Abstract
The Rh blood group antigens derive from 2 genes,RHD and RHCE, that are located at chromosomal position 1p34.1-1p36 (chromosome 1, short arm, region 3, band 4, subband 1, through band 6). In whites, a cde haplotype with a deletion of the whole RHD gene occurs with a frequency of approximately 40%. The relative position of the 2 RH genes and the location of the RHD deletion was previously unknown. A model has been developed for the RH locus using RHD- and RHCE-related nucleotide sequences deposited in nucleotide sequence databases along with polymerase chain reaction (PCR) and nucleotide sequencing. The open reading frames of bothRH genes had opposite orientations. The 3′ ends of the genes faced each other and were separated by about 30 000 base pair (bp) that contained the SMP1 gene. The RHD gene was flanked by 2 DNA segments, dubbed Rhesus boxes, with a length of approximately 9000 bp, 98.6% homology, and identical orientation. The Rhesus box contained the RHD deletion occurring within a stretch of 1463 bp of identity. PCR with sequence-specific priming (PCR-SSP) and PCR with restriction fragment length polymorphism (PCR-RFLP) were used for specific detection of the RHDdeletion. The molecular structure of the RH gene locus explains the mechanisms for generating RHD/RHCE hybrid alleles and the RHD deletion. Specific detection of theRHD− genotype is now possible.
The Rhesus D antigen (ISBT 004.001; RH1) is the most important blood group antigen determined by a protein. Anti-D remains the leading cause of hemolytic disease of the newborn.1,2 Depending on the population, 3%-25% of whites lack the antigen D.3 Anti-D immunizations can occur readily in D-negative recipients.4
The antigens of the Rh blood group are carried by proteins coded by 2 genes, RHD and RHCE, that are located at chromosomal position 1p34.1-1p36 (chromosome 1, short arm, region 3, band 4, subband 1, through band 6),5,6 probably within less than a 450 000–base pair (bp) distance.7 Both genes encompass 10 exons, and their structures are highly homologous. Until recently, the relative orientation of the genes, their distance, and the possibility of other interspersed genes were unknown.8 Very recently, Okuda et al9 reported a sequence of about 11 000 bp, which was thought to represent the DNA segment between RHD andRHCE.
In whites, the vast majority of D-negative haplotypes is due to a deletion of the RHD gene. This deletion spans the whole RHDgene because RHD-specific sequences ranging from exon 1 to the 3′ untranslated region are absent.10 The exact extent of the deletion was uncertain, leaving open the possibility that neighboring genes were also affected.
Identification of the RHD gene as the molecular basis of the D antigen enabled the RhD phenotype prediction by DNA typing.8 11 However, because the structure of the prevalent D-negative haplotype is unknown, a specific detection of theRHD deletion remained impossible, and discrimination ofRHD+/RHD+ homozygous individuals fromRHD+/RHD− heterozygous individuals relied on indirect methods. This discrimination is of particular clinical interest in D-negative mothers with an anti-D: the risk of an affected child is 100% with anRHD+/RHD+ father, but it is only 50% with anRHD+/RHD− father.
Several indirect approaches have been applied to determine zygosity: (1) A simple guess based on the phenotype is correct in about 95% of all cases; (2) determination of the D antigen density, which can be confounded by factors such as the presence of the C antigen; and (3) several methods involving the parallel quantitative amplification ofRHD- and RHCE-specific sequences.12,13These elaborate techniques may not be practical in routine laboratories, however. In addition, several investigators identified polymorphisms in the RHCE gene or neighboring sequences that were genetically linked to lack of the RHDgene.7 14-16 This indirect approach relied on the linkage disequilibrium associating RHD− with a polymorphism.
The most direct approach would be polymerase chain reaction (PCR) amplification spanning the RHD deletion site. Such an assay was not available because the structure of theRHD locus in D-positive and D-negative individuals was incompletely understood. We developed and proved a model of theRH gene locus, identified the RHD deletion site in the prevalent D-negative haplotypes in whites, and devised PCR methods for the discrimination ofRHD+/RHD+ andRHD+/RHD− individuals. Thus, direct testing for the presence of the RHDdeletion is now routinely feasible.
Materials and methods
Blood samples and DNA isolation
Blood samples anticoagulated with ethylenediamine tetraacetic acid (EDTA) or citrate were collected from white blood donors. DNA was isolated by a modified salting-out procedure as described previously.10
Yeast artificial chromosome DNA
DNA from the yeast artificial chromosome (YAC) 38A-A10 (UK HGMP Resource Center, Cambridge, England) was isolated after a single growth phase by standard methods.17 We confirmed that this YAC contained RH DNA. Furthermore, shotgun cloning experiments indicated that some of its insert probably derived from the X chromosome (data not shown).
DNA database searches
Identification of an RHD-specific sequence in theRHD promoter
An approximately 2000-bp RHD promoter sequence was established by chromosomal walking (GenomeWalker kit; Clontech, Heidelberg, Germany). D-positive and D-negative samples were amplified using primers re04 and re11d (Table 1), andRHD- and RHCE-specific sequences were established for 1200 bp 5′ of the start codon by sequencing with internal primers. A short deletion in the RHD gene was identified and used to develop the RHD-specific primer re011d. The 1200-bp sequence, including the RHD promoter, was deposited at EMBL under accession number AJ252314.
Primers
| Primer . | Nucleotide sequence . | Localization . | Position, range . |
|---|---|---|---|
| rb10b | GGCTAAATATTTTGATGACCAAGTT | RHD cDNA | 1194 to 1217 |
| re011d | GCAGCCAACTTCCCCTGTG | RHDpromoter | −883 to −901 |
| re014 | GCTCTACCTTGGTCACCTCC | dJ469D22 | 52189 to 52209 |
| re04 | AGGTCACATCCATTTATCCCACTG | dJ469D22 | 53968 to 53945 |
| re11d | AGAAGATGGGGGAATCTTTTTCCT | dJ469D22 | 51193 to 51216 |
| re96 | TTGTGACTGGGCTAGAAAGAAGGTG | dJ469D22 | 242 to 216 |
| rea7 | TGTTGCCTGCATTTGTACGTGAG | RHD cDNA | 1311 to 1333 |
| rend31 | TTCTGTCTGGGTTGGGGAGGG | dJ465N24 | 128649 to 128629 |
| rend32 | GGAGGGGTTAATATGGGTGGC | dJ465N24 | 127355 to 127375 |
| rend8b1 | TTTGTCCTGGTTGCCTGTGGTC | dJ465N24 | 69296 to 69274 |
| rend8b2 | CAAATCCTGTTGACTGGTCTCGG | dJ465N24 | 68451 to 68473 |
| rend9a1 | AACGGCTCCATCACCCCTAAAG | dJ465N24 | 50008 to 49987 |
| rend9a2 | CCCACTCCTAGATACCAACCCAAG | dJ465N24 | 49059 to 49083 |
| rey14a | CTTTATGCACTGCCTCGTTGAATC | dJ469D22 | 56792 to 56769 |
| rey14b | TTGACTGGTGTGGTTGCTGTTG | dJ469D22 | 55863 to 55884 |
| rey15a | GCAGAAAGGGGAGTTGATGCTG | dJ469D22 | 55416 to 55395 |
| rey7 | CTGACAAAGTTGAGAGCCCACTG | dJ469D22 | 62324 to 62346 |
| rey8 | TTAAGCCTACATCCACATGCTGAG | dJ469D22 | 62854 to 62831 |
| rez2 | CCTTGGTCTGCCAGAATTTTCA | RHD cDNA | 2738 to 2717 |
| rez4 | GTTTGGCATCATAGGAGATTTGGC | dJ465N24 | 120101 to 120124 |
| rez7 | CCTGTCCCCATGATTCAGTTACC | dJ465N24 | 124831 to 124854 |
| rh7 | ACGTACAAATGCAGGCAAC | RHDcDNA | 1330 to 1312 |
| rnb31 | CCTTTTTTTGTTTGTTTTTGGCGGTGC | downstream Rhesus box | 1330 to 1312 |
| rr4 | AGCTTACTGGATGACCACCA | RHD cDNA | 1541 to 1522 |
| sf1 | GACTGGGGGGAAAAGCGCAATAC | SMP1 cDNA | 142 to 164 |
| sf1c | GTATTGCGCTTTTCCCCCCAGTC | SMP1cDNA | 164 to 142 |
| sf3 | TGACTTGCTCTCATCCCACATG | SMP1 cDNA | 1696 to 1717 |
| sm19 | GGGCTTGAAGCAAGTAAATGGAAG | SMP1 intron 1 | −58 to −35 |
| sr1 | GCTATCAATATTTTCTTGGTTACAGACAC | SMP1 cDNA | 2172 to 2144 |
| sr3 | GTTCACTGCCATAAGTCTTCAGTGC | SMP1 cDNA | 575 to 551 |
| sr3kp | TGGCCGCACTGAAGACTTATGG | SMP1 cDNA | 546 to 567 |
| sr45 | CAGCTGCATCTATGATAATCCACC | SMP1cDNA | 224 to 243 |
| sr47 | ATGGACAAGTCCGAGGTGATAG | SMP1 cDNA | 315 to 344 |
| sr47c | ATCACCTCGGACTTGTCCATTC | SMP1 cDNA | 342 to 321 |
| sr5 | GCAATCAGAGATCCAAAGGCCAAC | SMP1 cDNA | 428 to 405 |
| sr5c | GTTGGCCTTTGGATCTCTGATTGC | SMP1cDNA | 405 to 428 |
| sr55 | GACATAGTATACCCTGGAATTGCTGT | SMP1 cDNA | 472 to 497 |
| sr55c | ACAGCAATTCCAGGGTATACTATGTC | SMP1 cDNA | 497 to 472 |
| sr9 | CTCCCCCGATTTTAGCCAAGAA | SMP1 cDNA | 27 to 6 |
| Primer . | Nucleotide sequence . | Localization . | Position, range . |
|---|---|---|---|
| rb10b | GGCTAAATATTTTGATGACCAAGTT | RHD cDNA | 1194 to 1217 |
| re011d | GCAGCCAACTTCCCCTGTG | RHDpromoter | −883 to −901 |
| re014 | GCTCTACCTTGGTCACCTCC | dJ469D22 | 52189 to 52209 |
| re04 | AGGTCACATCCATTTATCCCACTG | dJ469D22 | 53968 to 53945 |
| re11d | AGAAGATGGGGGAATCTTTTTCCT | dJ469D22 | 51193 to 51216 |
| re96 | TTGTGACTGGGCTAGAAAGAAGGTG | dJ469D22 | 242 to 216 |
| rea7 | TGTTGCCTGCATTTGTACGTGAG | RHD cDNA | 1311 to 1333 |
| rend31 | TTCTGTCTGGGTTGGGGAGGG | dJ465N24 | 128649 to 128629 |
| rend32 | GGAGGGGTTAATATGGGTGGC | dJ465N24 | 127355 to 127375 |
| rend8b1 | TTTGTCCTGGTTGCCTGTGGTC | dJ465N24 | 69296 to 69274 |
| rend8b2 | CAAATCCTGTTGACTGGTCTCGG | dJ465N24 | 68451 to 68473 |
| rend9a1 | AACGGCTCCATCACCCCTAAAG | dJ465N24 | 50008 to 49987 |
| rend9a2 | CCCACTCCTAGATACCAACCCAAG | dJ465N24 | 49059 to 49083 |
| rey14a | CTTTATGCACTGCCTCGTTGAATC | dJ469D22 | 56792 to 56769 |
| rey14b | TTGACTGGTGTGGTTGCTGTTG | dJ469D22 | 55863 to 55884 |
| rey15a | GCAGAAAGGGGAGTTGATGCTG | dJ469D22 | 55416 to 55395 |
| rey7 | CTGACAAAGTTGAGAGCCCACTG | dJ469D22 | 62324 to 62346 |
| rey8 | TTAAGCCTACATCCACATGCTGAG | dJ469D22 | 62854 to 62831 |
| rez2 | CCTTGGTCTGCCAGAATTTTCA | RHD cDNA | 2738 to 2717 |
| rez4 | GTTTGGCATCATAGGAGATTTGGC | dJ465N24 | 120101 to 120124 |
| rez7 | CCTGTCCCCATGATTCAGTTACC | dJ465N24 | 124831 to 124854 |
| rh7 | ACGTACAAATGCAGGCAAC | RHDcDNA | 1330 to 1312 |
| rnb31 | CCTTTTTTTGTTTGTTTTTGGCGGTGC | downstream Rhesus box | 1330 to 1312 |
| rr4 | AGCTTACTGGATGACCACCA | RHD cDNA | 1541 to 1522 |
| sf1 | GACTGGGGGGAAAAGCGCAATAC | SMP1 cDNA | 142 to 164 |
| sf1c | GTATTGCGCTTTTCCCCCCAGTC | SMP1cDNA | 164 to 142 |
| sf3 | TGACTTGCTCTCATCCCACATG | SMP1 cDNA | 1696 to 1717 |
| sm19 | GGGCTTGAAGCAAGTAAATGGAAG | SMP1 intron 1 | −58 to −35 |
| sr1 | GCTATCAATATTTTCTTGGTTACAGACAC | SMP1 cDNA | 2172 to 2144 |
| sr3 | GTTCACTGCCATAAGTCTTCAGTGC | SMP1 cDNA | 575 to 551 |
| sr3kp | TGGCCGCACTGAAGACTTATGG | SMP1 cDNA | 546 to 567 |
| sr45 | CAGCTGCATCTATGATAATCCACC | SMP1cDNA | 224 to 243 |
| sr47 | ATGGACAAGTCCGAGGTGATAG | SMP1 cDNA | 315 to 344 |
| sr47c | ATCACCTCGGACTTGTCCATTC | SMP1 cDNA | 342 to 321 |
| sr5 | GCAATCAGAGATCCAAAGGCCAAC | SMP1 cDNA | 428 to 405 |
| sr5c | GTTGGCCTTTGGATCTCTGATTGC | SMP1cDNA | 405 to 428 |
| sr55 | GACATAGTATACCCTGGAATTGCTGT | SMP1 cDNA | 472 to 497 |
| sr55c | ACAGCAATTCCAGGGTATACTATGTC | SMP1 cDNA | 497 to 472 |
| sr9 | CTCCCCCGATTTTAGCCAAGAA | SMP1 cDNA | 27 to 6 |
For the RHD promoter and the RHD cDNA, the positions refer to the distance from A of the start codon. For introns, they refer to the distance from the intron/exon junction. For all other sequences, including SMP1 cDNA, they refer to the distance from the start of the published sequences. The mismatches in primers rey14b, rnb31, and sf3 were inadvertently introduced. Primers re11d, re014, and re04 do not exactly match dJ469D22 because they were designed from our raw sequences covering the 5′ flanking region of RHD.
PCR
If not mentioned otherwise, PCR reactions comprised 60°C annealing, a 10-minute extension at 68°C, and denaturation at 92°C using the expand long-template or the expand high-fidelity PCR systems (Boehringer Mannheim, Mannheim, Germany) and the listed primers (Table 1). We used 3 PCR reactions to bridge gaps in the 3′ flanking regions of the RH genes. PCR 1 was completed using primers rea7 and rend31; PCR 2, rend32 and sf1c; and PCR 3, rea7 and sf3. The structure of the 5′ flanking regions was confirmed with PCR amplifications involving sense primers rend32, rey14a, and rey15a and antisense primers re011d and re014. The intron 9 size was estimated to be about 9000 bp, based on PCR amplifications using rb10b and rr4 for RHD (re96 and rh7 for RHCE).
Nuleotide sequencing
Nucleotide sequencing was performed with a DNA sequencing unit (Prism BigDye terminator cycle-sequencing ready reaction kit and ABI 373A; Applied Biosystems, Weiterstadt, Germany).
Evaluation of the genomic structure of SMP1
The sizes of the SMP1 introns were estimated by PCR amplicons obtained with primers rend32, sr9, sf1c, sf1, sm19, sr45, sr47, sr47c, sr5, sr5c, sr55, sr55c, sr3, sr3kp, and rea7. The positions of the intron/exon junctions and the absence of additional introns were determined by nucleotide sequencing.
Long-range PCR-SSP to specifically detect the RHDdeletion
PCR was performed using the expand long-template PCR system with buffer 3 and primers rez4 (5′ of upstream Rhesus box) and sr9 (SMP1 exon 1). Annealing was at 60°C, with a 20-minute extension at 68°C. PCR amplicons were resolved using a 1% agarose gel.
PCR-RFLP to detect the RHD deletion
PCR was performed using the expand high fidelity PCR system and primers rez7 (nonspecific, 5′ of Rhesus boxidentity region) and rnb31 (specific for downstream Rhesus box, 3′ of downstream Rhesus box identity region). Annealing was at 65°C, and extension was for 10 minutes at 68°C. PCR amplicons were digested with PstI for 3 hours at 37°C, and fragments were resolved using a 1% agarose gel.
Sequencing of the Rhesus boxes
The Rhesus boxes were amplified and sequenced using internal primers in 2 overlapping fragments with PCR primer pairs rez4/rend31 and rend32/re011d (upstream Rhesus box), rea7/rend31 and rend32/sr9 (downstream Rhesus box), and rez4/rend31 and rend32/sr9 (hybrid Rhesus box ofRHD−).
Results
DNA database searches and analysis
The high throughput sequences of the GenBank and the chromosome 1 database of the Sanger Center were screened for nucleotide sequences homologous to RHD or RHCE cDNA. We identified the 84 810-bp genomic clone dJ469D22 (GenBank accession number AL031284), the 129 747-bp genomic clone dJ465N24 (GenBank accession numberAL031432), and the 2234-bp SMP1 cDNA (GenBank accession numberAF081282). The genomic clone dJ469D22 represented a major fragment of the RHCE gene, starting 33 340-bp 5′ of the RHCEstart codon and ending 1142-bp 3′ of exon 9. In dJ465N24, an internal stretch of 1418 bp located between positions 120 158 and 121 568 was 96% homologous to the 3′ end of the RHDcDNA. The 3′ end of the SMP1 cDNA was complementary to the 3′ end of the RHCE cDNA, with an overlap of 58 bp.
The RH gene locus
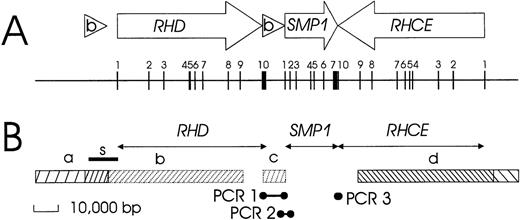
We derived a physical structure of the RH gene locus (Figure1) by reviewing 3′ and 5′ flanking regions and analyzing YAC 38A-A10, as described in the paragraphs that follow.
Schematic structure of the RH gene locus.
(A) The positions and orientations of the genes and the Rhesus boxes are indicated by open arrows and triangles, respectively. The exons are shown as vertical bars, and their exon numbers are indicated. The 2 RH genes have opposite orientation, face each other with their 3′ ends, and are separated by about 30 000 bp. A third gene, SMP1, has the same orientation as RHD and is positioned between RHD and RHCE. The RHDgene is flanked on both sides by the 2 highly homologous Rhesus boxes, which are noted by (b). All exons are shorter than 200 bp, with the exception of the RHD and SMP13′ terminal exons. (B) Data used to establish this structure include the extension of genomic sequences represented in cDNA (horizontal arrows) and identities and homologies to genomic clones as noted: bar a, identity with dJ465N24; bar b, homology of RHD to dJ469D22; bar c, homology of RHD 3′ part to dJ465N24; and bar d, identity with dJ469D22. The positions of 3 bridging PCR reactions are indicated. The correct position of a nucleotide stretch previously reported by Okuda et al9 as a “spacer” sequence between RHD and RHCE is indicated by the bar labeled with the letter “s.”
Schematic structure of the RH gene locus.
(A) The positions and orientations of the genes and the Rhesus boxes are indicated by open arrows and triangles, respectively. The exons are shown as vertical bars, and their exon numbers are indicated. The 2 RH genes have opposite orientation, face each other with their 3′ ends, and are separated by about 30 000 bp. A third gene, SMP1, has the same orientation as RHD and is positioned between RHD and RHCE. The RHDgene is flanked on both sides by the 2 highly homologous Rhesus boxes, which are noted by (b). All exons are shorter than 200 bp, with the exception of the RHD and SMP13′ terminal exons. (B) Data used to establish this structure include the extension of genomic sequences represented in cDNA (horizontal arrows) and identities and homologies to genomic clones as noted: bar a, identity with dJ465N24; bar b, homology of RHD to dJ469D22; bar c, homology of RHD 3′ part to dJ465N24; and bar d, identity with dJ469D22. The positions of 3 bridging PCR reactions are indicated. The correct position of a nucleotide stretch previously reported by Okuda et al9 as a “spacer” sequence between RHD and RHCE is indicated by the bar labeled with the letter “s.”
3′ flanking region.
The 3′ flanking region of RHD was highly homologous to the 3′ part of the genomic clone dJ465N24 (Figure 1B, region c). This homology continued beyond the end of the RHD cDNA and extended for at least 8000 bp, as proven by the fact that it was possible to obtain PCR amplicons (Figure 1B, PCR 1). Sequences homologous to the 3′ part of genomic clone dJ465N24 were neighboring to the 5′ region of the SMP1 gene (Figure 1B, PCR 2). The 3′ end of the SMP1 gene occurred immediately adjacent to the RHCE gene, as indicated by the complementarity of the 3′ ends of the respective cDNAs and confirmed by PCR (Figure 1B, PCR 3). Further details of the RHD 3′ flanking region (Rhesus box) and the SMP1 gene are described in subsequent paragraphs.
5′ flanking region.
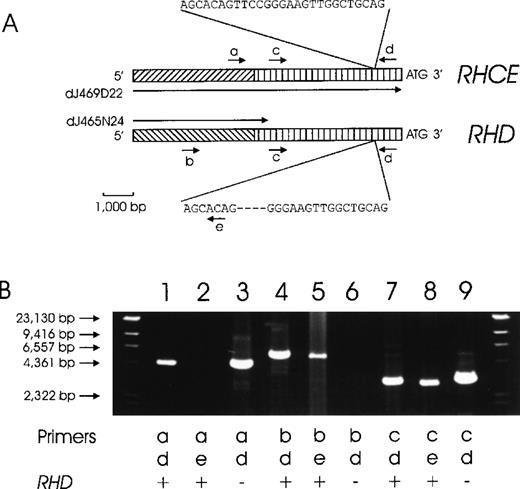
The genomic clone dJ469D22 comprised the 33 340-bp 5′ flanking region of RHCE. For RHD, a 466-bp homology between the 3′ end of clones dJ465N24 and dJ469D22 indicated that clone dJ465N24 might represent the 5′ flanking sequence of RHD. We proved this assumption by PCR (Figure2).
Chromosomal organization of the DNA regions located 5′ to the RHD and RHCE genes.
(A) The proposed structure of the RHCE and RHD 5′ flanking regions is depicted. A total of 4941 bp immediately 5′ of the ATG start codons are homologous between the RHCE andRHD genes (vertically hatched bars). No homology is present beyond this homology region (diagonally hatched bars). We used 2 genomic clones, dJ469D22 and dJ465N24, for primer design. DJ469D22 comprises the full length of the depicted RHCE region, whereas dJ465N24 extends only 466 bp into the homology region. The positions of several PCR primers are indicated: a, rey14a; b, rend32; c, rey15a; d, re014; and e, re011d. (B) This proposed structure is supported by several PCR reactions. Forward priming was done with primer a (RHCE specific, lane 1-3), primer b (RHD specific, lane 4-6), and primer c (RHCE and RHD homology region, lane 7-9). Amplicons were lacking for primer a with RHD-specific reverse primer e (lane 2) and for primer b withRHD− DNA (lane 6). The other 7 PCR reactions yielded amplicons of the predicted sizes in accordance with the genomic structure shown in panel A.
Chromosomal organization of the DNA regions located 5′ to the RHD and RHCE genes.
(A) The proposed structure of the RHCE and RHD 5′ flanking regions is depicted. A total of 4941 bp immediately 5′ of the ATG start codons are homologous between the RHCE andRHD genes (vertically hatched bars). No homology is present beyond this homology region (diagonally hatched bars). We used 2 genomic clones, dJ469D22 and dJ465N24, for primer design. DJ469D22 comprises the full length of the depicted RHCE region, whereas dJ465N24 extends only 466 bp into the homology region. The positions of several PCR primers are indicated: a, rey14a; b, rend32; c, rey15a; d, re014; and e, re011d. (B) This proposed structure is supported by several PCR reactions. Forward priming was done with primer a (RHCE specific, lane 1-3), primer b (RHD specific, lane 4-6), and primer c (RHCE and RHD homology region, lane 7-9). Amplicons were lacking for primer a with RHD-specific reverse primer e (lane 2) and for primer b withRHD− DNA (lane 6). The other 7 PCR reactions yielded amplicons of the predicted sizes in accordance with the genomic structure shown in panel A.
YAC 38A-A10.
This YAC had been known to contain RHCE exons 2-10 andRHD exons 1-107 and was thus expected to contain the DNA segments interspersed between RHD and RHCE. We checked for the presence of DNA segments representative of different parts of the RH locus (Table 2), and the results were concordant with the proposed structure of theRH locus shown in Figure 1A.
The presence of RHD flanking sequences in YAC 38A-A10
| Primer . | Predicted position . | Amplicon size, bp . | Amplicons obtained? . | |||
|---|---|---|---|---|---|---|
| Genomic DNA . | YAC 38A-A10 . | |||||
| Sense . | Antisense . | RHD+ . | RHD− . | |||
| rend9a1 | rend9a2 | RHD 5′ flanking region about 85 000 bp from ATG | 948 | yes | yes | yes |
| rend8b1 | rend8b2 | RHD 5′ flanking region about 50 000 bp from ATG | 845 | yes | yes | yes |
| rea7 | rez2 | RHD3′ flanking region about 1500 bp from STOP | 1412 | yes | no | yes |
| rend32 | sr9 | RHCE 3′ flanking region about 20 000 bp from STOP | 1989 | yes | yes | yes |
| sr1 | sf3 | RHCE 3′ flanking region about 1000 bp from STOP | 477 | yes | yes | yes |
| rey14b | rey14a | RHCE 5′ flanking region about 5300 bp from ATG | 929 | yes | yes | no |
| rey7 | rey8 | RHCE 5′ flanking region about 10 000 bp from ATG | 530 | yes | yes | no |
| Primer . | Predicted position . | Amplicon size, bp . | Amplicons obtained? . | |||
|---|---|---|---|---|---|---|
| Genomic DNA . | YAC 38A-A10 . | |||||
| Sense . | Antisense . | RHD+ . | RHD− . | |||
| rend9a1 | rend9a2 | RHD 5′ flanking region about 85 000 bp from ATG | 948 | yes | yes | yes |
| rend8b1 | rend8b2 | RHD 5′ flanking region about 50 000 bp from ATG | 845 | yes | yes | yes |
| rea7 | rez2 | RHD3′ flanking region about 1500 bp from STOP | 1412 | yes | no | yes |
| rend32 | sr9 | RHCE 3′ flanking region about 20 000 bp from STOP | 1989 | yes | yes | yes |
| sr1 | sf3 | RHCE 3′ flanking region about 1000 bp from STOP | 477 | yes | yes | yes |
| rey14b | rey14a | RHCE 5′ flanking region about 5300 bp from ATG | 929 | yes | yes | no |
| rey7 | rey8 | RHCE 5′ flanking region about 10 000 bp from ATG | 530 | yes | yes | no |
SMP1 gene
The genomic structure of the SMP1 gene was evaluated by PCR using internal primers and nucleotide sequencing (Figure3). We identified 6 introns. Exon 1 contained 5′ untranslated sequences only and was separated from the Rhesus box by 15 bp. The long 3′ untranslated sequence of exon 7 overlapped with RHCE exon 10. The total gene size was estimated to be 20 000 bp, therefore resulting, in conjunction with the downstream Rhesus box, in approximately a 30 000-bp distance between RHD and RHCE (Figure 1).
Chromosomal organization of the SMP1 gene.
The SMP1 gene has 7 exons, and the positions and approximate sizes of the introns are shown. The start of the published cDNA (GenBank accession number AF081282) is separated by 15 nt from the downstream Rhesus box. Exon 1 contains only a 5′ untranslated sequence, and the SMP1 start codon is located in exon 2. Exon 7 contains 16 codons and 1656-bp 3′ untranslated sequences and is contiguous with the 3′ untranslated sequence ofRHCE exon 10.
Chromosomal organization of the SMP1 gene.
The SMP1 gene has 7 exons, and the positions and approximate sizes of the introns are shown. The start of the published cDNA (GenBank accession number AF081282) is separated by 15 nt from the downstream Rhesus box. Exon 1 contains only a 5′ untranslated sequence, and the SMP1 start codon is located in exon 2. Exon 7 contains 16 codons and 1656-bp 3′ untranslated sequences and is contiguous with the 3′ untranslated sequence ofRHCE exon 10.
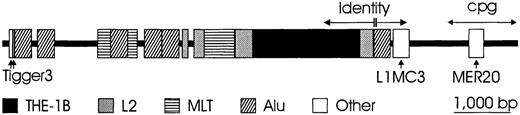
Rhesus boxes
Two DNA segments of approximately 9000 bp, located 5′ and 3′ of the RHD gene, were designated “Rhesus boxes.” They were highly homologous and had identical orientation (Figure 4). The upstreamRhesus box (5′ of RHD) was approximately 9142-bp long and ended approximately 4900-bp 5′ of the RHD start codon. The downstream Rhesus box (3′ of RHD) was 9145-bp long and originated 104 bp after the RHD stop codon. The Rhesus boxes exactly embraced the part ofRHD with homology to RHCE. The central portion of both Rhesus boxes contained a nearly complete remnant of a transposon-like human element (THE-1B). However, the single open reading frame usually found in the THE-1B element was abolished due to several nucleotide aberrations occurring in bothRhesus boxes in parallel, including a nonsense mutation in codon 4. While there was an overall 98.6% homology between bothRhesus boxes, a 1463-bp “identity region” located between positions 5701 and 7163 bp was completely identical, with the single exception of a 4-bp T insertion in a poly T tract.
Chromosomal organization of the Rhesus boxes.
The physical extension of the upstream Rhesus box (5′ toRHD) is 9145 bp (darkened bar). About 63% of the boxes' nucleotide sequence consists of repetitive DNA; the types of the repeat families are indicated. The overall homology between the upstream and downstream Rhesus box is 98.6%, but within a 1463-bp identity region (horizontal arrows), there is only a single 4-bp insertion (double vertical line). A CpG island (double-headed arrow) is located at the 3′ end and is in the downstream Rhesus box(3′ to RHD) adjacent to the SMP1 promoter.
Chromosomal organization of the Rhesus boxes.
The physical extension of the upstream Rhesus box (5′ toRHD) is 9145 bp (darkened bar). About 63% of the boxes' nucleotide sequence consists of repetitive DNA; the types of the repeat families are indicated. The overall homology between the upstream and downstream Rhesus box is 98.6%, but within a 1463-bp identity region (horizontal arrows), there is only a single 4-bp insertion (double vertical line). A CpG island (double-headed arrow) is located at the 3′ end and is in the downstream Rhesus box(3′ to RHD) adjacent to the SMP1 promoter.
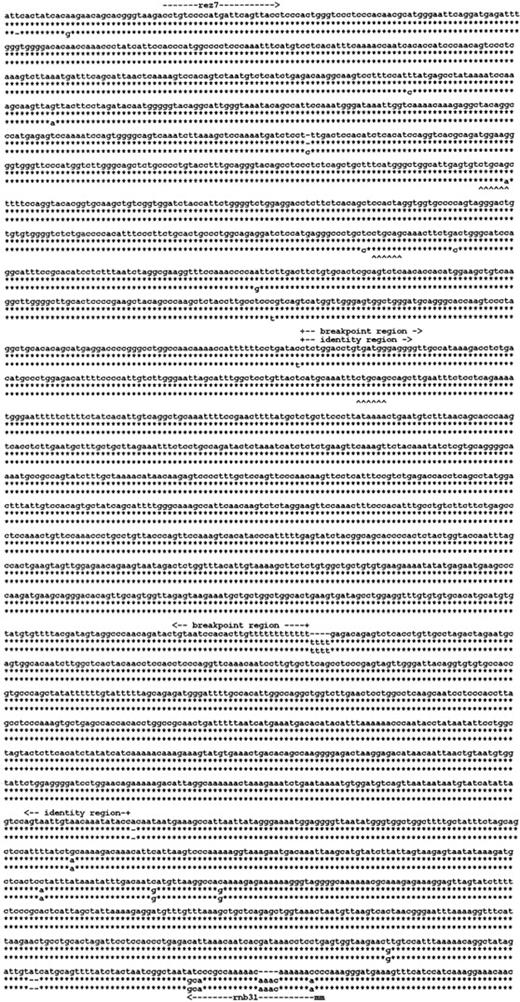
Localization of the RHD gene deletion in RHD−haplotypes
We reasoned that the homology of the 2 Rhesus boxesmay have been instrumental in the RHD deletion mechanism in the common RHD− haplotypes. We determined the nucleotide sequence of the Rhesus box inRHD− DNA (Figure5). The single Rhesus box detected in the RHD− haplotypes had a hybrid structure. The 5′ end of this Rhesus box represented an upstream Rhesus box, and the 3′ end represented a downstream Rhesus box. We determined that the 903-bp breakpoint region of the RHD deletion was located in the identity region of the Rhesus boxes (Figure 4, arrow pointing to left).
RHD gene deletion in the Rh-negative haplotypes.
Three of the 3100-bp segments of the Rhesus boxes are shown. The upper line indicates the nucleotide sequence of the upstreamRhesus box in D-positive haplotypes; the lower line indicates the nucleotide sequence of the downstream Rhesus box in D-positive haplotypes. The middle line gives the nucleotide sequence of the single Rhesus box carried by Rh-negative haplotypes. The asterisk symbols denote identical nucleotides. TheRHD deletion occurred in a 903-bp segment of absolute identity that was part of a 1463-bp identity region. The positions of primers rez7 and rnb31 are shown (m indicates mismatch).PstI restriction sites are indicated by upward carets. The 3 Rhesus boxes were deposited at EMBL under accession numbers AJ252311 (upstream Rhesus box), AJ252312(downstream Rhesus box), and AJ252313 (hybrid Rhesus box).
RHD gene deletion in the Rh-negative haplotypes.
Three of the 3100-bp segments of the Rhesus boxes are shown. The upper line indicates the nucleotide sequence of the upstreamRhesus box in D-positive haplotypes; the lower line indicates the nucleotide sequence of the downstream Rhesus box in D-positive haplotypes. The middle line gives the nucleotide sequence of the single Rhesus box carried by Rh-negative haplotypes. The asterisk symbols denote identical nucleotides. TheRHD deletion occurred in a 903-bp segment of absolute identity that was part of a 1463-bp identity region. The positions of primers rez7 and rnb31 are shown (m indicates mismatch).PstI restriction sites are indicated by upward carets. The 3 Rhesus boxes were deposited at EMBL under accession numbers AJ252311 (upstream Rhesus box), AJ252312(downstream Rhesus box), and AJ252313 (hybrid Rhesus box).
Specific detection of the RHD deletion by PCR
We developed 2 PCR-based methods for specific detection of theRHD gene deletion occurring in the prevalentRHD− haplotypes (Figure6). These techniques allowed the ready and direct detection of the common RHD−haplotypes, even if they were in trans toRHD+ haplotypes. We applied PCR-RFLP to a larger number of samples (Table 3). As expected, all 33 samples with known genotype were correctly typed. In 68 additional samples representative of the most common phenotypes, our results were consistent with the known haplotype frequencies in the population.
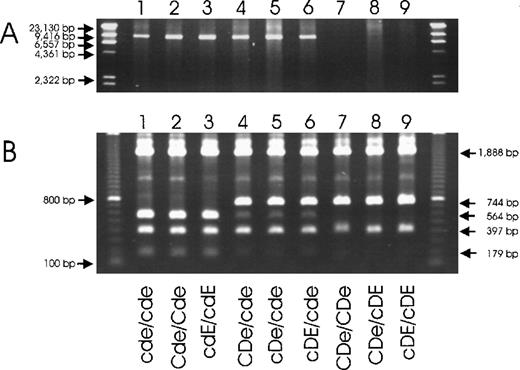
Two technical procedures for specific detection of theRHD deletion in the common RHD−haplotypes.
(A) A long-range PCR amplification with primers located in non-Rhesus box sequences and (B) PCR-RFLP with primers located in the Rhesus boxes are shown. The deduced genotypes are indicated. The primers of the long-range PCR were located 5′ of the upstream Rhesus box (primer rez4) and in SMP1exon 1 (primer sr9). RHD− haplotypes were detected specifically (panel A, lanes 1-6). DNA homozygous for theRHD gene was negative because PCR cannot amplify the 70 000-bp DNA stretch of the RHD gene. For the PCR-RFLP method, the PCR amplicons (primer rez7 and rnb31) were digested withPstI. In D-negative haplotypes, there are 3PstI sites in the amplicon (Figure 5) resulting in fragments of 1888 bp, 564 bp, 397 bp, and 179 bp (lanes 1-3). The downstream Rhesus box of D-positive haplotypes lacks 1PstI site, resulting in fragments of 1888 bp, 744 bp, and 397 bp (lanes 7-9).RHD+/RHD− heterozygotes show both fragments of 744 bp and 564 bp (lanes 4-6). The 564-bp fragment appears weaker because heterodimers are not cut by PstI. Primer rnb31 does not amplify the upstream Rhesus box of D-positive haplotypes.
Two technical procedures for specific detection of theRHD deletion in the common RHD−haplotypes.
(A) A long-range PCR amplification with primers located in non-Rhesus box sequences and (B) PCR-RFLP with primers located in the Rhesus boxes are shown. The deduced genotypes are indicated. The primers of the long-range PCR were located 5′ of the upstream Rhesus box (primer rez4) and in SMP1exon 1 (primer sr9). RHD− haplotypes were detected specifically (panel A, lanes 1-6). DNA homozygous for theRHD gene was negative because PCR cannot amplify the 70 000-bp DNA stretch of the RHD gene. For the PCR-RFLP method, the PCR amplicons (primer rez7 and rnb31) were digested withPstI. In D-negative haplotypes, there are 3PstI sites in the amplicon (Figure 5) resulting in fragments of 1888 bp, 564 bp, 397 bp, and 179 bp (lanes 1-3). The downstream Rhesus box of D-positive haplotypes lacks 1PstI site, resulting in fragments of 1888 bp, 744 bp, and 397 bp (lanes 7-9).RHD+/RHD− heterozygotes show both fragments of 744 bp and 564 bp (lanes 4-6). The 564-bp fragment appears weaker because heterodimers are not cut by PstI. Primer rnb31 does not amplify the upstream Rhesus box of D-positive haplotypes.
PCR-RFLP for the specific detection of RHDdeletion
| Phenotype . | Known genotype . | Samples tested, n . | Number of samples with RHD genotype, n . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Determined . | Expected . | ||||||||
| +/+ . | +/− . | −/− . | +/+ . | +/− . | −/− . | P . | |||
| Known genotype | |||||||||
| ccddee | cde/cde | 14 | 0 | 0 | 14 | 0 | 0 | 14 | NA |
| 3-150 | 5 | 0 | 0 | 5 | 0 | 0 | 5 | NA | |
| 3-150 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | NA | |
| D variants | 3-151 | 9 | 0 | 9 | 0 | 0 | 9 | 0 | NA |
| 3-152 | 4 | 4 | 0 | 0 | 4 | 0 | 0 | NA | |
| Common phenotypes | |||||||||
| 10 | 1 | 9 | 0 | 0.5 | 9.5 | 0 | >.4 | ||
| 10 | 0 | 10 | 0 | 0.3 | 9.7 | 0 | >.5 | ||
| 10 | 1 | 9 | 0 | 0.5 | 9.5 | 0 | >.4 | ||
| 10 | 9 | 1 | 0 | 9.5 | 0.5 | 0 | >.4 | ||
| 12 | 11 | 1 | 0 | 11 | 1 | 0 | >.5 | ||
| 10 | 10 | 0 | 0 | 9.2 | 0.8 | 0 | >.4 | ||
| 6 | 5 | 1 | 0 | 5.8 | 0.2 | 0 | >.1 | ||
| Phenotype . | Known genotype . | Samples tested, n . | Number of samples with RHD genotype, n . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Determined . | Expected . | ||||||||
| +/+ . | +/− . | −/− . | +/+ . | +/− . | −/− . | P . | |||
| Known genotype | |||||||||
| ccddee | cde/cde | 14 | 0 | 0 | 14 | 0 | 0 | 14 | NA |
| 3-150 | 5 | 0 | 0 | 5 | 0 | 0 | 5 | NA | |
| 3-150 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | NA | |
| D variants | 3-151 | 9 | 0 | 9 | 0 | 0 | 9 | 0 | NA |
| 3-152 | 4 | 4 | 0 | 0 | 4 | 0 | 0 | NA | |
| Common phenotypes | |||||||||
| 10 | 1 | 9 | 0 | 0.5 | 9.5 | 0 | >.4 | ||
| 10 | 0 | 10 | 0 | 0.3 | 9.7 | 0 | >.5 | ||
| 10 | 1 | 9 | 0 | 0.5 | 9.5 | 0 | >.4 | ||
| 10 | 9 | 1 | 0 | 9.5 | 0.5 | 0 | >.4 | ||
| 12 | 11 | 1 | 0 | 11 | 1 | 0 | >.5 | ||
| 10 | 10 | 0 | 0 | 9.2 | 0.8 | 0 | >.4 | ||
| 6 | 5 | 1 | 0 | 5.8 | 0.2 | 0 | >.1 | ||
The expected number ofRHD+/RHD+ andRHD+/RHD− samples are based on known genotypes or the haplotype frequencies in the local population.33 NA indicates not applicable. Probabilities were calculated based on confidence limits of binomial distribution.
Indicates RHD− in PCR.
Indicates RHD+/RHD−because a weak or partial D phenotype would be masked in aRHD+/RHD+ genotype. These samples were weak D type 1 (n = 2), type 2 (n = 2), type 3 (n = 2), type 4 (n = 2), and DVII(n = 1).
Indicates the presence of two RHD genes differing in their polymorphic HaeIII site in intron 3,34 as demonstrated by PCR-RFLP.
Discussion
The 2 genes, RHD and RHCE, had opposite orientation and faced each other with their 3′ ends. The RHD gene was surrounded by 2 highly homologous Rhesus boxes. The physical distance between RHD and RHCE was 30 000 bp and was filled with a Rhesus box and the SMP1 gene. The breakpoints of the RHD deletion in the prevalentRHD− haplotypes were located in the 1463-bp identity region of the Rhesus boxes. We established technical procedures for specifically detecting theRHD gene deletion in the common RHD−haplotypes.
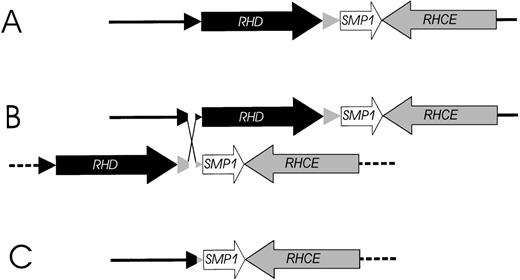
Based on the structure of the RH gene locus (Figure 1), we propose a parsimonious model for the RHD gene deletion event (Figure 7). The RHD deletion may be explained by unequal crossing-over triggered by the highly homologousRhesus boxes embracing the RHD gene. The 903-bp breakpoint region in the Rhesus boxes was located in a 1463-bp stretch of 99.9% homology resembling a THE-1B and an L2 repetitive DNA element (Figure 4). Interestingly, the DNA segment with more than 60 000 bp, which was deleted in theRHD− haplotype, consisted only of and contained all sequences that were duplicated in the RHD+ haplotype.
Model of the proposed mechanism causing the prevalentRHD− haplotypes in whites.
(A) The physical structure of the RHD and RHCE gene locus. (B) An unequal crossing-over between the upstream and downstreamRhesus boxes can be triggered by their high homology. The breakpoint region in the Rhesus boxes was found to be of 100% homology for 903 bp (Figure 5). (C) Resolving the crossed-over chromosome yields the RH gene structure of the extant RHD−haplotype.
Model of the proposed mechanism causing the prevalentRHD− haplotypes in whites.
(A) The physical structure of the RHD and RHCE gene locus. (B) An unequal crossing-over between the upstream and downstreamRhesus boxes can be triggered by their high homology. The breakpoint region in the Rhesus boxes was found to be of 100% homology for 903 bp (Figure 5). (C) Resolving the crossed-over chromosome yields the RH gene structure of the extant RHD−haplotype.
Previously, the discrimination of RHD homozygote individuals from RHD heterozygote individuals was difficult because the prevalent RHD− allele could not be detected specifically.8,12 Our results provide the basis for detecting the prevalent RHD− haplotypes, and hence, true RHD genotyping is now feasible.8 We describe a PCR-RFLP method and a long-range PCR method using eitherRhesus box sequences or Rhesus box flanking sequences. By using the same DNA stretches or combinations thereof, other methods, such as PCR-SSO or biochips, can be developed.
RHD genotyping by detection of the prevalentRHD− haplotype may not detect someRHD+ D-negative alleles. In whites, such alleles are exceedingly rare,8 but they may occur due to nonsense mutations (eg, RHD(Q41X)),20deletions (eg, RHD(488del4)21), orRHD-CE-D hybrid genes (eg, RHD-CE(2-9)-D22and RHD-CE(4-7)-D).23 In contrast, in Africans, there are 3 prevalent D-negative alleles: (1) theRHD deletion; (2) an RHD pseudogene designatedRHDΨ, which can be specifically detected by a 37-bp insertion in exon 4;24 and (3) an RHD-CE-D hybrid gene25,26 associated with the CdeS haplotype. Based on the data given by Singleton et al,24 these alleles represent 43%, 43%, and 15% of the D-negative alleles in the black population, respectively. Hence, all 3 alleles must be specifically detected for a reliable RHD genotyping in Africans and any other population harboring these alleles. A similar situation is present in the Japanese. In this population, theRHD deletion occurs with a frequency of 94% among D-negative alleles, and additional detection of the underlying cause of the RHD+ D-negative alleles associated with RHD(G314V)27 may be warranted.
The opposite orientation of the 2 RH genes explained the different character of hybrid genes in the MNS and Rh blood group: The glycophorin genes encoding the MNSs antigens occur in the same orientation,28 and many recombinations may be explained as an unequal crossing-over resulting in single hybrid genes.29 In the RH locus, the inversely oriented sequences are unlikely to trigger unequal crossing-over, and if this event occurred, no functional hybrid gene would result. Our conclusion that unequal crossing-over at the RH gene locus was unlikely may explain that most RH hybrid genes are of either theRHD-CE-D or RHCE-D-CE type and involve stretches of homologous DNA positioned in cis, as noted by us previously.30 Currently, the RH gene system is the only well investigated gene locus where the 2 genes have opposite orientation, rendering it a model system for the evolution of neighboring, oppositely oriented genes that are frequent throughout genomes.
Surprisingly, our data show that 3 genes are located at theRH locus: RHD, RHCE, and SMP1. The nucleotide sequence of the latter gene has been deposited in the GenBank as a putative member of an 18-kd small membrane protein family, and its function is as yet unknown. The gene shows homology to an open reading frame on chromosome 21.31 Its position between bothRH genes implies that any polymorphism of theSMP1 gene would be tightly linked to a specific RHhaplotype. It might be anticipated that functionally relevant mutations of the SMP1 gene may cause selection pressure for or against specific RH haplotypes. Such factors might explain some previously unresolved issues of RHhaplotype distribution, such as the high frequency ofRH− in the European population. Screening for polymorphisms in SMP1 appears to be necessary to further understand the RH locus.
While the molecular mechanism resulting in the prevalentRHD− haplotype is apparent, it is less clear how the much older duplication event gave rise to the structure of theRH genes in RHD+ individuals. The duplication of the Rhesus box and the RH genes probably occurred as a single event, because the overall homology of the 2Rhesus boxes is very similar to that of the RHgenes. It is tempting to speculate that the RHD duplication originated in a causal connection with the insertion of the near full-length transposon-like THE-1B in duplicate. However, the open reading frame of the THE-1B probably was nonfunctional at the time of the duplication. Further characterization of related RH gene loci, including those of the monkey species, will be helpful to resolve the duplication mechanism. Although the structure of the RHlocus in RHD− haplotypes is probably very similar to the structure predating the RH gene duplication, the “original” RH gene at the current RH chromosomal position is not extant in the human species. The number of RHgenes is variable among primates,32 and unequal crossing-over may explain both the loss of RHD and the possible generation of more than 2 RH genes in some species.
Acknowledgments
We thank Marianne Lotsch, Anita Hacker, Sabine Kaiser, Katharina Schmid, and Sabine Zahn for expert technical assistance and Bernd Widder for supplying 4 cDE/cDe samples identified in his thesis work.
Supported by Project 531 and Project 442 from the Universitätsklinikum Ulm, Institut Ulm, Ulm, Germany, and by the DRK-Blutspendedienst Baden-Württemberg, Stuttgart, Germany.
The nucleic acid sequence data were deposited in the European Molecular Biology Laboratory, Heidelberg, Germany; GenBank, National Center for Biotechnology Information, Bethesda, MD; and the DNA Data Bank of Japan, National Institute of Genetics, Mishima, Japan, under accession numbers AJ252311, AJ252312, AJ252313, and AJ252314.
Reprints:Willy A. Flegel, Abteilung Transfusionsmedizin, Universitätsklinikum Ulm, and DRK-Blutspendedienst Baden-Württemberg, Institut Ulm, Helmholtzstrasse 10, D-89081 Ulm, Germany; e-mail:waf@ucsd.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal