Abstract
AML1 is a transcriptional activator that is essential for normal hematopoietic development. It is the most frequent target for translocations in acute leukemia. We recently identified 3 patients in whom pancytopenia developed almost 50 years after high-level radiation exposure from nuclear explosions during or after World War II. In all 3 patients, acute myeloid leukemia (AML) eventually developed that had similar characteristics and clinical courses. Cytogenetics from the 3 patients revealed a t(1;21)(p36;q22), a t(18;21)(q21;q22), and a t(19;21)(q13.4;q22). By fluorescent in situ hybridization (FISH), all 3 translocations disrupted the AML1 gene. Two of theseAML1 translocations, the t(18;21) and the t(19;21), have not been reported previously. It is possible that the AML1 gene is a target for radiation-induced AML.
Exposure to a wide variety of environmental agents can induce acute myeloid leukemia (AML). Chemical exposure to organic compounds such as benzene or petroleum products can increase the risk of developing AML.1,2 Exposure to ionizing radiation also carries an increased risk of developing AML.3-7 There is a also a well-described increase in the incidence of AML in the survivors of atomic explosions.5 The incidence and rapidity of onset appear to be directly proportional to the dose of radiation.5 6
The clinical characteristics of the AML seen in patients exposed to nuclear explosions are typical of most secondary leukemias.3-7 Although the incidence of radiation-induced acute leukemias peaks 10 years after exposure, it still remains high over the entire life span of the person exposed.
Although the clinical behavior of the radiation-induced leukemias is well described, the molecular mechanism behind their origin is not well understood.8 We describe here 3 patients with AML after exposure to nuclear explosions who have translocations in theAML1 gene by fluorescent in situ hybridization (FISH).
Study design
Bone marrow aspirates were obtained from the 3 patients on their presentation with AML and shipped to the laboratory by overnight courier. The specimens were processed for GTG-banded cytogenetic analysis according to standard procedures as we previously described.9
FISH analysis was performed on metaphase cells with whole chromosome paints 19 (Oncor, Gaithersburg, MD) and 21 (Cambio, Cambridge, England). FISH was also performed using the D18Z1 centromere probe for chromosome 18 (Oncor), and TEL/AML1 ES (Vysis, Downer's Grove, IL) dual color translocation probe. The chromosome 18, 19, and 21 probes were labeled with biotin or digoxigenin and detected by fluorescein isothiocyanate (FITC)-conjugated avidin or rhodamine-conjugated antidigoxigenin antibodies. Probe labeling and hybridization conditions are as previously described.9
The TEL/AML1 ES probes are 350 and 500 kilobase (kb) cosmid contigs designed to detect the TEL/AML1 fusion, resulting from a translocation between chromosomes 12 and 21, respectively. These probes are directly labeled with SpectrumGreen (TEL) and SpectrumOrange (AML1). They were used in this analysis to determine whether the AML1 gene was disrupted. The TEL portion of the probe served as an internal control for the quality of the FISH. Metaphase chromosomes were counterstained with DAPI and captured using a PSI imaging system with MacProbe 3.4 software (Perceptive Systems, Houston, TX).
Results and discussion
Three patients with AML who had experienced radiation exposure were identified during cytogenetic analysis as having unusual translocations involving chromosome 21q22. The clinical behavior of each AML was similar.
Case 1
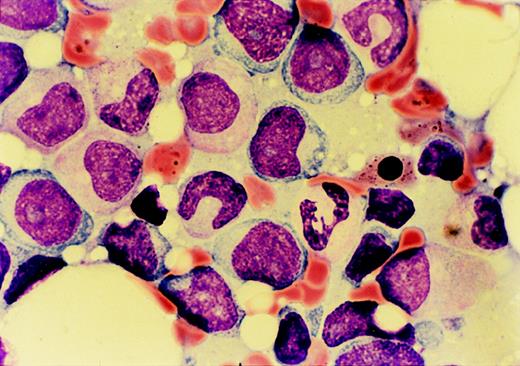
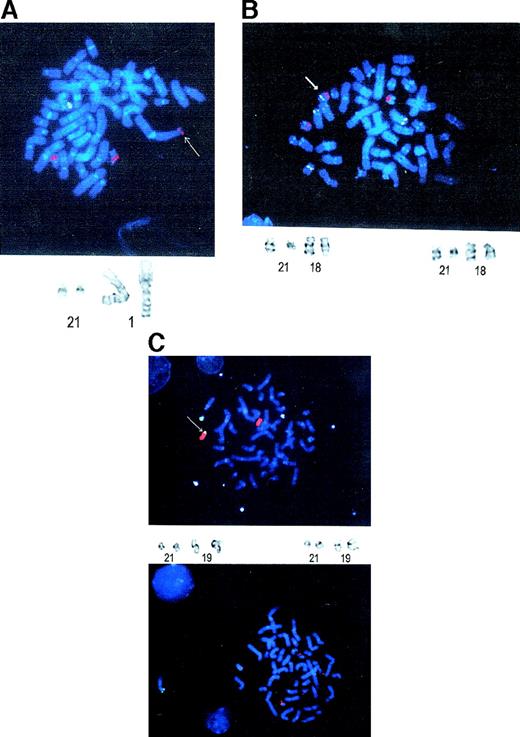
The patient was a 72-year-old man who presented in March 1997 with fatigue and weight loss. A complete blood count revealed pancytopenia. A bone marrow biopsy specimen was most consistent with refractory anemia with excess blasts in transformation (Figure1). The patient stated that he had lived immediately downwind of the Nevada aboveground nuclear weapon test site during atomic weapon testing in the late 1940s. He had personally observed multiple explosions from close proximity without shielding. One month after presentation, a bone marrow biopsy specimen revealed AML-M2. Cytogenetics found a t(1;21)(p36;q22) as the lone abnormality. FISH revealed that the AML1 gene was translocated to chromosome 1p36 (Figure 2A). The patient was treated with standard dose cytarabine for 5 days and idarubicin for 2 days. He achieved a complete remission by marrow morphology, but remained pancytopenic and transfusion dependent after chemotherapy. He suffered from multiple infections requiring hospitalization. Six months later, he died from progressive AML.
Marrow aspirate morphology on presentation of case 1 with refractory anemia with excess blasts in transformation.
Dysplastic granulocytic cells, a ringed sideroblast, and numerous myeloblasts are seen.
Marrow aspirate morphology on presentation of case 1 with refractory anemia with excess blasts in transformation.
Dysplastic granulocytic cells, a ringed sideroblast, and numerous myeloblasts are seen.
FISH analysis of the 3 radiation-induced AML cases.
(A) Case 1. The 3 red signals represent the 2 AML1 genes on chromosome 21q22 and the translocation to chromosome 1p36. The green signal represents TEL on chromosome 12, which serves as an internal control for quality of the FISH. An arrow denotes the translocatedAML1 gene. A partial karyotype is below the FISH photomicrograph, showing the reciprocal translocation between chromosomes 1 and 21. (B) Case 2. The 3 red signals represent the 2AML1 alleles on 21q22 and the translocation to chromosome 18q21. Besides the 2 green TEL signals on chromosome 12, chromosome 18 was labeled with a centromeric probe. An arrow denotes the chromosome 18 with the AML1 translocation. A partial karyotype showing the chromosome 18 and 21 translocations is below the photomicrograph. (C) Case 3. The upper photomicrograph shows 3 green signals representing chromosome 21 paint, whereas the red represents chromosome 19 paint. There is extra chromosome 21q material on chromosome 19q13. An arrow denotes the AML1 translocation to chromosome 19. The lower photomicrograph shows the FISH analysis with the TEL-AML1 probe. The 3 red signals indicate the split AML1 gene. A partial karyotype showing the translocation between chromosomes 19 and 21 is also present.
FISH analysis of the 3 radiation-induced AML cases.
(A) Case 1. The 3 red signals represent the 2 AML1 genes on chromosome 21q22 and the translocation to chromosome 1p36. The green signal represents TEL on chromosome 12, which serves as an internal control for quality of the FISH. An arrow denotes the translocatedAML1 gene. A partial karyotype is below the FISH photomicrograph, showing the reciprocal translocation between chromosomes 1 and 21. (B) Case 2. The 3 red signals represent the 2AML1 alleles on 21q22 and the translocation to chromosome 18q21. Besides the 2 green TEL signals on chromosome 12, chromosome 18 was labeled with a centromeric probe. An arrow denotes the chromosome 18 with the AML1 translocation. A partial karyotype showing the chromosome 18 and 21 translocations is below the photomicrograph. (C) Case 3. The upper photomicrograph shows 3 green signals representing chromosome 21 paint, whereas the red represents chromosome 19 paint. There is extra chromosome 21q material on chromosome 19q13. An arrow denotes the AML1 translocation to chromosome 19. The lower photomicrograph shows the FISH analysis with the TEL-AML1 probe. The 3 red signals indicate the split AML1 gene. A partial karyotype showing the translocation between chromosomes 19 and 21 is also present.
Case 2
The patient was a 73-year-old man who presented with pneumonia in June 1998. He was found to be pancytopenic at that time. A bone marrow biopsy specimen revealed refractory anemia with excess blasts in transformation. The patient photographed Ground Zero in Hiroshima as a member of the armed forces during World War II 2 weeks after the nuclear explosion. The marrow blasts were CD34+/CD33+/CD13+, but were negative for lymphoid antigens. Cytogenetics found a clonal t(18;21)(q21;q22) with an added trisomy 8 in a minority of those t(18;21) cells. FISH analysis revealed that the AML1 gene was translocated to chromosome 18 (Figure 2B). He refused chemotherapy and was treated with transfusions, granulocyte colony-stimulating factor (G-CSF), erythropoietin, and antibiotics. He died from progressive AML 13 months after diagnosis.
Case 3
The patient was a 74-year-old man who presented with fatigue and light-headedness in January 1998. An initial complete blood count revealed pancytopenia. A bone marrow aspirate revealed AML-M2. The patient had worked for a trucking company that delivered supplies and removed debris from Ground Zero in the Nevada aboveground nuclear weapon test site after multiple explosions. Flow cytometry found that the AML was CD34+/CD33+/CD13+. Cytogenetics showed a t(19;21)(q13;q22),-X,-7, add (14)(q32),-16. FISH analysis found that the AML1 gene was split by this translocation (Figure 2C). The patient was treated with standard dose cytarabine for 7 days and idarubicin for 3 days. He achieved a complete remission by marrow morphology. However, all 3 lineages were dysplastic. He remained pancytopenic and transfusion dependent after chemotherapy. He eventually died from recurrent AML 1.5 years later.
Discussion
The AML1 gene codes for the CBFα2 subunit of core binding factor, a heterodimer transcription factor complex composed ofCBFα and CBFβ subunits. Genes for 3 α and 1 β subunits have been identified (CBFΑ1, CBFΑ2,CBFΑ3, and CBFΒ).10,11 TheCBFΒ gene maps to 16p13 and is involved in the AML-M4 associated with inv(16) and t(16;16).10 11
The AML1 gene has had 10 distinct chromosomal translocations implicated in leukemia to date. Six fusion genes have been identified from these translocations: EAP, MDS1, EVI1, ETO, TEL, and MTG16.10-12 EAP, MDS-1, and EVI-1 all occur in the t(3;21) from alternative splicing.12 The genes at 5q13, 17q22, 1p36, 14q22, 15q22, and 12q24 have yet to be determined.13In this study, we have identified 2 other chromosomal regions involved in AML1 disruption, at chromosomes 18q21 and 19q13.4. All the abnormalities seen in the current study are associated with secondary AML, as are many of the previously described but uncloned translocations listed above.12 The relatively good prognosis seen in AML1-ETO is not seen in the other AML1 gene disruptions in AML.
In one study, ionizing radiation was used to induce translocations in hematopoietic cell lines,14 then clones were assayed by polymerase chain reaction (PCR) for the presence of known fusion messenger RNAs (mRNAs). Surprisingly, AML1-ETO was found to a greater extent than any other known fusion translocation. In addition, cytogenetic analysis of radiation-induced AML-M2 in Japan from 1963 to 1988 found that chromosome 21 was involved in 6 of 25 cases.15 Three of these were t(8;21), 2 were t(16;21), and 1 was t(3;21).
The finding that 2 of the 3 patients here had additional cytogenetic abnormalities, all had myelodysplasia, all had similar types of radiation exposure, and all had a similar clinical syndrome imply that radiation played a role in their AML. The long latency may be due to the necessity of another mutation for leukemogenesis or a damaged quiescent stem cell that was not activated until much later in life.
The data cited above and described here indicate that the AML1 gene may be a significant target for damage induced by radiation and that its disruption may be a key step in some radiation-induced leukemias.
Acknowledgments
C. Lytle, D. Crenshaw, J. Velasco, and S. Roherty are acknowledged for their superb cytogenetics technical support.
R.H. is supported by NIH RO1 HL48914, PO1 CA74295, and a Translational Research Award from the Leukemia Society of America.
Reprints:Robert Hromas, Indiana University Cancer Center, R4-202, 1044 W Walnut St, Indianapolis, IN 46202; e-mail:rhromas@iupui.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal