA and cDNA samples stored in −80°C for up to 2 months showed no degradation of either gene's transcripts.
AML1-MTG8 quantitation
As we showed previously,10 this method is linear over a wide range of AML1-MTG8 transcripts levels. The RT-PCR method has a sensitivity level of 1 leukemic cell in 106 normal cells (10−6). This method can detect as few as 3 copies of the AML1-MTG8 transcript. In this study, the level ofAML1-MTG8 transcripts was quantified in sequential BM and PB samples from 21 patients with t(8;21) AML; 4 other patients were tested at relapse only. Five of the 21 patients who had remission subsequently had relapse.
Levels at presentation
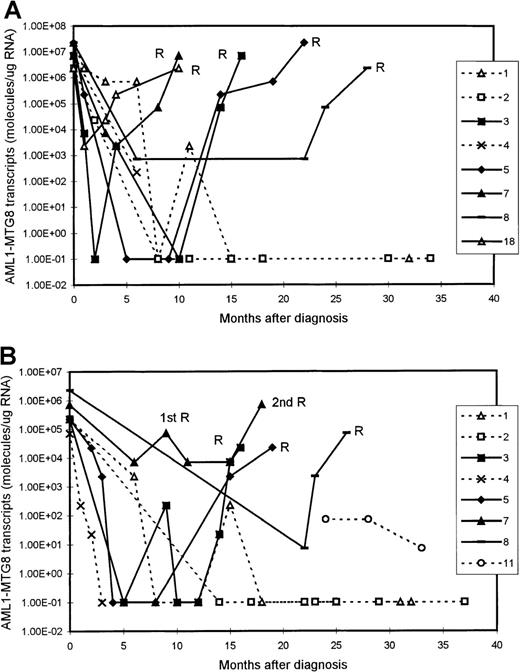
We examined 11 patients at presentation of AML (10 BM samples and 8 PB samples). The levels of AML1-MTG8 transcripts at presentation were in the range of 2.27 × 106 to 2.27 × 107 molecules/μg of RNA (median, 1.49 × 107 molecules/μg of RNA) in the BM samples and 0.71 × 105 to 2.27 × 106molecules/μg of RNA (median, 2.27 × 105molecules/μg of RNA) in the PB samples (Figure1 and Figure2). There was no difference in the level of fusion transcripts at presentation between patients who subsequently had a durable remission and those who had relapse.
Serial quantitation of AML1-MTG8 transcripts in 9 patients with t(8;21) acute myeloid leukemia (AML).
Results with bone marrow (BM) (A) and peripheral blood (PB) (B) samples are shown. Patients 3, 5, 7, 8, and 18 had a relapse (R), whereas patients 1, 2, 4, and 11 are in continuous remission. Levels of 1.00E-01 are reverse transcriptase-polymerase chain reaction (RT-PCR) negative.
Serial quantitation of AML1-MTG8 transcripts in 9 patients with t(8;21) acute myeloid leukemia (AML).
Results with bone marrow (BM) (A) and peripheral blood (PB) (B) samples are shown. Patients 3, 5, 7, 8, and 18 had a relapse (R), whereas patients 1, 2, 4, and 11 are in continuous remission. Levels of 1.00E-01 are reverse transcriptase-polymerase chain reaction (RT-PCR) negative.
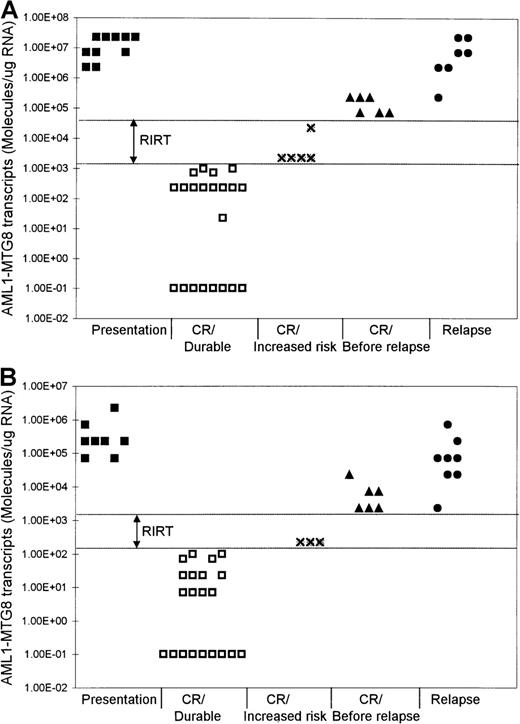
Levels of AML1-MTG8 transcripts at different phases of t(8;21) AML.
Results with BM (A) and PB (B) samples are shown. RIRT indicates relapse increased-risk thresholds. Levels higher than the upper limit of these thresholds are indicative of relapse within 3 to 6 months. Levels of 1.00E-01 are RT-PCR negative.
Levels of AML1-MTG8 transcripts at different phases of t(8;21) AML.
Results with BM (A) and PB (B) samples are shown. RIRT indicates relapse increased-risk thresholds. Levels higher than the upper limit of these thresholds are indicative of relapse within 3 to 6 months. Levels of 1.00E-01 are RT-PCR negative.
Levels at remission
Serial quantitation of MRD by RT-PCR analysis in 21 patients showed a gradual reduction in the level of AMLl-MTG8 transcripts as remission was induced. In both BM and PB, we detected a reduction of approximately 2 to 3 log in the level of AML1-MTG8 after the induction chemotherapy. This was followed by a further reduction of 2 to 3 log after consolidation/intensification treatment (Figure 1). There were no differences in the kinetics of molecular responses between patients receiving different chemotherapy regimens.
Although the level of AML1-MTG8 transcripts became undetectable in a number of patients during durable remission, most patients continued to express detectable levels of transcripts of this fusion gene, even during long-term remission. In all patients in stable remission, the level of AML1-MTG8 was found to be ≤ 1 × 103 molecules/μg of RNA in the BM samples (median, 227) and ≤ 1 × 102molecules/μg of RNA in the PB samples (median, 7.1) (Figure 1 and Figure 2). Five patients had been in remission for > 3 years, 2 patients > 5 years, 2 patients > 7 years, and 2 patients > 10 years. BM samples from patients in remission were both morphologically and karyotypically normal. Karyotype was established by conventional cytogenetic analysis. There was no difference in disease prognosis between patients who had negative results during remission and those who had AML1-MTG8 levels of up to 103and 102 molecules/μg of RNA in BM and PB, respectively.
Prerelapse levels
In 5 of the patients who were tested serially, we detected a significant increase in the levels of AML1-MTG8 transcripts 3 to 6 months before the onset of hematologic relapse; the values were 0.71 × 105 to 2.27 × 105molecules/μg of RNA in BM samples (5 patients; median, 1.49 × 105 molecules/μg of RNA) and 2.27 × 103 to 2.27 × 104molecules/μg of RNA in PB samples (4 patients; median, 7.1 × 103 molecules/μg of RNA) (Figure 1 and Figure 2). BM examination yielded both morphologically and karyotypically normal results at the time of the detection of the marked increase in the level of AML1-MTG8 transcripts. Three of 5 patients who had levels in BM that were intermediate between the durable-remission and prerelapse levels (2.27 × 103to 2.27 × 104 molecules/μg of RNA; median, 2.27 × 103 molecules/μg of RNA) subsequently had relapse. Similarly, 2 of 3 patients with levels in PB that were intermediate between durable-remission and prerelapse levels (2.27 × 102 molecules/μg of RNA; median, 2.27 × 102 molecules/μg of RNA) had relapse later.
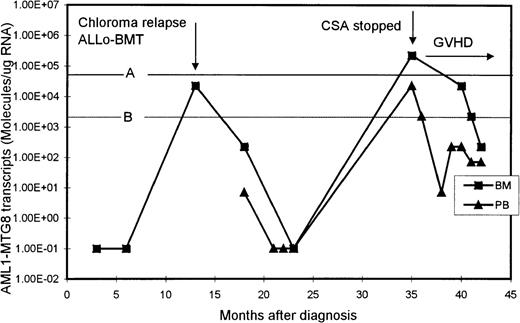
One patient initially presented with an extramedullary paraspinal chloroma and had susequent development of AML with t(8;21). After intensive chemotherapy, his AML1-MTG8 transcript levels became undetectable but increased significantly (2.27 × 104 molecules/μg of RNA in BM) when he had chloroma relapse, without bone marrow involvement (Figure 3). He then received an allogeneic BM transplant from his histocompatible sister, after which the level of AML1-MTG8 transcripts became undetectable again. His posttransplantation course was complicated by chronic graft-versus-host disease (GVHD) of the liver, which was controlled with cyclosporine. However, 22 months after transplantation, the level of AML1-MTG8 transcripts was found, on routine monitoring, to have risen to 2.27 × 105 and 2.27 × 104 molecules/μg of RNA in BM and PB, respectively. These levels were strongly suggestive of imminent hematologic relapse, although the BM was normal on morphologic assessment and karyotypic examination revealed only female cells. The cyclosporine therapy was stopped, and acute skin and liver GVHD (grade III) promptly developed. The patient was treated with prednisolone alone, which brought the GVHD under control. Subsequently, the levels of AML1-MTG8 transcripts in both BM and PB decreased and remained below the critical thresholds for relapse (Figure3). The patient remained in complete clinical remission 10 months after he stopped taking cyclosporine.
Serial quantitation of AML1-MTG8 transcripts in BM and PB in a patient presenting with chloroma and AML.
A and B are the upper limits of RIRT in BM and PB, respectively. Levels of 1.00E-01 are RT-PCR negative.
Serial quantitation of AML1-MTG8 transcripts in BM and PB in a patient presenting with chloroma and AML.
A and B are the upper limits of RIRT in BM and PB, respectively. Levels of 1.00E-01 are RT-PCR negative.
Relapse levels
We examined 9 patients at relapse, 5 of whom were also examined before the onset of hematologic relapse (see above). The levels of fusion transcripts in these 5 patients at the onset of hematologic relapse increased further, to 2.27 × 106 to 2.27 × 107 molecules/μg of RNA in BM (5 patients) and to 0.71 × 105 to 0.71 × 106molecules/μg of RNA in PB (4 patients) (Figure 1 and Figure 2). Similar levels (2.27 × 105 to2.27 × 107 molecules/μg of RNA in BM and 2.27 × 103 to 2.27 × 105molecules/μg of RNA in PB) were detected in 4 other patients tested only during hematologic relapse. The median values for relapse levels were 0.71 × 107 molecules/μg of RNA in BM and 0.71 × 105 molecules/μg of RNA in PB.
Critical threshold levels
We found that patients in durable remission had levels ofAML1-MTG8 transcripts ≤ 1 × 103molecules/μg of RNA in BM and ≤ 1 × 102molecules/μg of RNA in PB. These results identified threshold levels, termed relapse increased-risk thresholds (RIRT), of > 103to < 0.71 × 105 molecules/μg of RNA in BM and of > 102 to < 2.27 × 103molecules/μg of RNA in PB (Figure 2). Levels above the upper limit of these thresholds are indicative of hematologic relapse within 3 to 6 months. Patients who have AML1-MTG8 levels within these thresholds' limits during remission are at increased risk of relapse.
Discussion
The t(8;21) is one of the most common translocations in AML. It is associated with a relatively good prognosis, with most patients achieving complete hematologic remission with induction chemotherapy. Chemotherapy alone can result in a cure rate of approximately 60% and is thus offered as first-line treatment for AML.17,18 The aim of MRD monitoring is to assess the effectiveness of treatment and predict relapse at an early stage, possibly allowing preemptive therapy in an attempt to improve clinical outcome. Most published studies have shown that qualitative RT-PCR was able to detect AML1-MTG8transcripts in most patients in long-term remission.2-7Therefore, qualitative RT-PCR methods have limited value in monitoring MRD in patients with t(8;21) AML.
We have developed a sensitive competitive RT-PCR method to quantitate the number of AML1-MTG8 transcripts in PB and BM samples from patients with t(8;21) AML at different stages of the disease. This method has a sensitivity level of 10−6 and can detect as few as 3 copies of the AML1-MTG8 fusion transcripts. To quantitate fusion transcripts accurately, a suitable control gene must be used to estimate the amount of amplifiable RNA in each sample. The control gene should have a degradation rate similar to that of the gene of interest. Our study found that ABL and AML1-MTG8have equal degradation rates: 0.5 log after 24 hours of incubation at room temperature and 1 log after 48 hours. This finding shows that the effect of RNA degradation of up to 48 hours on the final estimation of the level of AML1-MTG8 transcripts could be eliminated by quantitation of ABL as a control gene. It also shows that this protocol could be used in multicenter studies in which samples are transported between different centers by mail at room temperature.
In this study, the level of AML1-MTG8 transcripts was expressed as the number of molecules per microgram of RNA after the necessary correction based on the level of ABL transcript detected was done. Although the level of AML1-MTG8 transcripts could also be expressed as a ratio of AML1-MTG8/ABL transcripts, we thought that expressing it as the number of molecules of the fusion transcripts per microgram of RNA was preferable because various research groups use different control genes for their studies, which will produce different ratios depending on the control gene used.
We estimated the levels of residual leukemic cells at different stages of t(8;21) AML and showed individual patients' response to treatment. At presentation, BM samples had levels of 2.27 × 106 to 2.27 × 107molecules/μg of RNA and PB samples had levels of 0.71 × 105 to 2.27 × 106molecules/μg of RNA. We detected a reduction of approximately 2 to 3 log in leukemic cells after completion of successful induction chemotherapy and a further 2- to 3-log reduction after consolidation/intensification treatment. Our data show thatAML1-MTG8 transcripts levels of up to 1 × 103 molecules/μg of RNA in BM and 1 × 102 molecules/μg of RNA in PB are consistent with durable remission. On the other hand, levels ≥ 0.71 × 105 molecules/μg of RNA in BM and ≥ 2.27 × 103 molecules/μg of RNA in PB were predictive of relapse within 3 to 6 months. All 5 patients with such levels had relapse within 3 to 6 months. It is noteworthy that in all 5 of these patients, BM examination yielded both morphologically and karyotypically normal results at the time the marked increase inAML1-MTG8 transcripts level was detected.
Our data also show that patients with levels between 2.27 × 103 and 2.27 × 104molecules/μg of RNA in BM and of 2.27 × 102molecules/μg of RNA in PB are at increased risk of hematologic relapse. Three of 5 patients with these intermediate levels in BM and 2 of 3 patients with these intermediate levels in PB had subsequent hematologic relapse. Our data identified RIRT of > 103 to < 0.71 × 105 molecules/μg of RNA in BM and > 102 to < 2.27 × 103molecules/μg of RNA in PB. Patients with fusion levels below the lower limit of these thresholds can have long-term remissions. On the other hand, levels higher than the upper limit of these thresholds are indicative of hematologic relapse within 3 to 6 months. We propose that patients with fusion levels within the limits of these thresholds can be considered to be at increased risk of relapse and that they therefore require more frequent monitoring to assess the kinetic behavior of their residual disease.
Therefore, serial quantitation of AML1-MTG8 transcripts can identify patients at high risk of relapse and allows establishment of critical thresholds, which are predictive of imminent hematologic relapse. It also offers a window of opportunity of between 3 and 6 months, when preemptive therapy, including allogeneic BM transplantation, to prevent hematologic relapse may be given. Our data also show that it is important to serially monitor AML1-MTG8levels frequently, perhaps at 2- to 3-month intervals in the first 2 years after achievement of remission, when most relapses occur. In subsequent years, monitoring could be done at longer intervals.
The clinical usefulness of quantitative monitoring of AML1-MTG8transcripts is well illustrated in a patient in our study who presented with chloroma and AML. In this patient, relapse of the chloroma, without bone marrow involvement, was associated with a marked increase in BM level of AML1-MTG8 transcripts. The patient was treated successfully with allogeneic BM transplantation. However, on routine testing 22 months after transplantation, both the BM and PB levels ofAML1-MTG8 transcripts had risen to values that our data indicated were predictive of imminent hematologic relapse. The patient's cyclosporine therapy was stopped, and subsequent monitoring showed a prompt decrease in the AML1-MTG8 transcripts levels to below the critical relapse thresholds. It is likely that this patient would have had a clinical relapse within 3 to 6 months after the high levels of AML1-MTG8 transcripts were detected. We postulate that when the cyclosporine therapy was stopped, an enhanced graft-versus-leukemia effect was induced, along with GVHD, resulting in a prompt decrease in the levels of residual leukemia and a continuing clinical remission.
Therefore, in patients with AML who have undergone allograft transplantation, a strategy for quantitative monitoring of MRD, where a specific molecular marker is available, may identify those at high risk of relapse. This may allow appropriate clinical interventions, such as reduction in immunosuppression or use of donor lymphocyte infusions, to prevent the onset of hematologic relapse. Such an approach, using quantitative assays for BCR-ABL transcripts, has already been proposed for patients with chronic myeloid leukemia.19
Another important finding of this study is that PB samples could be used in our RT-PCR method to estimate the level of MRD . The levels detected in PB were 1 to 2 log lower than those detected in BM. However, we were able to detect changes in the level ofAML1-MTG8 transcripts in PB because of the sensitivity of our method. We showed that changes in the level of AML1-MTG8transcripts in BM were closely mirrored by those detected in PB and that both accurately reflect MRD status. The advantages of using PB for monitoring MRD are its easy access and avoidance of the inconvenience of BM sampling.
In conclusion, we have shown that a sensitive competitive RT-PCR method for quantitation of AML1-MTG8 transcripts can detect important changes in the level of this fusion transcript, thereby allowing accurate prediction of relapse at an early stage and application of preemptive clinical intervention. Having a sensitive method also allows the use of PB for MRD monitoring in t(8;21) AML. Although we used competitive RT-PCR in our study, we believe that highly sensitive and reliable techniques for quantitation of AML1-MTG8 transcripts that are based on other principles, such as real-time PCR, may provide alternative approaches for monitoring MRD in t(8;21) AML.
Acknowledgment
We thank N. Kamada for providing the Kasumi-l cell line.
Reprints:K. Tobal, University Department of Hematology, Manchester Royal Infirmary, Manchester, UK; email:ktobal@labmed.cmht.nwest.rhs.uk.
Supported by a grant from the Leukaemia Research Fund (LRF), UK, and by the Manchester Leukas-Aid Research Charity. J.N. was an LRF Research Fellow.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal