To the editor:
C-kit mutations in core binding factor leukemias
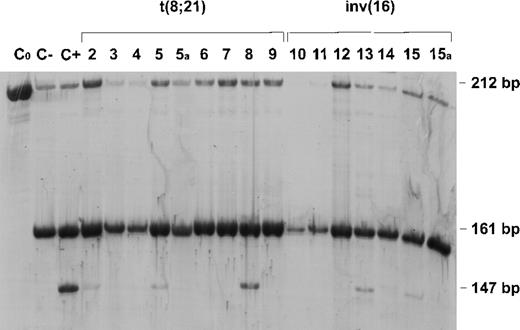
Positivity for CD117(c-kit) expression is present in 80% of acute myeloblastic leukemia (AML) cases,1 and has been associated with the expression of CD33, a marker of myeloid precursors, and CD13, a myeloid-associated antigen.2 Combined positivity for the stem cell antigens CD34, CD117, and HLA-DR characterizes AML-M2 with t(8;21) and AML-M4Eo with inv(16), currently designated core binding factor (CBF) leukemias.3-5 According to a recent study, the PEBP2β/MYH11 fusion transcript is involved in CD34+/c-kit+ immature cells and may have leukemogenic potential only in these cells.6 The same authors pointed out that the disruption of AML1 and PEBP2β genes encoding the α and β subunits of a single transcriptional complex (CBF) affects the same differentiation stage of the myeloid cells. On the basis of published data concerning c-kit (CD117) expression in CBF leukemias,6-8 and our finding of the Asp816Tyr activating c-kit mutation in an AML-M2 patient with t(8;21),9 we recruited 15 patients with either AML-M2 with t(8;21) or AML-M4Eo with inv(16) to screen for c-kit mutations. Mutation screening was targeted on exon 17 as codon 816 was found to be affected by Asp816Val—first detected in the mast cell leukemia cell line HMC-110 and the alternative mutation Asp816Tyr.9 As can be seen in the Table, c-kit mutations at the critical codon 816 were found in 4 AML-M2 and 2 AML-M4Eo patients. Only patient 1 was found to carry the Asp816Tyr mutation which, as previously reported, was present in 100% of blasts.9 The application of DGGE/CDGE to screen for the presence of the Asp816Tyr mutation did not reveal any other carrier among the patients recruited for this study. The presence of the Asp816Val mutation, which creates an additional HinfI site, leads to a new DNA fragment that is detected by HinfI digestion of c-kit exon 17 PCR products. As a result of HinfI digestion of the mutated allele, the 147 bp DNA fragment was apparent in AML patients 2, 5, 8, 13 and 15 (Figure1). The molecular test at remission in patients 5 and 15 is also shown. There was a remarkable variability in the intensity of the 147 bp band due to the different expansion of the mutated leukemic clone among the patients. During follow-up monitoring using the same HinfI assay in patients 5 and 15, the mutated allele in purged bone marrow DNA at remission was undetectable (lanes 5a and 15a), indicating a contraction of the mutant clone together with disease. The targeted selection of AML patients with antigenic and karyotypic features of CBF leukemias thus allowed us to identify 6 out of 15 patients carrying either a D816V or a D816Y c-kit mutation. These results confirm the strict correlation between the stage of myelomonoblast differentiation and the susceptibility to c-kit activation. Our previous analysis of 12 unselected AML patients did not lead to the detection of any c-kit mutation, which is in line with the results of the study reported by Ferrao,11 who found that only one AML-M2 patient out of 33 AML cases carried the D816V mutation. Mutation screening of all 21 c-kit exons has recently disclosed new c-kit mutations in AML,12 which underlines their relevance in CBF leukemias.
Immunophenotypic, cytogenetic and molecular findings of core binding factor leukemia patients
| BMMNC Asp816Val and Asp816Tyr Status . | Patient Sex/Age . | Surface Antigens . | FAB . | Karyotype . | ||||
|---|---|---|---|---|---|---|---|---|
| CD13 . | CD33 . | CD34 . | CD117 . | HLADR . | ||||
| Asp816Val | 3 F/28 | 36, 7 | ND | 46, 9 | ND | 47, 4 | AML-M2 | 46,XX,t(8;14;21) |
| and | 4 F/72 | 48, 6 | 68 | 29 | 21, 9 | 60 | AML-M2 | 46,XX,t(8;21)(q22;q22) |
| Asp816Tyr | 6 M/26 | 88, 9 | 88, 7 | 87, 4 | 87, 2 | 95 | AML-M2 | 45,X,-Y,t(8;21)(q22;q22){15};46XYt(8;21)(q22;q22){17};46XY{5} |
| negative | 7 F/31 | 78 | 72 | 68 | 50 | 60 | AML-M2 | 46,XX,t(8;21)(q22;q22){40} |
| 9 M/32 | 21 | 53, 5 | 56, 8 | 57 | ND | AML-M2 | 45,X,-Y,t(8;21)(q22;q22) | |
| 10 F/42 | 79, 7 | 28, 9 | 58, 1 | ND | 69, 4 | AML-M4Eo | 46,XX,inv(16)(p13;q22){20} | |
| 11 M/45 | 80 | 71, 4 | 65, 2 | ND | 78, 3 | AML-M4Eo | 47,XY,inv(16)(p13;q22),+21{20} | |
| 12 F/59 | 84 | 79 | 1 | 60 | 61 | AML-M4Eo | 46,XX,inv(16)(p13;q22){19};46XX{1} | |
| 14 F/42 | 81, 1 | 73 | 30, 7 | 20 | 75, 3 | AML-M4Eo | 46,XX,inv(16)(p13;q22){32};46XX{2} | |
| Asp816Val | 2 M/65 | 15, 3 | 21, 4 | 52, 9 | ND | ND | AML-M2 | 47,XY,+4,t(8;21)(q22;q22) |
| positive | 5 M/16 | 77, 2 | 62, 1 | 62, 9 | 39, 7 | 73, 2 | AML-M2 | 46,XY;46,XY,t(8;21)(q22;q22);47,XY,+13,t(8;21)(q22;q22) |
| 8 M/40 | 93, 7 | 95 | 33, 5 | 48 | 91, 2 | AML-M2 | 46,XY,t(2;8;21)(q24;q22;q22) | |
| 13 M/55 | 89, 8 | 56, 5 | 61, 7 | 40 | 10, 5 | AML-M4Eo | 46,XY;inv(16)(p13;q22) | |
| 15 M/53 | 81, 1 | 78, 6 | 66, 9 | 73, 8 | 44 | AML-M4Eo | 46,XY,inv(16)(p13;q22) | |
| Asp816Tyr | 1 M/47 | 87 | 70 | 87 | 23 | 94 | AML-M2 | 47,XY,+4,t(8;21)(q22;q22) |
| positive | ||||||||
| BMMNC Asp816Val and Asp816Tyr Status . | Patient Sex/Age . | Surface Antigens . | FAB . | Karyotype . | ||||
|---|---|---|---|---|---|---|---|---|
| CD13 . | CD33 . | CD34 . | CD117 . | HLADR . | ||||
| Asp816Val | 3 F/28 | 36, 7 | ND | 46, 9 | ND | 47, 4 | AML-M2 | 46,XX,t(8;14;21) |
| and | 4 F/72 | 48, 6 | 68 | 29 | 21, 9 | 60 | AML-M2 | 46,XX,t(8;21)(q22;q22) |
| Asp816Tyr | 6 M/26 | 88, 9 | 88, 7 | 87, 4 | 87, 2 | 95 | AML-M2 | 45,X,-Y,t(8;21)(q22;q22){15};46XYt(8;21)(q22;q22){17};46XY{5} |
| negative | 7 F/31 | 78 | 72 | 68 | 50 | 60 | AML-M2 | 46,XX,t(8;21)(q22;q22){40} |
| 9 M/32 | 21 | 53, 5 | 56, 8 | 57 | ND | AML-M2 | 45,X,-Y,t(8;21)(q22;q22) | |
| 10 F/42 | 79, 7 | 28, 9 | 58, 1 | ND | 69, 4 | AML-M4Eo | 46,XX,inv(16)(p13;q22){20} | |
| 11 M/45 | 80 | 71, 4 | 65, 2 | ND | 78, 3 | AML-M4Eo | 47,XY,inv(16)(p13;q22),+21{20} | |
| 12 F/59 | 84 | 79 | 1 | 60 | 61 | AML-M4Eo | 46,XX,inv(16)(p13;q22){19};46XX{1} | |
| 14 F/42 | 81, 1 | 73 | 30, 7 | 20 | 75, 3 | AML-M4Eo | 46,XX,inv(16)(p13;q22){32};46XX{2} | |
| Asp816Val | 2 M/65 | 15, 3 | 21, 4 | 52, 9 | ND | ND | AML-M2 | 47,XY,+4,t(8;21)(q22;q22) |
| positive | 5 M/16 | 77, 2 | 62, 1 | 62, 9 | 39, 7 | 73, 2 | AML-M2 | 46,XY;46,XY,t(8;21)(q22;q22);47,XY,+13,t(8;21)(q22;q22) |
| 8 M/40 | 93, 7 | 95 | 33, 5 | 48 | 91, 2 | AML-M2 | 46,XY,t(2;8;21)(q24;q22;q22) | |
| 13 M/55 | 89, 8 | 56, 5 | 61, 7 | 40 | 10, 5 | AML-M4Eo | 46,XY;inv(16)(p13;q22) | |
| 15 M/53 | 81, 1 | 78, 6 | 66, 9 | 73, 8 | 44 | AML-M4Eo | 46,XY,inv(16)(p13;q22) | |
| Asp816Tyr | 1 M/47 | 87 | 70 | 87 | 23 | 94 | AML-M2 | 47,XY,+4,t(8;21)(q22;q22) |
| positive | ||||||||
ND, not determined; BMMNC, bone marrow mononuclear cell.
HinfI assay for detection of Asp816Val c-kit mutation.
Digestion of the 212 bp exon 17 polymerase chain reaction (PCR) product gives rise to a 161 bp DNA fragment r: when an A-T transition is present, an additional HinfI site is created leading to a 147 bp DNA fragment. This additional fragment was present in patients 2, 5, 8, 13 and 15 as well as in a patient with systemic mastocytosis included as positive control (lane C+). The negative controls were undigested (lane C0) and digested (lane C−) DNAs from normal subjects. Molecular assay at follow up (5a and 15a) is also shown for patients 5 and 15.
HinfI assay for detection of Asp816Val c-kit mutation.
Digestion of the 212 bp exon 17 polymerase chain reaction (PCR) product gives rise to a 161 bp DNA fragment r: when an A-T transition is present, an additional HinfI site is created leading to a 147 bp DNA fragment. This additional fragment was present in patients 2, 5, 8, 13 and 15 as well as in a patient with systemic mastocytosis included as positive control (lane C+). The negative controls were undigested (lane C0) and digested (lane C−) DNAs from normal subjects. Molecular assay at follow up (5a and 15a) is also shown for patients 5 and 15.
A direct role of the mutant c-kit in ligand-independent mast cell growth, tumorigenesis in vivo, and mast cell differentiation has been demonstrated in murine systems using either retroviral infection of hematopoietic progenitor cells with KIT D814V,13 or the transfection of KIT D814Y cDNA into the murine IL3-dependent mast cell line IC2.14,15 With regards to human mutations, only the functional effects of D816V have been investigated. Transfection of D816V KIT cDNA into myb-transformed granulocyte-macrophage colony stimulating factor (GM-CSF)-dependent early murine myeloid cells has been shown to lead to factor independence, increased survival of immature and mature cells, tumorigenicity, and a differentiation resembling that previously noted only in the IC-2 mast cell line.11
It is worth noting that 3 of the 6 patients positive for c-kit mutations, namely patient 1 with t(8;21) AML-M2, and patients 13 and 15 both with inv(16) AML-M4Eo, showed bone marrow mast cell (BMMC) involvement. Mast cell differentiation was massive in patient 1, who carried the Asp816Tyr mutation in 100% of the blasts.9 16 Patients 13 and 15 carrying the Asp816Val mutation had clusters of BMMCs characterized by polar nuclei, a reduced number of small granules, and large empty vacuoles. The remaining three patients positive for Asp816Val did not show any significant increase in BMMCs. The data provided here show that c-kit mutations are not such a rare event in CBF leukemias that display a highly significant correlation for CD117 positivity. In order to assess the true frequency of c-kit mutations in these AML subtypes, scanning of the whole c-kit gene should be performed on a larger sample of patients. Further studies will help elucidate whether c-kit mutations are a secondarily acquired event, as suggested by their occurrence in a small proportion of blast cells, orwhether in a few cases they have a primary involvement in leukemogenesis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal