FVII is a vitamin-K-dependent plasma glycoprotein. It is synthesized in the liver and circulates in the blood as an inactive zymogen.1 Upon vascular injury and the presence of tissue factor (TF), FVII is complexed to TF and is cleaved to its active form, FVIIa. The VIIa/TF complex then cleaves and activates both factors X and IX to initiate the coagulation process. Deficiency of factor VII results in a defect in the initiation of coagulation by the extrinsic pathway. Hereditary factor VII deficiency is a rare autosomal recessive bleeding disorder with variable clinical expressions.2 It is estimated to occur in 1 out of 500 000 persons. The severity of bleeding is variable. The patients may have symptoms such as easy bruising, epistaxis, gingival hemorrhage, increased menstrual blood loss, and cerebral hemorrhage. The clinical symptoms of patients can be improved by transfusion of fresh plasma or blood but not by the administration of vitamin K or hemostat. The FVII gene is located on 13q34-qter and contains 9 exons and 8 introns.3,4Characterization of FVII gene mutations will provide insight into the understanding of function/structure correlation of FVII and the heterogeneity of the deficiency. More than 50 mutations responsible for FVII deficiency have been identified so far.5 Those include missense mutation, base deletion, splicing site mutation, and promotor mutation. In this short report, we describe a novel missense mutation from a Chinese patient of FVII deficiency with mild symptoms.
The female patient, a 53-year-old, was born of parents with a known consanguinity in Zhousan, Zhenjian, China. She has been easy to bruise and has had gingival hemorrhaging since her childhood, with no liver disease but a history of increased menstrual blood loss. The administration of hemostat and vitamin K fails to improve the clinical symptoms. After obtaining consent, blood specimens were drawn from the patient and her family members. The patient has a normal platelet count (149 × 109/L) and coagulation screen results (ACL3000 plus, Instrumentation Laboratory, Lexington, MA): prothrombin time (PT) 48 seconds (normal range 11 to 15); activated prothrombin time (APTT) 46 seconds (normal range 35 to 47); thrombin time (TT) 15 seconds (normal range 10 to 16); and factor VII coagulant activity (FVII: C) is 3% of the normal level. By use of enzyme-linked immunoabsorbent assay (Stago, Paris, France), approximately 55% of the normal pooled plasma level of FVII was measured in the plasma of the patient.
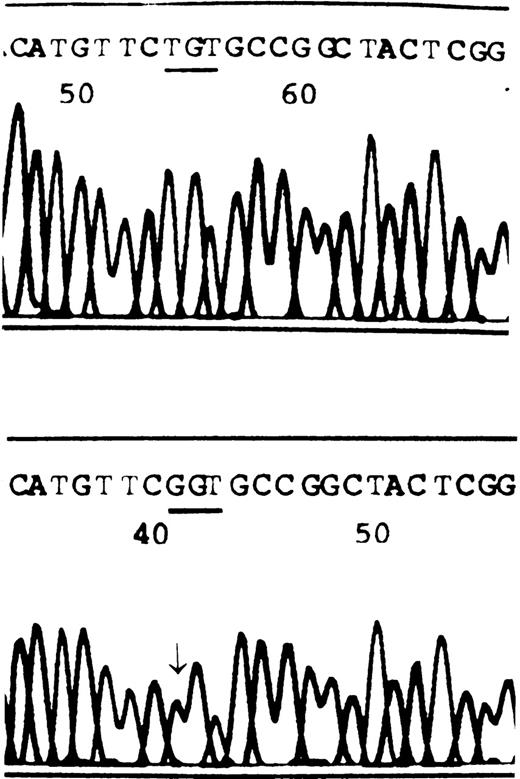
For gene analysis, DNA was prepared by using a Qiamp Blood Kit (Qiagen, Hilden, Germany). Primers6 for polymerase chain reaction (PCR) amplification of FVII genomic DNA fragments were provided by F. Bernardi. Target sequences were amplified in 25 μL of 10 × PCR buffer 2.5 μL and 1 U Taq polymerase (both were purchased from Sangon, Toronto, Canada), dNTP 200 μmol/L, each primer 0.5 μmol/L, and approximately 200 ng genomic DNA. The samples were subjected to predenaturation at 94°C for 2 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, and primer extension at 72°C for 45 seconds. After the final cycle, an additional keep at 72°C for 7 minutes was performed. Amplified PCR fragments were purified from agarose gels using a gel extraction kit (Viogene, Sunnyvale, CA). Direct sequencing of PCR-amplified products was performed using Dye Terminates Ready Reaction Kit (Perkin-Elmer, Foster City, CA), as recommended by the manufacturer's instructions. The result shows that the sequence of the normal subject was identical to the FVIIgene coding sequence published, while a T → G nucleotide change was revealed at site 10913 for FVII from the patient (Figure1). This base substitution caused a TGT → GGT missense mutation at codon 329 of exon 8, leading to a cysteine residue substituted by glycine in FVII protein.
The partial sequences of FVII from a healthy subject and the patient.
Healthy subject, A; patient, B. The arrow indicates a homozygous T → G transition at the first position of codon 329. This missense mutation results in replacement of cysteine by glycine.
The partial sequences of FVII from a healthy subject and the patient.
Healthy subject, A; patient, B. The arrow indicates a homozygous T → G transition at the first position of codon 329. This missense mutation results in replacement of cysteine by glycine.
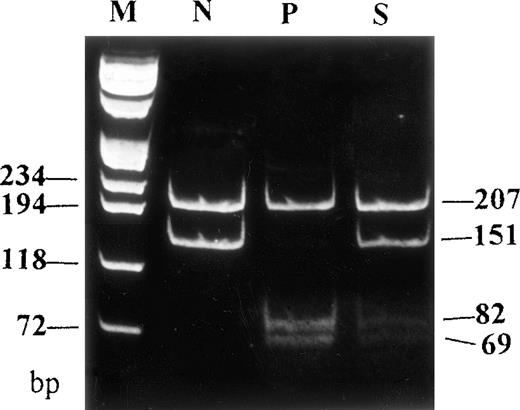
In order to confirm this mutation obtained by sequencing and screen the patient's family members and healthy individuals for the same nucleotide change, PCR products, amplified by using sense primer (location from 10813 to 10849) and antisense primer (location from 11185 to 11203),6 were digested with endonuclease HgiC I, under the manufacturer's (MBI, Fermentas, Italy) suggested condition. Digested DNA was electrophoresed on a 12% polyacriminate gel. Since the nucleotide change generates another HgiC I recognition site in the 358 bp PCR product, 82 and 69 bp fragments would be observed in a mutant digestion pattern instead of 151 bp in the normal pattern. The digestion result shows 207, 82, and 69 bp in the patient, while 207 and 151 bp in normal controls (n = 12), and a heterozygous digestion pattern (207, 151, 82, and 69 bp) in the patient's son (Figure 2). A heterozygous digestion pattern for the mutation in the patient's 2 daughters, and healthy digestion patterns in the patient's husband and her 3 grandchildren were also observed (data not shown).
Genomic DNA fragments of FVII amplified by PCR were digested with HgiC I endonuclease and electrophoresed on a 12% polyacriminate gel.
Another HgiC I endonuclease restriction site was generated by a T → G mutation at codon 329 of the FVII gene, leading to the occurrence of 3 fragments (207, 82, and 69 bp) in the patient, instead of 2 fragments (207 and 151 bp) in the healthy subject. A heterozygous restriction pattern was shown in the patient's son. M indicates DNA markers (πx174/Hae III); N, normal subject; P, patient; S, patient's son.
Genomic DNA fragments of FVII amplified by PCR were digested with HgiC I endonuclease and electrophoresed on a 12% polyacriminate gel.
Another HgiC I endonuclease restriction site was generated by a T → G mutation at codon 329 of the FVII gene, leading to the occurrence of 3 fragments (207, 82, and 69 bp) in the patient, instead of 2 fragments (207 and 151 bp) in the healthy subject. A heterozygous restriction pattern was shown in the patient's son. M indicates DNA markers (πx174/Hae III); N, normal subject; P, patient; S, patient's son.
Previously, Bernardi et al6 found a missense mutation C310F in 2 unrelated pedigrees. It was proposed that such a mutation would destroy the formation of a disulphide bond between Cys310 and Cys329, which is highly conserved in serine proteases and is critical to maintain the structure and function of FVII. Although C329Q mutant factor VII was not expressed and characterized in vitro, we predict it to be the causative mutation for the phenotype of the patient. More than one-half the mutations found so far in FVIIwere located in exon 8.5 The mutant allele identified herein provided more evidence of exon 8 being the hot region of mutation and further demonstrates the heterogeneity of the FVII deficiency.
Acknowledgments
Our thanks to Dr Francesco Bernardi (Istituto Chimica Biologica, Universita' di ferrara, Ferrara, Italy) for providing the primers for amplification of FVII DNA fragments and helpful discussion. We are grateful to the patient and her family members for their generous collaboration.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal