The type V (for variable) promyelocytic leukemia retinoic acid receptor (PML-RAR) transcript, found in approximately 8% of adult patients with acute promyelocytic leukemia (APL), is defined molecularly by truncation of PML exon 6 and frequent insertion of genetic material from RAR intron 2. To more fully characterize the molecular features of PML-RAR V-type transcripts and to determine whether V-form APL patients have a distinct clinical presentation or prognosis, we analyzed 18 adult V-form APL patients enrolled on Intergroup protocol 0129 (INT-0129). Truncations in PML exon 6 ranged from 8 to 146 nucleotides, and 3 to 127 extra nucleotides (1 to 42 extra amino acids) were inserted at the PML exon 6/RAR exon 3 junction in 13 cases. No distinguishing morphologic, cytogenetic, or immunophenotypic features of V-form blasts were identified. A total of 5 of 7 patients induced with ATRA and 8 of 11 patients who received chemotherapy for induction achieved complete remission (CR). Six patients have relapsed, 4 after chemotherapy induction and 2 after ATRA. Nine patients (50%) are alive, 6 in continuous CR, 2 after salvage therapy for relapsed or refractory disease, and 1 after alternative treatment following early removal from protocol. Although the failure rate for V-form APL patients was high (61%), the low power of the current study to detect clinically significant differences precludes a meaningful comparison of clinical outcomes between the 18 V-form cases and non–V-form adult APL patients enrolled on INT-0129.
Acute promyelocytic leukemia (APL) accounts for approximately 10% of all cases of acute myeloid leukemia (AML). It is highly curable with combined differentiation/cytotoxic therapy1 and is perhaps the best understood subtype of AML at the molecular level. The defining genetic feature of APL is disruption of the retinoic acid receptor (RAR)α gene, at 17q12, and fusion of RARα with 1 of 4 partner genes.2 In more than 99% of APL cases, RARα is fused to a gene termed PML, located at 15q22, and the resulting promyelocytic leukemia (PML)-RARα fusion protein is felt to be responsible for the development of APL. At the messenger RNA (mRNA) level, different numbers of PML 5′ exons are fused with RARα exons 3 through 9 to create 3 structurally distinct PML-RARα fusion transcripts, the so-called L (long), V (variable), and S (short) isoforms (Figure1A). The oncogenic potential of all 3 PML-RARα isoforms derives from their ability to inhibit, in a dominant negative fashion, both PML-dependent– and RARα-dependent signaling pathways. This inhibition of native PML and RARα function leads to a block of myeloid differentiation, inhibition of apoptosis, and ultimately to the APL phenotype.
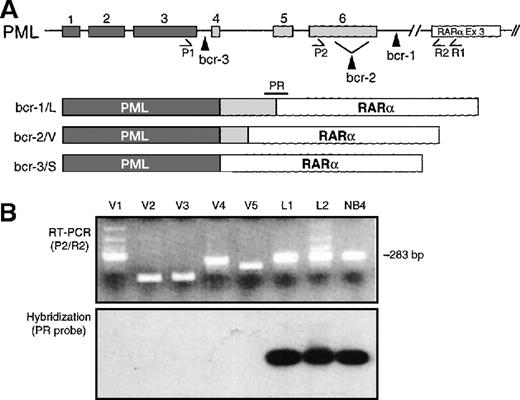
Schematic representation of PML breaksites and PML-RARα fusion mRNAs.
(A) The 3 PML breakpoint cluster regions (bcr-1, -2, and -3) are indicated by vertical arrowheads, and the resultant L, V, and S PML-RARα isoforms are diagrammed. PML exons are indicated by numbered, shaded rectangles. The positions of primers used to amplify PML-RARα mRNA are indicated below their respective exons. (B) PCR products were electrophoresed and visualized by staining with ethidium bromide and then transferred to nylon membranes and hybridized with junctional probe PR. In L-form APL, as well as in the NB4 cell line, the PML breaksite is in intron 6 (bcr-1), and full-length PML exon 6 is fused in-frame to exon 3 (Ex 3) of RARα. In such cases, primers P2 and R2 generate a 283-bp chimeric PCR product that is detected by hybridization to the junctional oligonucleotide probe, designated PR (lanes L1, L2, NB4). V-form cases, by definition, lack variable amounts of terminal exon 6 sequence and give variable-sized bands with primer set P2/R2 (panel B, lanes V1-V5). In most cases, due to loss of exon 6 sequence, the amplicon is smaller than 283 bp (ie, V2, V3, V5); however, due to insertion of genomic DNA from RARα intron 2, the amplified fragment can be larger (case V1) or essentially the same size (case V4) as the 283-bp L-form fragment. These latter V-form cases are distinguished from L-form cases by lack of hybridization to the PR probe, and/or by sequencing the P2/R2 amplicon.
Schematic representation of PML breaksites and PML-RARα fusion mRNAs.
(A) The 3 PML breakpoint cluster regions (bcr-1, -2, and -3) are indicated by vertical arrowheads, and the resultant L, V, and S PML-RARα isoforms are diagrammed. PML exons are indicated by numbered, shaded rectangles. The positions of primers used to amplify PML-RARα mRNA are indicated below their respective exons. (B) PCR products were electrophoresed and visualized by staining with ethidium bromide and then transferred to nylon membranes and hybridized with junctional probe PR. In L-form APL, as well as in the NB4 cell line, the PML breaksite is in intron 6 (bcr-1), and full-length PML exon 6 is fused in-frame to exon 3 (Ex 3) of RARα. In such cases, primers P2 and R2 generate a 283-bp chimeric PCR product that is detected by hybridization to the junctional oligonucleotide probe, designated PR (lanes L1, L2, NB4). V-form cases, by definition, lack variable amounts of terminal exon 6 sequence and give variable-sized bands with primer set P2/R2 (panel B, lanes V1-V5). In most cases, due to loss of exon 6 sequence, the amplicon is smaller than 283 bp (ie, V2, V3, V5); however, due to insertion of genomic DNA from RARα intron 2, the amplified fragment can be larger (case V1) or essentially the same size (case V4) as the 283-bp L-form fragment. These latter V-form cases are distinguished from L-form cases by lack of hybridization to the PR probe, and/or by sequencing the P2/R2 amplicon.
Although all 3 PML-RARα isoforms lead to APL in humans, there are clinical and biochemical differences among them.3-6 The type V PML-RARα isoform, which is the subject of this report, is found in approximately 8% of adult patients with APL3 but is significantly more common in pediatric APL.7,8 This PML-RARα V isoform results from either an aberrant splicing event or from a genomic break in PML exon 6 (or, rarely, in PML exon 5); the molecular consequence, in both cases, is loss of coding DNA from the distal region of PML exon 6 (or loss of all of exon 6), often with insertion of genomic DNA from RARα intron 2. The presence of potential phosphorylation sites and of a caspase cleavage site9 in the distal region of PML exon 6 lends scientific support to the hypothesis that PML exon 6 truncations may influence the function of the PML-RARα oncoprotein and thereby influence response to treatment. Indeed, a recent study reported that the extent of truncation of PML exon 6 correlated with APL blast sensitivity to all-trans retinoic acid (ATRA) in vitro.10 Because adult V-form APL cases are rare, and because of the suggestion that some V-form APL blasts may be ATRA-resistant, we present here the clinical and molecular features, and response to therapy, of 18 adult V-form APL patients who were enrolled on Intergroup protocol 0129 (INT-0129).1 This is the largest combined molecular and clinical study of the rare V-form subtype of APL, and our results suggest that, although such patients have a high failure rate, they nevertheless have a long-term survival that does not appear to differ significantly from that of other adult APL cases.
Materials and methods
Patients
INT-0129 was a comparison of induction therapy of APL with ATRA alone versus chemotherapy with daunorubicin and ara-C (DA), with a subsequent randomization of patients achieving complete remission (CR) to 1 year of maintenance ATRA or observation.1 A total of 401 patients were enrolled, and 350 were evaluable; 230 evaluable adult patients (82% of evaluable adult patients) enrolled on INT-0129 by Cancer and Leukemia Group B (CALGB), the Southwest Oncology Group (SWOG), and the Eastern Cooperative Oncology Group (ECOG) had reverse-transcription polymerase chain reaction (RT-PCR) performed on a pretreatment bone marrow or blood sample. Among these 230 patients, 18 (8%) expressed the PML-RARα V isoform and are described in detail here. The clinical data in this report are based on information received at the ECOG data management center as of January 23, 1999.
Flow cytometry, cytogenetics, and molecular analyses
Flow cytometric and cytogenetic analyses were performed by laboratories affiliated with the respective cooperative groups. Karyotypes were reported according to guidelines of the International System for Human Cytogenetic Nomenclature,11 and immunophenotypic analyses were carried out using standard antibody combinations.12 The details of the RT-PCR procedure for detection of PML-RARα and documentation of the V isoform have been previously published.3
Statistics
The coding of CR is based on remission status on the initial induction regimen. One patient randomized to ATRA who failed to achieve a CR after 6 weeks of treatment is coded as a nonresponder even though CR was achieved after cross over to DA. For purposes of this report, 1 patient randomized to ATRA who received only DA induction is analyzed with the DA arm. Disease-free survival (DFS) is calculated from the date of induction CR to documented relapse or to censoring. DFS was censored if the subject was in continuing remission at the last update. However, if off-study treatment was initiated in a patient in CR, DFS was censored at that date. Data regarding off-study treatment (ie, bone marrow transplant) were not available in most instances. Univariate analyses testing the association of categorical variables were performed with the Fisher exact test. DFS and survival curves were estimated by the method of Kaplan and Meier.13
Results
Molecular analysis
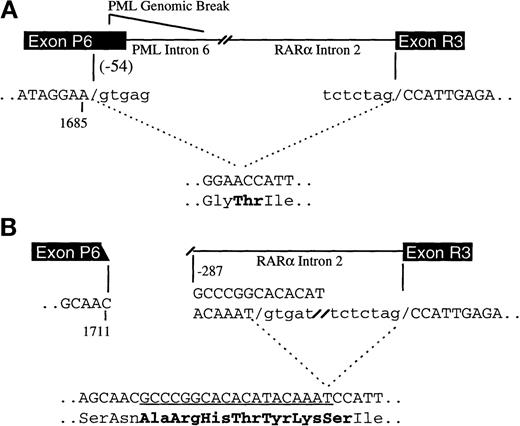
Figure 1 shows the position of primers used in RT-PCR to identify APL patients with the PML-RARα V isoform. The initial PCR was performed with primers from PML exon 3 and RARα exon 3 (P1/R1), followed by a nested PCR with primers from the proximal regions of PML exon 6 and RARα exon 3 (P2/R2; Figure1A). Electrophoretic analysis, hybridization with a junctional probe (Figure 1B) and, ultimately, sequencing of PCR products allowed determination of the extent of PML exon 6 truncation and of the nature of inserted sequences, if any. Table 1presents detailed molecular data for the 18 INT-0129 V-form APL patients. In 4 cases (C902, E003, E011, and E801) and in 4 of 12 previously described V-form cases,10 14-16 PML exon 6 was truncated at nucleotide 1685, 54 base pairs (bp) upstream of the normal exon 6/intron 6 boundary; there were no inserted bases in any of these 8 cases. Because a 5′ splice site donor consensus sequence (gtgag) follows nucleotide 1685, it is likely that the PML genomic breakpoint in these cases is 3′ to nucleotide 1685, possibly in PML intron 6, and that the truncated PML exon 6-RARα exon 3 fusion results from an aberrant splicing event. This proposed event, diagrammed in Figure 2A, results in the loss of 54 nucleotides (18 amino acids) from PML exon 6 and in the production of a novel threonine residue at the PML-RARα junction (Figure 2A and Table 1).
Molecular data of V-form APL cases on Intergroup 0129
| Case . | Exon 6 Break* . | Inserted Nucleotides & Amino Acids† . |
|---|---|---|
| C132 | −146 | gaggagtccttctgcaggaagaggagagattgcca |
| ArgSerProSerAlaGlyArgGlyGluIleAla | ||
| C205 | −142 | tcctcccctcttctctctctagcca |
| SerSerProLeuLeuSerLeuAla | ||
| S448 | −78 | ctatggacacacaggttggagcca |
| MetAspThrGlnValGlyAla | ||
| S444 | −77 | tagtcttagagcca |
| SerLeuArgAla | ||
| E076 | −61 | gtctgccatcctaaccttccatcttggcaaggggcactgggtcct |
| ValCysHisProAsnLeuProSerTrpGlnGlyAlaLeuGlyPro | ||
| tatggggttgttgtcctggccccagacacttggctgtcatctttg | ||
| TyrGlyValValValLeuAlaProAspThrTrpLeuSerSerLeu | ||
| aggctttcatccccaggagtggaggggagaagctgctctgcca | ||
| ArgLeuSerSerProGlyValGluGlyArgSerCysSerAla | ||
| C902 | −54 | gaacca |
| E003 | Thr | |
| E011 | ||
| E801 | ||
| E036 | −37 | cccgaaggactggacacacaggttggagcca |
| ProGluGlyLeuAspThrGluValGlyAla | ||
| E086 | −30 | gcagagcca |
| ArgAla | ||
| S386 | −28 | aacgcccggcacacatacaaatcca |
| AsnAlaArgHisThrTyrLysSer | ||
| E805 | −24 | acggagccacca |
| GlyArgThr | ||
| C179 | −23 | cgtactctttcttagagcca |
| ValLeuPheLeuArgAla | ||
| E027 | −21 | tgggagcca |
| GlyAla | ||
| E091 | −12 | gcgcca |
| Ala | ||
| C183 | −10 | gccaccccgctccggactccgctttggaatggctcaaaccactcca |
| AlaThrProLeuArgThrProLeuTrpAsnGlySerAsnHisSer | ||
| E070 | −8 | cggagtttgggcca |
| GlyValTrpAla |
| Case . | Exon 6 Break* . | Inserted Nucleotides & Amino Acids† . |
|---|---|---|
| C132 | −146 | gaggagtccttctgcaggaagaggagagattgcca |
| ArgSerProSerAlaGlyArgGlyGluIleAla | ||
| C205 | −142 | tcctcccctcttctctctctagcca |
| SerSerProLeuLeuSerLeuAla | ||
| S448 | −78 | ctatggacacacaggttggagcca |
| MetAspThrGlnValGlyAla | ||
| S444 | −77 | tagtcttagagcca |
| SerLeuArgAla | ||
| E076 | −61 | gtctgccatcctaaccttccatcttggcaaggggcactgggtcct |
| ValCysHisProAsnLeuProSerTrpGlnGlyAlaLeuGlyPro | ||
| tatggggttgttgtcctggccccagacacttggctgtcatctttg | ||
| TyrGlyValValValLeuAlaProAspThrTrpLeuSerSerLeu | ||
| aggctttcatccccaggagtggaggggagaagctgctctgcca | ||
| ArgLeuSerSerProGlyValGluGlyArgSerCysSerAla | ||
| C902 | −54 | gaacca |
| E003 | Thr | |
| E011 | ||
| E801 | ||
| E036 | −37 | cccgaaggactggacacacaggttggagcca |
| ProGluGlyLeuAspThrGluValGlyAla | ||
| E086 | −30 | gcagagcca |
| ArgAla | ||
| S386 | −28 | aacgcccggcacacatacaaatcca |
| AsnAlaArgHisThrTyrLysSer | ||
| E805 | −24 | acggagccacca |
| GlyArgThr | ||
| C179 | −23 | cgtactctttcttagagcca |
| ValLeuPheLeuArgAla | ||
| E027 | −21 | tgggagcca |
| GlyAla | ||
| E091 | −12 | gcgcca |
| Ala | ||
| C183 | −10 | gccaccccgctccggactccgctttggaatggctcaaaccactcca |
| AlaThrProLeuArgThrProLeuTrpAsnGlySerAsnHisSer | ||
| E070 | −8 | cggagtttgggcca |
| GlyValTrpAla |
Extent of PML exon 6 truncation (0 = no loss of PML exon 6 sequence).
Inserted or novel nucleotides and amino acids (in 3-letter code) are shown in bold type, preceded by the last 3 nucleotides of truncated PML exon 6 and followed by the first 3 nucleotides (cca) of RARα exon 3. In cases with no insertions (C902, E003, E011, E801, and E091), the junctional amino acid is in italics.
Proposed mechanisms of PML exon 6 truncations in V-form APL.
In (A), represented by cases C902, E003, E011, and E801, the PML exon 6 fusion site is 54 bp 5′ of the normal PML exon 6/intron 6 boundary (at nucleotide 1685), but the PML genomic breakpoint is 3′ of nucleotide 1685, either in the distal region of PML exon 6 or in intron 6. The loss of 54 nucleotides from exon 6 occurs as a result of an aberrant splicing event. The cryptic 5′ splice donor site (gtgag) is shown. Only the relevant PML and RARα exons are shown for clarity. In (B), the molecular events responsible for case S386 are diagrammed. The PML genomic break is in exon 6 at nucleotide 1711, and the RARα break is in intron 2 at bp –287 relative to the exon 3 boundary. The processed mRNA contains a 19-nucleotide insertion from RARα intron 2 (underlined) that results from use of a cryptic 5′ splice donor site (gtgat) 19 bp downstream from the breaksite. Genomic sequence that is spliced out is shown in lowercase type. Six novel amino acids and a new junctional amino acid (Ser) are incorporated in the final PML-RARα protein and are shown in bold.
Proposed mechanisms of PML exon 6 truncations in V-form APL.
In (A), represented by cases C902, E003, E011, and E801, the PML exon 6 fusion site is 54 bp 5′ of the normal PML exon 6/intron 6 boundary (at nucleotide 1685), but the PML genomic breakpoint is 3′ of nucleotide 1685, either in the distal region of PML exon 6 or in intron 6. The loss of 54 nucleotides from exon 6 occurs as a result of an aberrant splicing event. The cryptic 5′ splice donor site (gtgag) is shown. Only the relevant PML and RARα exons are shown for clarity. In (B), the molecular events responsible for case S386 are diagrammed. The PML genomic break is in exon 6 at nucleotide 1711, and the RARα break is in intron 2 at bp –287 relative to the exon 3 boundary. The processed mRNA contains a 19-nucleotide insertion from RARα intron 2 (underlined) that results from use of a cryptic 5′ splice donor site (gtgat) 19 bp downstream from the breaksite. Genomic sequence that is spliced out is shown in lowercase type. Six novel amino acids and a new junctional amino acid (Ser) are incorporated in the final PML-RARα protein and are shown in bold.
In 13 of the remaining 14 V-form INT-0129 cases, and in 6 additional cases reported previously,15,16 the PML breaksite is within PML exon 6, and there are variable numbers of additional nucleotides inserted between the PML exon 6/RARα exon 3 junction (Table 1). By comparison to DNA sequence deposited in GenBank, the origin of the inserted sequences could be ascertained in 5 of the 8 cases in which there were more than 10 inserted nucleotides. The inserted DNA in cases C205 and S386 originates from the distal portion of RARα intron 2, 19, and 287 nucleotides 5′ of the intron 2/exon 3 boundary, respectively (GenBank accession AF088 890.1). Cases S448 and E036 contain an identical stretch of 18 nucleotides that maps to the proximal region of RARα intron 2313 nucleotides from the exon 2/intron2 boundary (GenBank AF088 889.1). The RARα intron 2 breaksites in these 2 cases are 7 bp apart, and the resulting PML-RARα isoforms contain an identical stretch of 6 novel amino acids (AspThrGlnValGlyAla; Table 1). The inserted sequence in case C183 is homologous (39 out of 40 identical bases) to exon 1 of mouse and rat RARα isoform 2,17 mapping the genomic breakpoint in this case to exon 1 of human RARα isoform 2. Thirteen amino acids from this exon are included in the resulting C183 PML-RARα fusion protein. A diagram of the molecular events responsible for these cases, using S386 as an example, is shown in Figure 2B. Case E091 is anomalous in that it does not contain additional inserted sequence, nor does it appear to have a cryptic splice donor site.
Patient data
The 18 V-form patients included 10 women and 8 men (Table2). The median age was 33.5, and the median white blood cell count at presentation was 4.05 × 109/L (range, 0.70 × 109/L to 68.6 × 109/L). Eight of 18 (44%) V-form patients had white cell counts >5 × 109L, and 5 (28%) had white cell counts >10 × 109L. For comparison, 35 of 92 (38%) S-form, and 25 of 121 (21%) L-form cases, had white cell counts >5 × 109L (P = .04, L vs V). Three patients had an ECOG performance status ≥2 (Table 2).
Pretreatment demographic, hematologic, and cytogenetic characteristics of 18 V-form APL patients on Intergroup protocol 0129
| UPN . | Age . | Gender . | PS . | WBC . | Blood P/B% . | Marrow P/B% . | Karyotype . |
|---|---|---|---|---|---|---|---|
| C132 | 22 | F | 0 | 1.8 | 36 | 55 | 46,XX,t(15;17)(q22;q12)[20] |
| C205 | 23 | F | 1 | 4.8 | 36 | 47 | 46,XX,t(15;17)(q22;q12)[20] |
| S448 | 35 | M | 2 | 7.4 | 73 | 83 | 46,XY,add(7)(q31),t(15;17)(q22;q12)[20] |
| S444 | 25 | F | 0 | 2.0 | 0 | 38 | 46,XX/46,XX,t(15;17)(q22;q12)[20] |
| E076 | 19 | M | 0 | 7.3 | 83 | NA | 46,XY,del(9)(q12q22),t(15;17)(q22;q12)[15]/46,XY[5] |
| C902 | 25 | M | 0 | 0.9 | 0 | 92 | 46,XY,t(15;17)(q22;q12)[19]/46,XY[1] |
| E003 | 66 | F | 3 | 63.1 | 59 | 84 | 46,XX[8] |
| E011 | 32 | F | 0 | 77.4 | 95 | 91 | 46,XX[97]/47,XX,+13[3] |
| E801 | 36 | F | 0 | 0.7 | 23 | 90 | NA |
| E036 | 31 | M | 1 | 2.3 | 1 | NA | 46,XY,t(15;17)(q22;q12)[9]/46,XY[11] |
| E086 | 29 | F | 1 | 68.6 | 85 | 90 | 46,XX,t(15;17)(q22;q12)[17]/46,XX[18] |
| S386 | 41 | F | 0 | 1.1 | 0 | 34 | NA |
| E805 | 19 | F | 0 | 3.3 | NA | 75 | 46,XX,t(15;17)(q22;q12)[20] |
| C179 | 37 | M | 1 | 11.0 | 77 | 88 | 47,XY,+8,t(15;17)(q22;q12)[17]/46,XY,der(15)t(8;15)(q13;p11), t(15;17)(q22;q12),der(22)t(8;22)(q13;p11)[12]/46,XY[1] |
| E027 | 44 | M | 0 | 5.7 | 56 | NA | 46,XY,t(15;17)(q22;q12)[20] |
| E091 | 57 | M | 3 | 1.7 | 0 | 80 | 46,XY,t(15;17)(q22;q12)[17]/46,XY[3] |
| C183 | 41 | F | 0 | 12.7 | 50 | NA | NA |
| E070 | 43 | M | 0 | 0.9 | 30 | 90 | 47,XY,+8,t(15;17)(q22;q12)[20] |
| UPN . | Age . | Gender . | PS . | WBC . | Blood P/B% . | Marrow P/B% . | Karyotype . |
|---|---|---|---|---|---|---|---|
| C132 | 22 | F | 0 | 1.8 | 36 | 55 | 46,XX,t(15;17)(q22;q12)[20] |
| C205 | 23 | F | 1 | 4.8 | 36 | 47 | 46,XX,t(15;17)(q22;q12)[20] |
| S448 | 35 | M | 2 | 7.4 | 73 | 83 | 46,XY,add(7)(q31),t(15;17)(q22;q12)[20] |
| S444 | 25 | F | 0 | 2.0 | 0 | 38 | 46,XX/46,XX,t(15;17)(q22;q12)[20] |
| E076 | 19 | M | 0 | 7.3 | 83 | NA | 46,XY,del(9)(q12q22),t(15;17)(q22;q12)[15]/46,XY[5] |
| C902 | 25 | M | 0 | 0.9 | 0 | 92 | 46,XY,t(15;17)(q22;q12)[19]/46,XY[1] |
| E003 | 66 | F | 3 | 63.1 | 59 | 84 | 46,XX[8] |
| E011 | 32 | F | 0 | 77.4 | 95 | 91 | 46,XX[97]/47,XX,+13[3] |
| E801 | 36 | F | 0 | 0.7 | 23 | 90 | NA |
| E036 | 31 | M | 1 | 2.3 | 1 | NA | 46,XY,t(15;17)(q22;q12)[9]/46,XY[11] |
| E086 | 29 | F | 1 | 68.6 | 85 | 90 | 46,XX,t(15;17)(q22;q12)[17]/46,XX[18] |
| S386 | 41 | F | 0 | 1.1 | 0 | 34 | NA |
| E805 | 19 | F | 0 | 3.3 | NA | 75 | 46,XX,t(15;17)(q22;q12)[20] |
| C179 | 37 | M | 1 | 11.0 | 77 | 88 | 47,XY,+8,t(15;17)(q22;q12)[17]/46,XY,der(15)t(8;15)(q13;p11), t(15;17)(q22;q12),der(22)t(8;22)(q13;p11)[12]/46,XY[1] |
| E027 | 44 | M | 0 | 5.7 | 56 | NA | 46,XY,t(15;17)(q22;q12)[20] |
| E091 | 57 | M | 3 | 1.7 | 0 | 80 | 46,XY,t(15;17)(q22;q12)[17]/46,XY[3] |
| C183 | 41 | F | 0 | 12.7 | 50 | NA | NA |
| E070 | 43 | M | 0 | 0.9 | 30 | 90 | 47,XY,+8,t(15;17)(q22;q12)[20] |
UPN indicates unique patient number; PS, performance status (ECOG); WBC, white blood cell count prior to any cytoreductive therapy (×103/L); P, promyelocytes; B, blasts; NA, not available.
Cytogenetic analysis was successfully completed in 15 patients (Table2). Although 13 patients had the t(15;17)(q22;q12), the other 2 patients lacked this abnormality—patient E003 had an apparently normal karyotype (although only 8 metaphases were available for analysis), and patient E011 had primarily normal metaphases with a minor clone showing trisomy 13. Four of the 13 t(15;17)-positive patients (30%) had 1 or more additional chromosomal abnormalities (Table 2), and all 4 either failed induction therapy or relapsed (Table3). Immunophenotypic information was available for most patients; no consistent differences in surface antigen expression could be detected between the V-form patients and APL patients described in other series. Two cases (E003 and E091) had relatively high CD11b expression (expression on 72% and 32% of blast cells, respectively), which is uncommon in APL. One patient (E003) expressed CD2, and another (C205) expressed CD34 at high levels. All cases examined were HLA-DR–negative, and no case expressed CD56.
Therapy and clinical outcome of 18 V-form APL patients on Intergroup protocol 0129
| UPN . | Induction Therapy . | CR . | Maintenance Arm . | Relapse . | CR Duration (Months) . | Status . | Survival (Months) . |
|---|---|---|---|---|---|---|---|
| C132 | DA | No | — | — | — | Dead | 19 |
| C205 | DA | Yes3-150 | ATRA | — | — | Alive | 51 |
| S448 | DA | Yes | Obs | Yes | 8 | Dead | 27 |
| S444 | ATRA | Yes | ATRA | No | 40 | Alive | 42 |
| E076 | ATRA | No | — | — | — | Dead | 35 |
| C902 | ATRA | Yes | Obs | No | 46 | Alive | 48 |
| E003 | ATRA3-151 | No | — | — | — | Dead | 27 |
| E011 | DA | Yes | ATRA | Yes | 8 | Dead | 11 |
| E801 | DA | Yes | ATRA | No | 48 | Alive | 49 |
| E036 | DA | Yes | — | No | 13-152 | Alive | 67 |
| E086 | DA3-153 | Yes | Obs | Yes | 16 | Alive | 52 |
| S386 | DA | Yes | Obs | Yes | 28 | Dead | 35 |
| E805 | DA | No | — | — | — | Dead | 14 |
| C179 | ATRA | Yes | —3-6 | Yes | 48 | Alive | 60 |
| E027 | DA | No | — | — | — | Dead | 3 |
| E091 | DA | Yes | Obs | No | 50 | Alive | 51 |
| C183 | ATRA | Yes | ATRA | No | 5 | Alive | 7 |
| E070 | ATRA | Yes | Obs | Yes | 10 | Dead | 13 |
| UPN . | Induction Therapy . | CR . | Maintenance Arm . | Relapse . | CR Duration (Months) . | Status . | Survival (Months) . |
|---|---|---|---|---|---|---|---|
| C132 | DA | No | — | — | — | Dead | 19 |
| C205 | DA | Yes3-150 | ATRA | — | — | Alive | 51 |
| S448 | DA | Yes | Obs | Yes | 8 | Dead | 27 |
| S444 | ATRA | Yes | ATRA | No | 40 | Alive | 42 |
| E076 | ATRA | No | — | — | — | Dead | 35 |
| C902 | ATRA | Yes | Obs | No | 46 | Alive | 48 |
| E003 | ATRA3-151 | No | — | — | — | Dead | 27 |
| E011 | DA | Yes | ATRA | Yes | 8 | Dead | 11 |
| E801 | DA | Yes | ATRA | No | 48 | Alive | 49 |
| E036 | DA | Yes | — | No | 13-152 | Alive | 67 |
| E086 | DA3-153 | Yes | Obs | Yes | 16 | Alive | 52 |
| S386 | DA | Yes | Obs | Yes | 28 | Dead | 35 |
| E805 | DA | No | — | — | — | Dead | 14 |
| C179 | ATRA | Yes | —3-6 | Yes | 48 | Alive | 60 |
| E027 | DA | No | — | — | — | Dead | 3 |
| E091 | DA | Yes | Obs | No | 50 | Alive | 51 |
| C183 | ATRA | Yes | ATRA | No | 5 | Alive | 7 |
| E070 | ATRA | Yes | Obs | Yes | 10 | Dead | 13 |
UPN indicates unique patient number; CR, complete remission; DA, daunorubicin and Ara-C; ATRA, all-trans retinoic acid; Obs, observation.
Because the CR for case C205 was undocumented, no CR duration is available, and this case is not included in the disease-free survival plot shown in Figure 3.
No CR after ATRA for 42 days; crossed over to DA and achieved CR.
Taken off protocol and received alternative consolidation. Censored at day 28 for CR duration.
Randomized to ATRA but received DA instead due to refractory leukocytosis.
Not registered to the maintenance arm.
Treatment and outcome
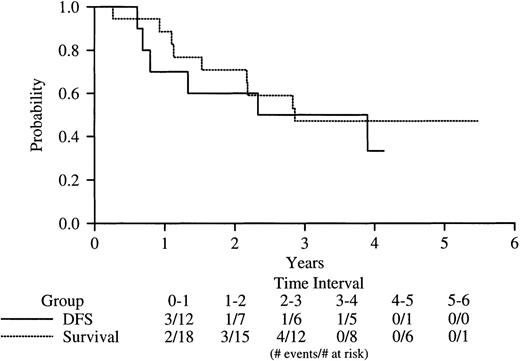
Among the 18 V-form patients, 11 received chemotherapy (DA) induction, and 7 received ATRA (Table 3). One patient (E086) was randomized to receive ATRA for induction but did not receive the drug and was given DA because of hydrea-resistant leukocytosis. Twelve of the 18 patients overall (67%) achieved a documented CR—5 of the 7 ATRA-treated patients and 7 of the 11 patients induced with DA (95% CI, 41%-87%). One additional patient (C205) achieved an undocumented CR. Six of the 13 CR patients (46%) have relapsed at a median of 20 months after attainment of CR—4 after induction with DA and 2 after induction with ATRA (Table 3). As of the date of this analysis (January 23, 1999), 9 of the 18 patients are alive. Figure 3 shows DFS and overall survival of the V-form cases. Of the total group of 18 V-form patients, 11 (61%) either failed induction therapy or relapsed after achieving CR. The median DFS for all patients is 36 months.
Kaplan-Meier analysis of DFS and overall survival of V-form APL cases on INT-0129.
DFS survival is indicated by the solid line, overall survival, by the dotted line.
Kaplan-Meier analysis of DFS and overall survival of V-form APL cases on INT-0129.
DFS survival is indicated by the solid line, overall survival, by the dotted line.
Influence of PML exon 6 break/fusion site on response to ATRA
In a previous study,10 blasts from V-form patients with large PML exon 6 truncations were found to be relatively resistant to ATRA in vitro. Thus, it was of interest to examine whether such “E6-short” (E6S) patients (defined here as patients with loss of ≥54 nucleotides from PML exon 6) had a poorer clinical response to ATRA than patients with loss of fewer than 54 nucleotides from PML exon 6 (“E6-long,” or E6L patients) or if E6S patients had an overall poorer prognosis. There were 9 patients in each subgroup. The median age was 25 (range, 19-66) for the E6S patients and 41 (range, 19-57) for the E6L group (P = .07). The median white cell counts were 4.8 × 109/L (range, 0.90 × 109/L to 77.4 × 109/L) and 3.3 × 109/L (range, 0.90 × 109/L to 68.6 × 109/L), respectively (P = 0.6). Of the 9 E6S patients, 3 failed induction, and 2 of these failures were in response to ATRA (patients E076 and E003; Table 3). Patient E076, whose blasts were relatively ATRA-resistant in vitro,10 received only 3 weeks of ATRA due to development of a fungal infection. Case E003, which has been discussed in detail elsewhere,10 received ATRA for 42 days without achievement of CR but attained CR after crossover to DA. At diagnosis, no t(15;17)-positive cells were seen out of 8 metaphases examined (material was not available for fluorescence in situ hybridization [FISH] analysis). At relapse, PML-RARα was not detected, suggesting that patient E003 may have had a chimeric form of APL from the outset. The other case whose blasts were tested and found to be ATRA-resistant in vitro (E011) received induction with DA (Table3), and thus the ATRA sensitivity of this case in vivo could not be assessed. In the E6L group, there were 2 induction failures, both in response to DA (of 6 who received DA induction), and all 3 E6L patients treated with ATRA achieved CR. A total of 4 and 5 patients of 9 are alive, respectively, in the E6S and E6L subgroups (Table3).
Discussion
Almost all cases of APL result from a chromosomal translocation that fuses the RARα gene, at 17q12, with the PML gene, located at 15q22. The genomic breakpoints in RARα are distributed within the second intron of that gene, in an apparent random distribution; although the issue has received limited attention, there is no compelling evidence for recombination “hotspots” flanking the RARα breakpoint sites. In contrast, the breakpoints in the PMLgene are found in 3 distinct breakpoint clusterregions: bcr-1, which encompasses PML intron 6; bcr-2, which includes PML exons 5 and 6; and bcr-3, which spans the small (about 1 kb) PML intron 3.14,15,18 The clustering of breakpoint sites in the PML genomic locus produces 3 distinct types of PML-RARα transcripts—the short (S), long (L) andvariable (V) isoforms—that appear to have subtly different behavior both in vitro5,6,10 and in vivo. For example, compared with L-form cases, both S- and V-isoform APL patients tend to have higher presenting white cell counts;3 in addition, S-form cases have more frequent secondary cytogenetic changes,4 and express a different profile of cell surface antigens, than L-form cases.8,19 The distal region of PML exon 6, which is variably lost in V isoform cases, and is always lost in S-form APL, is serine/proline–rich and thus likely to be heavily phosphorylated; it also contains a caspase cleavage site that has been shown to mediate retinoid-induced, caspase-mediated, degradation of PML-RARα.9 When overexpressed in vitro, the S isoform is resistant to retinoid-induced degradation,5 possibly due to loss of the PML exon 6 caspase cleavage site. Although no in vitro data are available regarding the specific issues of stability or function of the different V-form PML-RARα transcripts, it is possible that altered phosphorylation or loss (or altered function) of the caspase cleavage site imparts subtle biological differences to V-form APL blasts, as previously suggested in studies of leukemic samples from V-form patients.10 In addition, and although speculative, the insertion of entirely foreign amino acids in many V-form transcripts might be expected to generate a more robust immune response in these patients.
The rarity of adult V-form APL, and its frequent inclusion with L-form (bcr-1) patients in other series, has until now prevented a comprehensive description of such cases at both a clinical and molecular level and has precluded an analysis of response to therapy and outcome of V-form APL patients after modern retinoid-based therapy. The median age (33.5 years) of V-form APL patients in this study, while somewhat lower than generally seen in other adult APL series, was not statistically different from the median age of L- or S-form patients enrolled on INT-0129 from the 3 adult cooperative groups.3Nevertheless, a trend toward younger age of V-form cases is consistent with the more frequent occurrence of V-form APL in children, an observation that remains unexplained.7 8 As noted earlier, there was also a trend toward higher presenting WBC counts in V-form patients as compared with L-form patients. These 2 trends were the only distinguishing clinical features that tended to separate V-form APL patients from other patients with APL.
The striking difference occurs at a submicroscopic level and involves molecular events that produce individually unique PML-RARα isoforms. The analyses of V-isoform cases reported here and elsewhere10 14-16 document these molecular events and highlight 2 possible mechanisms of PML exon 6 truncations. In the first mechanism, diagrammed in Figure 2A, the distal region of exon 6 is lost due to an aberrant splicing event that removes the terminal 54 nucleotides. It is presumed, though not formally proven, that the PML genomic break in these cases is in the distal region of PML exon 6, or possibly in PML intron 6 (ie, in bcr-1). In the second mechanism, diagrammed in Figure 2B, the PML breaksite lies within exon 6 (or, in 1 case, in exon 5; reference 16). The truncated exon lacks an acceptable splice donor site, but the situation is remedied by use of an “unnatural” but functional splice donor site in RARα intron 2. The reading frame is preserved by inclusion of variable numbers of bases from RARα intron 2 in the final, processed PML-RARα transcript and inclusion of variable numbers of “foreign” amino acids in the final PML-RARα fusion protein. As discussed above, whether the presence of these additional amino acids affects the function of the resultant PML-RARα protein is unknown.
One of the purposes of this report was to follow up on a previous correlative laboratory observation that blasts from a subset of APL V-form patients, primarily those with the largest exon 6 deletions, were ATRA-resistant in vitro.10 In the current clinical study, 7 patients received induction therapy with ATRA alone, and 5 of these patients achieved CR. Of the 2 patients who failed ATRA therapy, 1 (E076) received an abbreviated 3-week course due to development of a fungal infection. The other patient (E003) appeared to be truly ATRA-resistant but had a highly unusual, possibly chimeric, type of leukemia. In this case, the PML-RARα–positive clone appears to have been successfully eradicated because, at relapse, the patient had no detectable PML-RARα–positive cells. The PML-RARα transcripts from patients E076 and E003 had relatively large exon 6 truncations (61 and 54 nucleotides, respectively), and blasts from both patients were relatively ATRA-resistant in vitro.10 Overall, however, too few patients with large PML exon 6 truncations were treated with ATRA alone on this study to adequately test the hypothesis that such patients as a group are ATRA-resistant in vivo. In this analysis of 18 V-form cases, which is the largest reported series to date, there was no suggestion that the extent of PML exon 6 truncation, or the type of insertion, had a significant influence on clinical outcome.
Our previous suggestion20 that V-form APL patients as a group might have a poor prognosis is both supported and challenged by the mature outcome data reported here. Although the incidence of treatment failure in the V-isoform patients was high (11 of 18, or 61%, either failed induction or relapsed), 9 of 18 (50%) remain alive—6 in continuous CR, 2 after salvage therapy, and 1 after receiving therapy off protocol. Because 8 of the 18 patients never received ATRA on protocol (although several of the 8 may have received ATRA for salvage therapy) and because the prognosis of APL patients treated with chemotherapy alone remains poor,1 the high failure rate of the V-form patients seen here may reflect their overall minimal exposure to ATRA. Despite the high early failure rate, the 50% overall survival of V-form cases cannot, given the small numbers of patients and the caveat just noted, be considered different from the survival of non–V-form adult APL patients enrolled on INT-0129. Thus, although it remains possible that V-form APL patients are a poor prognosis group, further study of larger numbers of uniformly treated patients will be required to confirm or refute this hypothesis. Such a study might most profitably be pursued in the pediatric population, where the incidence of V-form APL is significantly higher than in adults.
Acknowledgments
The authors thank the individuals at each participating CALGB, SWOG, and ECOG institution for their role in patient care and in procurement and analysis of specimens for this study. We specifically thank Drs David Head (SWOG), Frederick Davey (CALGB), and John M. Bennet (ECOG) for pathologic analysis and Drs Elizabeth Paietta (ECOG) and Carlton Stewart (CALGB) for immunophenotype data.
Supported by National Cancer Institute grants CA59518-04, CA23318, CA31946, CA77658, CA16058, and CA56771 and the Coleman Leukemia Research Fund.
Reprints:James L. Slack, Roswell Park Cancer Institute, Department of Medicine, Elm and Carlton Sts, Buffalo, NY 14263; e-mail:james.slack@roswellpark.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal