Abstract
BCL10 is directly involved in t(1;14)(p22;q32) of mucosa-associated lymphoid tissue (MALT) lymphoma. Wild-type BCL10 promoted apoptosis and suppressed malignant transformation in vitro, whereas truncated mutants lost the pro-apoptotic activity and exhibited gain of function enhancement of transformation. We studied 220 lymphomas for genomic BCL10 mutation by polymerase chain reaction–single-strand conformational polymorphism and DNA sequencing. Nineteen mutations were found in 13 lymphoma specimens, as follows: 8 of 120 (6.7%) mucosa-associated lymphoid tissue (MALT) lymphomas, 4 of 42 (9.5%) follicular lymphomas, and 1 of 23 (4.3%) diffuse large B-cell lymphomas. No mutations were found in 14 mantle cell lymphomas or 21 T-cell lymphomas. High-grade MALT lymphoma tended to show a slightly higher mutation frequency (2 of 25, 8%) than low-grade MALT tumor (6 of 95, 6.3%). Among low-grade gastric MALT lymphoma, mutations were found in 3 of 11 tumors that did not respond to Helicobacter pylori eradication therapy, but none were found in 22 tumors that regressed completely after H pylori eradication. All 14 potentially pathogenic mutations were distributed in the carboxyl terminal domain of BCL10. Deletion accounted for 10 of these mutations; 10 of 14 mutations caused truncated forms of BCL10. Western blot analysis of a mutant case confirmed the presence of truncated BCL10 products of anticipated size. Our results suggest that BCL10 mutation may play a pathogenic role in B-cell lymphoma development, particularly in aggressive and antibiotic unresponsive MALT lymphomas, and may further implicate the biologic importance of the carboxyl terminal of the molecule.
BCL10 is an apoptotic regulatory molecule identified through its direct involvement in t(1;14)(p22;q32) of mucosa-associated lymphoid tissue (MALT) lymphoma.1,2 The gene encodes a protein of 233 amino acids, with residues 13-101 forming a caspase recruitment domain (CARD), whereas the carboxy terminal 132 amino acids contain no known motifs.1-7 Wild-type BCL10 weakly promotes apoptosis by activating caspase-9, activates NF-κB, and suppresses in vitro transformation of rat embryonic fibroblasts by synergistic oncogenes.1,4,5 The pro-apoptotic activity of BCL10 requires an intact caspase recruitment domain and the carboxyl terminal domain, whereas NF-κB activation requires an intact caspase recruitment domain but not the full-length carboxyl terminal.1,4,6 The domain requirement for the tumor-suppressor activity of BCL10 is unclear. However, 2 truncated BCL10 mutants isolated from cDNA clones of a MALT lymphoma with t(1;14)(p22;q32) have been shown to enhance malignant transformation in in vitro assays.1 These data indicate that wild-type BCL10 is a tumor-suppressor gene and that mutations are necessary for oncogenic activity.
BCL10 maps within a region of chromosome 1p22 that is commonly deleted in a number of malignancies, including non-Hodgkin B-cell lymphoma,8 mesothelioma,9 and male germ cell tumors.10 Using fluorescent in situ hybridization techniques, it has been shown that hemizygous deletions of BCL10 occur at high frequency in a number of malignancies including mantle cell lymphoma and mesothelioma (Jadayel et al, unpublished observations, October 1999). These findings suggest that BCL10 mutations play an important role in tumorigenesis outside the context of the t(1;14)(p22;q32). However, studies of primary solid tumors at the genomic level failed to show frequent somatic mutations.11-16 In contrast, our previous preliminary study of primary B-cell lymphoma showed that BCL10 mutation occurs at the genomic level in some patients.1 The frequency of BCL10 mutation in different lymphoma subtypes and its possible role in lymphomagenesis remained unclear. We have now examined DNA samples from 220 patients with primary lymphoma of various subtypes for BCL10 gene mutation.
Materials and methods
Materials
Tissue blocks from 40 patients with normal tissue and 227 patients with lymphoma were retrieved from the Department of Histopathology, Royal Free and University College London Medical School. The lymphoma specimens were from 120 patients with MALT lymphoma (95 low grade, 25 high grade), 42 patients with follicular lymphoma, 14 patients with mantle cell lymphoma, 30 patients with diffuse large B-cell lymphoma, and 21 patients with T-cell lymphoma. Of the MALT lymphomas, 108 tumors were of gastric origin, and 33 low-grade gastric tumors had clinical follow-up data for Helicobacter pylorieradication therapy. These patients received similar anti-H pylori therapy consisting of a combination of amoxicillin, tinidazole, colloidal bismuth subcitrate, and omeprazole. In 22 patients the tumors regressed completely, 17,18 but 11 patients did not show any histologic response to the therapy (Table1). Three low-grade MALT lymphomas were known to harbor t(1;14)(p22;q32).19
Summary of 11 patients with Helicobacter pylori-eradication, nonresponsive MALT lymphoma
| Patients . | Sex . | Age (y) . | Diagnosis . | Follow-up period since H pylori eradication (mo) . |
|---|---|---|---|---|
| GB | F | 67 | B-MALT-L | 16 |
| MM | F | 59 | B-MALT-L | 6 |
| CL | F | 58 | B-MALT-L | 3 |
| GP | F | 62 | B-MALT-L | 3 |
| CS | M | 47 | B-MALT-L | 8 |
| TA | M | 34 | B-MALT-L | 10 |
| MR | M | 47 | B-MALT-L | 6 |
| LM | F | 71 | B-MALT-L | 12 |
| EA | M | 66 | B-MALT-L | 36 |
| CF | F | 55 | B-MALT-L | 24 |
| PP | M | 60 | B-MALT-L | 5 |
| Patients . | Sex . | Age (y) . | Diagnosis . | Follow-up period since H pylori eradication (mo) . |
|---|---|---|---|---|
| GB | F | 67 | B-MALT-L | 16 |
| MM | F | 59 | B-MALT-L | 6 |
| CL | F | 58 | B-MALT-L | 3 |
| GP | F | 62 | B-MALT-L | 3 |
| CS | M | 47 | B-MALT-L | 8 |
| TA | M | 34 | B-MALT-L | 10 |
| MR | M | 47 | B-MALT-L | 6 |
| LM | F | 71 | B-MALT-L | 12 |
| EA | M | 66 | B-MALT-L | 36 |
| CF | F | 55 | B-MALT-L | 24 |
| PP | M | 60 | B-MALT-L | 5 |
B-MALT-L, low-grade B-cell MALT lymphoma.
Frozen tissues were available from 142 patients, and formalin-fixed and paraffin-embedded tissues were available from the remaining patients. For all low-grade MALT lymphomas, DNA samples were prepared from tumor cells enriched by microdissection20 and used for mutation screening. For the remaining lymphomas, DNA samples were prepared from whole tissue sections in which tumor cells accounted for at least 60% of the total cell population, as estimated by examination of hematoxylin–eosin-stained sections. DNA samples from 3 specimens were also prepared from microdissected normal cells and were used to exclude germline mutations.
Southern blot analysis
High-molecular-weight DNA was digested with restriction enzymesPst1, EcoR1, and HindIII, respectively, separated on 0.7% agarose gels, and blotted to Hybond N+membranes (Pharmacia-Amersham, Amersham, UK). The membranes were hybridized with a BCL10 probe, and a 1.2-kb StyI fragment was isolated from the t(1;14)(p22;q32) breakpoint sequence and labeled with 32P-dCTP by the random hexamer method. The conditions for hybridization, washing, and autoradiography have been described elsewhere.21
PCR–SSCP analysis
The full coding sequence of the BCL10 gene was amplified by 5 overlapping polymerase chain reactions (PCR), with a single reaction spanning the coding exon 1 and with 2 reactions spanning each of the remaining 2 coding exons1; PCR was performed on a thermal cycler (Hybaid, Teddington, UK) using a “hot start–touch-down” program. Appropriate negative controls were included in each PCR experiment. For single-strand conformational polymorphism (SSCP) analysis, 2 μL PCR product was mixed with 4 μL sequencing loading buffer, denatured, and separated on Genphor electrophoresis system (Pharmacia-Amersham) under 15 W constant power for 2 to 4 hours at 5°C, and then it was visualized by silver staining.
Specimens showing abnormal SSCP patterns from the initial screening were reexamined by further duplicate SSCP analyses of PCR products derived from 2 additional separate PCR reactions. Those showing reproducible results were subjected to DNA sequencing. In 2 specimens, paired SSCP bands were excised from SSCP gels, reamplified, and sequenced because sequencing of primary PCR products had failed to identify mutations caused by the presence of normal alleles. Dilution experiments showed that the system could detect mutations in samples with the mutant allele presented as low as 5% of tumor cells (Diss et al, unpublished results, 1999).
DNA sequencing
Sequencing of the BCL10 coding region from specimens with abnormally migrating SSCP bands was carried out twice from separate PCR reactions in both directions using Big Dye DNA polymerase (Perkin-Elmer, Foster City, CA) according to the manufacturer's instructions. If reamplified PCR products from the abnormal SSCP bands were used, sequencing was performed on separate PCR products from paired bands of duplicate PCR–SSCP analyses.
Western blotting
Frozen tissue sections were homogenized in lysis buffer containing 50 mmol/L Tris-HCl (pH 8.0), 150 mmol/L NaCl, 0.02% sodium azide, 0.1% sodium dodecyl sulfate (SDS), 1% NP-40, 0.5% sodium deoxycholate, 100 μg/mL phenylmethylsulfonyl fluoride, and 1 μg/mL leupeptin. Protein extracts were mixed with SDS gel loading buffer, denatured, separated on 12% polyacrylamide gels, and electrotransferred onto nitrocellulose membranes. The membranes were sequentially incubated with a mouse BCL10 monoclonal antibody (clone 151), which recognizes amino acids 122-168,22 biotinylated rabbit antimouse immunoglobulin, and alkaline phosphatase-conjugated avidin and were visualized with 5-bromo-4-chloro-3-indolyl phosphate and nitro-blue tetrazolium.
Results
BCL10 gene rearrangement is a rare event in lymphomas
All BCL10 breakpoints reported to date have been clustered within the 5′ promoter region of the gene.1,2 To examine BCL10 gene rearrangement, Southern blot analysis was carried out in the 54 patients with B-cell lymphoma in which sufficient high-molecular-weight DNA was available, including 2 with low-grade MALT lymphoma with t(1;14)(p22;q32) shown previously by karyotyping.19 BCL10 gene rearrangement was found only in the 2 patients with t(1;14)(p22;q32) but not in the remaining patients, which included 41 with MALT lymphomas (31 low grade, 10 high grade) and 11 with diffuse large B-cell lymphomas. These findings indicate that rearrangements of this region of the BCL10 gene are uncommon in non-Hodgkin B-cell lymphoma and that they have not been significantly underestimated from cytogenetics analysis alone.23
BCL10 gene is mutated in B-cell lymphoma and is associated with MALT lymphoma progression
To exclude potential experimental artifacts from PCR reactions, particularly from bands excised from silver-stained gels, samples showing abnormal SSCP patterns from the initial screening were reexamined by further duplicate PCR amplification and SSCP analyses, and only those showing reproducible results were regarded as true positive. All sequencing was performed from both directions, and mutations were confirmed in both sequencing reactions.
In 40 normal DNA samples, several abnormal SSCP patterns were found in coding exon 1, whereas 1 abnormal SSCP pattern was observed in the 5′ region in coding exon 2 and in the 3′ region in coding exon 3, respectively, indicating the presence of polymorphisms. Sequencing of these samples identified 4 different nucleic acid changes, 3 in the coding sequence (codon 8 CTC > CTG, codon 29 TGT > TCT, and codon 213 GGA > GAA) and 1 in intron 1 (G > C) 11 bases downstream of coding exon 1. The polymorphism at codon 213 caused a GLY → GLU substitution and the remaining 2 in the coding region were silent. The polymorphisms at codon 8 and intron 1 were found alone or in combination and were responsible for the complex SSCP patterns observed for the exon 1 PCR product. These polymorphisms were identical and occurred at a frequency comparable to those reported previously.11, 12
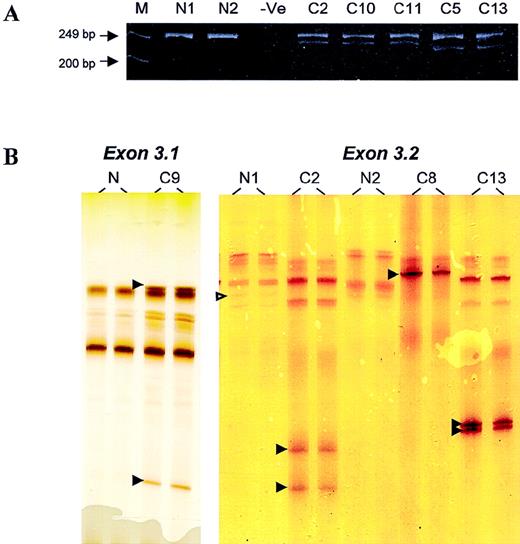
DNA samples from 220 lymphomas, including 133 from frozen and 87 from formalin-fixed, paraffin-embedded tumor tissues, were screened for BCL10 mutation by PCR–SSCP; this was followed by sequencing analysis of the positive specimens. In 5 specimens, novel SSCP bands ran much faster than the normal bands, indicating the possible involvement of deletion (Figure 1). Analysis of the PCR products on polyacrylamide gels showed bands of smaller size in addition to the expected PCR products (Figure 1).
PCR and PCR–SSCP analyses.
(A) Analysis of PCR products of BCL10 exon 3.2 on polyacrylamide gels. M, marker; N, normal DNA samples; −Ve, negative control. Patients C2, C10, and C11 showed a novel PCR product corresponding to a 17-bp deletion, whereas patients C5 and C13 showed a novel PCR band corresponding to a 28-bp deletion. (B) PCR–SSCP analysis of BCL10 exon 3.1 (left panel) and exon 3.2 (right panel). N, normal DNA samples; C, DNA samples from patients with lymphoma. Open arrowhead indicates a G > A polymorphism in codon 213. Filled arrowhead highlights abnormal migrating SSCP bands.
PCR and PCR–SSCP analyses.
(A) Analysis of PCR products of BCL10 exon 3.2 on polyacrylamide gels. M, marker; N, normal DNA samples; −Ve, negative control. Patients C2, C10, and C11 showed a novel PCR product corresponding to a 17-bp deletion, whereas patients C5 and C13 showed a novel PCR band corresponding to a 28-bp deletion. (B) PCR–SSCP analysis of BCL10 exon 3.1 (left panel) and exon 3.2 (right panel). N, normal DNA samples; C, DNA samples from patients with lymphoma. Open arrowhead indicates a G > A polymorphism in codon 213. Filled arrowhead highlights abnormal migrating SSCP bands.
In total, 19 mutations were identified in 13 patients (Table2, Figure 2). Three patients had multiple mutations ranging from 2 to 4, 14 mutations disrupted the amino acid sequence and may be potentially pathogenic, and the remaining 5 were silent. Of the 14 potentially pathogenic mutations, 11 were caused by deletions or insertions, and the remaining 3 were substitutions. To exclude the possibility that the substitution mutations identified were polymorphisms, DNA samples were prepared from microdissected normal cells of these patients and were analyzed in parallel with the corresponding tumor samples by PCR–SSCP. Mutation was detected in the tumor cells but not in the corresponding normal cells in each patient.
Summary of BCL10 mutations
| Lymphoma subtypes . | Patient . | Tissue type* . | Template DNA for sequencing† . | Alteration . | Coding exon . | Codon . | Mutations . | Predicted protein size . |
|---|---|---|---|---|---|---|---|---|
| MALT lymphoma | ||||||||
| Low grade | ||||||||
| H pylori non-res | 1 | P | p-PCR | 2-bp (GA) del | 3 | 140 | Truncation | 140 |
| 2 | P | s-PCR | 17-bp del | 3 | 174 | Truncation | 176 | |
| 3 | P | p-PCR | AAT > GAT | 3 | 173 | ASN > ASP | ||
| 1-bp (C) del | 3 | 185 | Truncation | 208 | ||||
| TCA > TCG | 3 | 189 | Silent | |||||
| CCA > CCG | 3 | 204 | Silent | |||||
| Others‡ | 4 | F | p-PCR | G > A | 3′ coding exon 2 splicing site | Truncation | 119 | |
| 5 | F + P | p-PCR | 28-bp del | 3 | 170 | Truncation | 169 | |
| 6 | F + P | p-PCR | 5-bp del, AGH > AGG | 3 | 218 | Truncation | ||
| 3 | 181 | Silent | ||||||
| High grade | 7 | F | p-PCR | 3-bp del | 3 | 210 | GLU del | 232 |
| 8 | P | s-PCR | ACC > GCC | 3 | 185 | THR > ALA | 233 | |
| GGA > GGG | 3 | 213 | Silent | |||||
| CCT > CCC | 3 | 175 | Silent | |||||
| Follicular lymphoma | 9 | P | p-PCR | TT ins | 3 | 143 | Truncation | 147 |
| 10 | F | p-PCR | 17-bp del | 3 | 174 | Truncation | 176 | |
| 11 | P | p-PCR | 17-bp del | 3 | 174 | Truncation | 176 | |
| 12 | F | p-PCR | 3-bp del | 3 | 210 | GLU del | 232 | |
| DLBCL | 13 | F | p-PCR | 28-bp del | 3 | 170 | Truncation | 169 |
| Lymphoma subtypes . | Patient . | Tissue type* . | Template DNA for sequencing† . | Alteration . | Coding exon . | Codon . | Mutations . | Predicted protein size . |
|---|---|---|---|---|---|---|---|---|
| MALT lymphoma | ||||||||
| Low grade | ||||||||
| H pylori non-res | 1 | P | p-PCR | 2-bp (GA) del | 3 | 140 | Truncation | 140 |
| 2 | P | s-PCR | 17-bp del | 3 | 174 | Truncation | 176 | |
| 3 | P | p-PCR | AAT > GAT | 3 | 173 | ASN > ASP | ||
| 1-bp (C) del | 3 | 185 | Truncation | 208 | ||||
| TCA > TCG | 3 | 189 | Silent | |||||
| CCA > CCG | 3 | 204 | Silent | |||||
| Others‡ | 4 | F | p-PCR | G > A | 3′ coding exon 2 splicing site | Truncation | 119 | |
| 5 | F + P | p-PCR | 28-bp del | 3 | 170 | Truncation | 169 | |
| 6 | F + P | p-PCR | 5-bp del, AGH > AGG | 3 | 218 | Truncation | ||
| 3 | 181 | Silent | ||||||
| High grade | 7 | F | p-PCR | 3-bp del | 3 | 210 | GLU del | 232 |
| 8 | P | s-PCR | ACC > GCC | 3 | 185 | THR > ALA | 233 | |
| GGA > GGG | 3 | 213 | Silent | |||||
| CCT > CCC | 3 | 175 | Silent | |||||
| Follicular lymphoma | 9 | P | p-PCR | TT ins | 3 | 143 | Truncation | 147 |
| 10 | F | p-PCR | 17-bp del | 3 | 174 | Truncation | 176 | |
| 11 | P | p-PCR | 17-bp del | 3 | 174 | Truncation | 176 | |
| 12 | F | p-PCR | 3-bp del | 3 | 210 | GLU del | 232 | |
| DLBCL | 13 | F | p-PCR | 28-bp del | 3 | 170 | Truncation | 169 |
P, paraffin-embedded tissue; F, frozen tissue.
p-PCR, primary PCR; s-PCR: secondary PCR from SSCP bands.
Low-grade MALT lymphoma without clinical data on H pyloritherapy.
Patients 1, 2, and 3 correspond to patients CF, GP, and CL in Table 1, respectively.
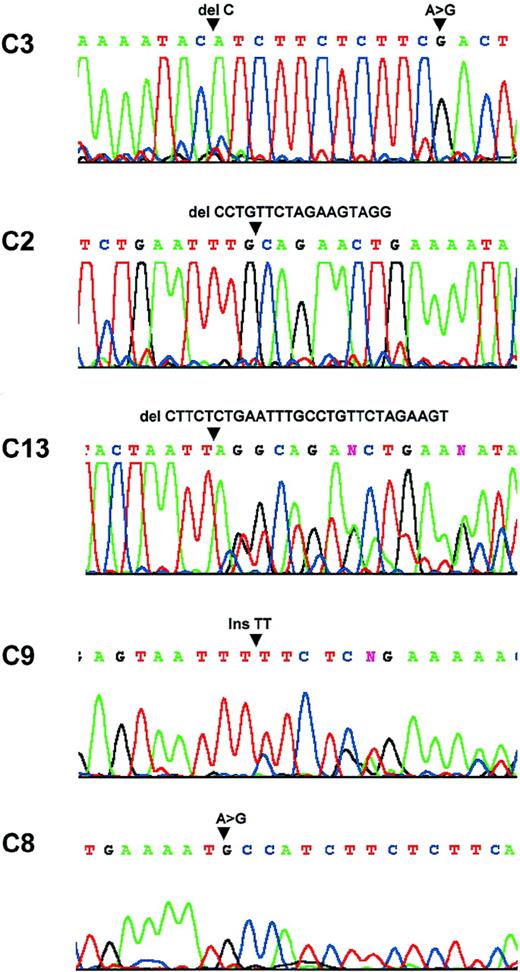
Sequence analysis.
Patient C3: a C deletion at nucleic acid 554 and a substitution at nucleic acid 567. Patient C2: a 17-bp deletion between nucleic acids 522 and 539. Patient C13: a 28-bp deletion between nucleic acids 508 and 536. Patient C9: a TT insertion at nucleic acid 428. Patient C8: a substitution at nuclei acid 553.
Sequence analysis.
Patient C3: a C deletion at nucleic acid 554 and a substitution at nucleic acid 567. Patient C2: a 17-bp deletion between nucleic acids 522 and 539. Patient C13: a 28-bp deletion between nucleic acids 508 and 536. Patient C9: a TT insertion at nucleic acid 428. Patient C8: a substitution at nuclei acid 553.
BCL10 mutations were found in 13 of 199 (6.5%) patients with B-cell lymphoma but not in 21 patients with T-cell lymphoma. Among the B-cell lymphomas, mutations were found in 8 of 120 (6.7%) patients with MALT lymphoma, 4 of 42 (9.5%) patients with follicular lymphoma, and 1 of 23 (4.3%) patients with diffuse large B-cell lymphoma, but they were not found in 14 patients with mantle cell lymphoma (Table3). Intriguingly, a mutation was found in only 1 of the 3 patients with MALT lymphoma with t(1;14)(p22;q32).
Incidence of BCL10 mutation in different subtypes of lymphoma
| Tumor type . | No. of cases . | Mutation frequency (%) . |
|---|---|---|
| B-cell lymphomas | 199 | 13 (6.5) |
| MALT | 120 | 8 (6.7) |
| Low grade | 95 | 6 (6.3) |
| H pylori sensitive | 22 | 0 |
| H pyloriresistant | 11 | 3 (27.3) |
| Others3-150 | 63 | 3 (4.7) |
| High grade | 25 | 2 (8.0) |
| Follicular | 42 | 4 (9.5) |
| Mantle cell | 14 | 0 |
| DLBCL | 23 | 1 (4.3) |
| T-cell lymphomas | 21 | 0 |
| Total | 220 | 13 (5.9) |
| Tumor type . | No. of cases . | Mutation frequency (%) . |
|---|---|---|
| B-cell lymphomas | 199 | 13 (6.5) |
| MALT | 120 | 8 (6.7) |
| Low grade | 95 | 6 (6.3) |
| H pylori sensitive | 22 | 0 |
| H pyloriresistant | 11 | 3 (27.3) |
| Others3-150 | 63 | 3 (4.7) |
| High grade | 25 | 2 (8.0) |
| Follicular | 42 | 4 (9.5) |
| Mantle cell | 14 | 0 |
| DLBCL | 23 | 1 (4.3) |
| T-cell lymphomas | 21 | 0 |
| Total | 220 | 13 (5.9) |
H pylori eradication therapy was not carried out.
There was a tendency for the mutation frequency to be slightly higher in high-grade (2 of 25, 8%) than in low-grade (6 of 95, 6.3%) MALT lymphomas. But the difference was not statistically significant, and a larger study is needed to prove this. Of the patients with low-grade MALT lymphoma, 33 with low-grade gastric tumors underwent clinical follow-up for H pylori eradication therapy. BCL10 mutations were found in 3 of 11 tumors that had not responded to H pylori eradication therapy, but not in the 22 cases that completely regressed after antibiotic therapy (follow-up period, 23 to 79 months). Furthermore, the other 3 instances of low-grade MALT lymphoma with BCL10 mutation (but lacking clinical data on H pylori eradication therapy) were all of gastric origin and showed bulky disease. In all 3 patients, the tumors had invaded the gastric serosa and spread to local lymph nodes. Patient 6 had t(1;14)(p22;q32) and showed splenic dissemination, whereas patient 4 showed tumor invasion to the pancreas. Therefore, in MALT lymphoma, BCL10 mutations appeared to be associated with advanced, aggressive, and antibiotic-unresponsive disease.
Mutations clustered in the carboxyl terminal and caused truncated forms of BCL10 in most patients
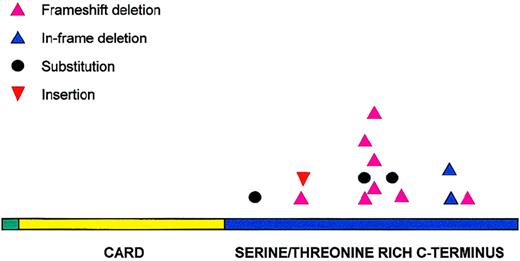
All 14 potentially pathogenic mutations were located in the carboxyl terminal of the molecule (Figure 3). Eleven of these mutations were distributed between amino acids 170 and 210. Codons 170, 174, 185, and 210 were targeted more than once and accounted for a total of 9 mutations. Deletion was the most common form of mutation and occurred in 10 patients, followed by substitution in 3 patients and insertion in 1 patient. Deletions were not restricted to particular lymphoma subtypes and varied in size, ranging from 1 to 28 nucleotides. Two regions appeared to be targeted frequently for deletion—amino acids 170-180 (17-bp deletion in 3 patients, 28-bp deletion in 2 patients), and amino acid 210 (3-bp deletion in 2 patients). The deletion in the former resulted in truncated BCL10 proteins, whereas deletion in the latter caused a deletion of 1 of the 3 consecutive glutamic acids in codons 210-212. In total, 11 of the 14 potentially pathogenic mutations altered the reading frame and caused truncated forms of BCL10.
Distribution of BCL10 mutations.
BCL10 mutations are clustered in the carboxy terminal domain.
Distribution of BCL10 mutations.
BCL10 mutations are clustered in the carboxy terminal domain.
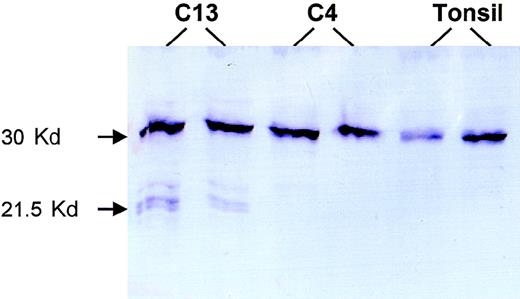
For 2 patients with truncating mutations, sufficient frozen tumor tissues were available, and Western blot analysis was performed to search for truncated forms of BCL10. Patient 13 had a 28-bp deletion between codons 170 and 179 and was predicted to produce a truncated BCL10 of 169 amino acids. Western blot analysis of this patient with mouse monoclonal antibody 151 (mapping to amino acids 122-168; Du et al, unpublished observations, 1999) showed truncated BCL10 of the anticipated size (Figure 4). Patient 4 had a substitution in the 3′ splicing site of the coding exon 2 that produced a truncated BCL10 of 119 amino acids, which was outside the recognition site of the above antibody. As expected, Western blot analysis of this patient showed only wild-type BCL10 (Figure 4).
Western blot analysis.
Patient C13 had a 28-bp deletion between codons 170 and 179 that caused a truncated BCL10 of 169 amino acids. Western blot analysis of this case shows 3 truncated forms of BCL10. The biochemical differences of these various forms are unclear, nor is it clear why the amount of the truncated protein is much less than that of the full-length BCL10 despite the prevalence of tumor cells in the fresh tissue examined. Patient C4 had a substitution at the 3′ coding exon 2 splicing site, which led to a truncated BCL10 product of 119 amino acids that was not recognized by the monoclonal antibody 151 (mapping to amino acids 122-168). Both this case and tonsil show only full-length BCL10.
Western blot analysis.
Patient C13 had a 28-bp deletion between codons 170 and 179 that caused a truncated BCL10 of 169 amino acids. Western blot analysis of this case shows 3 truncated forms of BCL10. The biochemical differences of these various forms are unclear, nor is it clear why the amount of the truncated protein is much less than that of the full-length BCL10 despite the prevalence of tumor cells in the fresh tissue examined. Patient C4 had a substitution at the 3′ coding exon 2 splicing site, which led to a truncated BCL10 product of 119 amino acids that was not recognized by the monoclonal antibody 151 (mapping to amino acids 122-168). Both this case and tonsil show only full-length BCL10.
Discussion
In our initial studies performed on cDNA preparations from a variety of tumors, it appeared that BCL10 underwent mutation at high frequency. However, a comparable rate of mutation has not been detected in genomic DNA from a wide range of tumors.11-16 It, therefore, seems that BCL10 undergoes what has been termed molecular misreading.24 The possible pathologic consequences of this phenomenon remain to be determined. Nevertheless, our initial study of 4 of 81 malignant cell lines and of some patients with non-Hodgkin B-cell lymphoma indicated that genomic BCL10 mutation could occur outside the context of the t(1;14)(p22;q32).1 These mutations consisted predominantly of deletions or insertions, or both, and were predicted to result in truncated protein; loss of the intact carboxy terminus was associated with the loss of pro-apoptotic functions and the acquisition of gain of function enhancement of transformation activity. The aim of this study was to determine the frequency and nature of BCL10 mutations in a wider panel of new patients with non-Hodgkin B-cell and T-cell lymphoma, with and without the t(1;14)(p22;q32).
Because others have reported the absence of BCL10 mutations in non-Hodgkin B-cell lymphoma, specifically in MALT lymphoma, care was taken to avoid any potential experimental artifacts, and different experimental approaches were used to confirm the mutations.
1. All mutations were confirmed by at least 2 separate PCR–SSCP and sequencing analyses. When possible, PCR–SSCP analysis was carried out from DNA samples isolated from fresh tumor tissues, and sequencing was performed from primary PCR products.
2. Patients with recurrent mutations were reexamined through fresh DNA samples prepared from the corresponding tumor tissues to ascertain that these mutations were not the result of PCR contamination. In each patient, the same abnormal SSCP pattern was seen after analysis of the fresh DNA samples (data not shown).
3. Large deletions (17-28 bp) were also verified by analysis of the corresponding PCR products on polyacrylamide gels. In each patient, a smaller PCR product of the expected size was observed.
4. Truncated forms of BCL10 protein were demonstrated in 1 patient with truncation mutations.
There were no differences in either the frequency or the pattern of BCL10 mutations between formalin-fixed, paraffin-embedded tissues and frozen tissues.
Our results indicate that BCL10 genomic mutation occurs in some, but not all, subtypes of B-cell lymphomas and is independent of t(1;14)(p22;q32). BCL10 mutation was found in only 1 of the 3 patients with translocation. This finding is intriguing in view of the pro-apoptotic activity of wild-type BCL10 and the overexpression of BCL10 in these patients.22 The lack of BCL10 genomic mutation in the patients with MALT lymphoma with t(1;14)(p22;q32) appears to contradict the expected role of BCL10 in the development of this tumor. Our study of BCL10 protein expression may shed light on these paradoxical findings.22 Using immunohistochemistry, we found that BCL10 protein was expressed only in the cytoplasm of the marginal zone B-cells, the normal cell counterpart of MALT lymphoma. However, the protein was highly expressed in both the nucleus and the cytoplasm in each of the 3 patients with t(1;14)(p22;q32). Thus, the altered cellular localization of BCL10 protein may be the underlying mechanism for BCL10-induced lymphomagenesis, though the causes for BCL10 nuclear localization are unclear.
Unlike solid tissue tumors in which BCL10 genomic mutations are rare,11-16 lymphomas generally showed a relatively high frequency of BCL10 mutation. The frequency of BCL10 genomic mutation varied among different lymphoma subtypes from 7% to 10% in patients with MALT and follicular lymphoma to 0% in patients with mantle cell and T-cell lymphoma. These findings suggest that the BCL10 gene, unlike other tumor suppressor genes such as p53, may be selectively targeted by different human tumors. The variation of BCL10 genomic mutation among various tumor types is in line with the heterogeneous expression of BCL10 protein in different normal tissues. By immunohistochemistry, the B-cell follicle was shown to express abundant BCL10 protein, whereas solid tissue, with the exception of that of the breast, did not express the protein.22 It is conceivable, therefore, that BCL10 may be differentially required for the physiology of various normal tissues and consequently targeted variably by different tumors.
The presence of BCL10 mutations in MALT and follicular lymphoma suggests a pathogenic role of BCL10 mutation in these lymphoma subtypes. This is further supported by the observation of an association of BCL10 gene mutation with disease progression in MALT lymphomas. There was a tendency for the mutation frequency to be higher in high-grade (8%) than low-grade (6.3%) MALT lymphomas. Our preliminary results also show that within low-grade gastric MALT lymphomas, BCL10 mutation was associated with aggressive and antibiotic-unresponsive forms.
BCL10 gene mutation in low-grade gastric MALT lymphomas that are H pylori therapy resistant may be clinically important. Approximately 30% of low-grade gastric MALT lymphomas have reached theH pylori-independent growth stage at diagnosis.25,26 With H pylorieradication therapy, up to 14 months' observation may be necessary to determine whether there is tumor regression after therapy or whether other forms of treatment are required.25 26 If the above preliminary finding is confirmed in a larger controlled study, BCL10 gene mutation could be used to identify prospectively those patients who would not benefit from anti-H pylori therapy.
Selection during tumor development results in clustering of mutations within domains critical to the function of a tumor suppressor or oncogene. The distribution of BCL10 mutations is also characteristic. No mutations were found in the caspase recruitment domain, which mediates BCL10 oligomerization and is crucial for both the pro-apoptotic and the NF-κB activation properties of the molecule.5,6 All 14 potentially pathogenic mutations were located in the carboxyl terminal domain, which may mediate the interaction with caspase-9 and is essential for the pro-apoptotic activity of the molecule.5 Ten of these mutations led to truncated forms of BCL10, and 6 of these were similar to the 2 truncated BCL10 mutants examined previously.1 These BCL10 mutants have been shown to lose the pro-apoptotic activity but to retain the NFκB activation of the wild-type BCL10. Moreover, these mutants have been shown to enhance the transformation of rat embryonic fibroblasts by synergistic oncogenes. Thus, the observed BCL10 mutations in the current study appear to disrupt the carboxyl terminal domain of the molecule, and at least some of these predicted alterations may abolish its interaction with caspase-9 and gain oncogenic activity.
The residues of the carboxyl terminal of BCL10 responsible for its interaction with caspase-9 are unclear. The finding of a recurrent 3-bp in-frame deletion in codon 210 suggests that this region could be important for BCL10 function. In particular, codons 210-212 encode 3 consecutive glutamic acids, which are negatively charged; moreover, codon 213 is a site of a polymorphism, which causes a change from glycine to glutamic acids. The biologic importance of this highly negatively charged region and the polymorphism at codon 213 remain to be determined.
In summary, our results show that BCL10 genomic mutation occurred in 7% to 10% of patients with MALT and follicular lymphoma and was associated with MALT lymphoma progression. Mutations clustered in the carboxyl terminal of the molecule, and the majority caused truncated forms of BCL10, further implicating the biologic importance of the region.
Supported by grants from the Cancer Research Campaign and the Leukaemia Research Fund.
Reprints:Ming-Qing Du, Department of Histopathology, University College London Medical School, University Street, London WC1E 6JJ, United Kingdom; e-mail: m.du@ucl.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal