Abstract
X-linked chronic granulomatous disease (CGD) derives from defects in the CYBB gene, which encodes the gp91-phox component of NADPH oxidase. We studied the molecular basis of the disease in a kindred with variant CGD, due to a single base substitution at the sixth position of CYBB first intron. The patients' phagocytes have been shown previously to greatly increase superoxide release in response to interferon-gamma (IFN-γ) in vitro and in vivo. We examined CYBB gene expression in an Epstein-Barr virus (EBV)-transformed B-cell line from 1 patient in this kindred. These cells showed markedly decreased levels of CYBB transcripts in total RNA (5% of normal) and nuclear RNA (1.4% of normal), despite equal CYBB transcription rates in the CGD and control cells. Incubation with IFN-γ produced a 3-fold increase in CYBBtotal messenger RNA (mRNA) levels in the patient's cells, and decreased nuclear transcripts to undetectable levels. Reverse transcriptase–polymerase chain reaction analysis of RNA splicing revealed a preponderance of unspliced CYBB transcripts in the patient's nuclear RNA. In vitro incubation with IFN-γ increased by 40% the ratio of spliced relative to unspliced CYBB mRNA in nuclei from the CGD B-cell line. Total RNA harvested from the same patient's monocytes, on and off therapy with IFN-γ, showed a similar improvement in splicing. We conclude that IFN-γ partially corrects a nuclear processing defect due to the intronic mutation in theCYBB gene in this kindred, most likely by augmentation of nuclear export of normal transcripts, and improvement in the fidelity of splicing at the first intron.
Chronic granulomatous disease (CGD) is a hereditary disorder of host defense due to defective activity of a phagocyte NADPH oxidase that generates superoxide and related toxic oxygen metabolites necessary for microbial killing.1,2 Patients usually present early in life with multiple, sometimes fatal, pyogenic infections.3
The NADPH oxidase enzyme system responsible for superoxide generation forms a small transmembrane electron transport system that results in the oxidation of NADPH on the cytoplasmic surface and the generation of superoxide on the outer surface of the membrane, which becomes the inner surface of the phagosome on invagination during phagocytosis.2,4 The terminal electron donor to oxygen is a unique, low midpoint potential flavocytochrome, termed cytochromeb558 for its absorption peak at 558 nm.5 The heterodimeric molecule combines a 91-kd glycoprotein, termed gp91-phox, for phagocyte oxidase, and a 22-kd nonglycosylated polypeptide (termed p22-phox).6,7 The CYBB gene that encodes gp91-phox was one of the first to be identified by positional cloning6 after chromosomal localization to Xp21.1; it encompasses 13 exons spanning approximately 30 kilobases (kb) of genomic DNA.8 CGD kindreds with defects in the gp91-phox component thus show X-linked inheritance and, in most cases, the cytochrome b558 is reduced or absent in their phagocytes. The rarer autosomal recessive forms9 of CGD derive from defects in genes encoding p22-phox and 2 cytosolic components of the oxidase complex: p47-phox and p67-phox.2 4
Diverse molecular defects producing X-linked CGD have been identified within the coding region and intron boundaries of the CYBBgene; such mutations include large multigene deletions, smaller deletions and insertions, missense and nonsense substitutions, and splicing defects.9,10 Splice site mutations occur at or near the splice junction and, in most cases, result in a typical CGD phenotype with no NADPH oxidase expression.9-11 However, they can also lead to the less common and clinically less severe “variant” form of CGD, usually characterized by a uniform population of neutrophils that exhibit a low level of oxidase activity, roughly proportional to the level of cytochromeb558 expressed.9,10,12 13
We investigated the molecular basis of the variant X-linked CGD phenotype and response to interferon gamma (IFN-γ) in a kindred with a genomic DNA mutation recently identified as a single base substitution at the sixth position of CYBB gene first intron: ATT/gtaagt→ ATT/gtaagc.9,14 This kindred has previously been shown to have phagocytes that are unusually responsive to IFN-γ, which nearly corrects the oxidase defect in vitro and in vivo.15 16 The current studies indicate that the dramatic effect of IFN-γ treatment in this kindred is largely due to posttranscriptional molecular mechanisms, including an increase of nuclear export of CYBB transcripts and improvement in the fidelity of splicing at the first intron.
Materials and methods
Subjects
Peripheral blood samples for cell culture and monocyte preparation were obtained from the patient and from healthy volunteers. The University of Massachusetts Medical Center Committees on Protection of Human Subjects in Research approved all procedures and consent forms.
Cell preparation
Epstein-Barr virus (EBV)-transformed B lymphocytes were developed from peripheral blood mononuclear cells of healthy donors and from the patient with variant X-linked CGD. To initiate B-lymphocyte cultures, peripheral blood was fractionated by Ficoll-Hypaque centrifugation17 and the mononuclear cells were cultured with supernatants from B95-8, an EBV-producer cell line.18,19 After EBV-transformation, B lymphocytes from healthy donors or from the patient with CGD were cultured in RPMI 1640 medium supplemented with 10% heat inactivated fetal bovine serum, 2 mmol/L L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin, at 37°C in a humid atmosphere with 5% CO2. To examine the effects of cytokines, EBV-transformed B lymphocytes were cultured for 7 days with human recombinant IFN-γ (100 U/mL), tumor necrosis factor-alpha (TNF-αb 1000 U/mL), alone or in combination. These cytokines were chosen because they have well-characterized receptors and transduction mechanisms in B lymphocytes,20,21 and are strong stimulators of the NADPH oxidase system in phagocytic cell lines.22-24 In addition, the phagocytes of this CGD kindred show a dramatic response to IFN-γ both in vitro and in vivo.15 16 Cell counts and viability, monitored on a daily basis, were always above 90%.
Structure of RNA transcripts
The kindred's entire CYBB gene coding region and proximal 5′ flanking region has previously been shown to have a normal structure by the analysis of single strand conformation polymorphisms9 and by complementary DNA (cDNA) sequencing (R. A. B. Ezekowitz and P. E. Newburger, unpublished data). We restudied the patient's transcripts in the regions neighboring the affected intron, using polymerase chain reaction (PCR) amplification of cDNA produced by reverse transcription (RT) of total RNA, isolated by the guanidine HCl method26 from the CGD and B-cell lines. Reverse transcription was performed with SuperScript II RT (GIBCO BRL, Gaithersburg, MD) from random hexamer primers. The cDNA was amplified by a 2-stage nested PCR, with forward primers corresponding to nucleotides 3→34 (5′→3′) and 32→64 (5′→3′) and reverse primers corresponding to nucleotides 572→540 (3′→5′) and 537→503 (3′→5′) ofCYBB cDNA sequence (GenBank accession number x04011). The resultant PCR products were analyzed on agarose gels stained with ethidium bromide, extracted from gels, subcloned into pBluescript (Stratagene, La Jolla, CA), and sequenced by the ABI PRISM Dye Terminator Cycle Sequencing method (Perkin Elmer, Foster City, CA).
Gene expression studies
Northern blot analysis of total cell RNA, extracted from EBV-transformed B lymphocytes by the guanidine HCl method,26 was performed according to standard procedures.27 Hybridization probes were full-length cDNAs for the human CYBB6 and NCF128genes, encoding gp91-phox and p47-phox, respectively. The procedures for sequential cycles of filter stripping and reprobing were performed as previously described.29 Equal loading of lanes was demonstrated by the examination of gels after ethidium bromide staining and by rehybridization with a 5.8-kilobase (kb)HindIII restriction fragment of rat 18S ribosomal cDNA.30 Positive control RNA was obtained from HL-60 cells differentiated with dimethyl formamide, and negative control RNA from HeLa cells.24,31 Levels of message in CGD cells were measured quantitatively by a computer analysis of phosphorimager data and compared with normal cells. We have found that the oxidase component p47-phox is expressed in parallel with gp91-phox in EBV-transformed B cells, phagocyte cell lines, and monocyte-derived macrophages22 32; so levels of the p47-phox transcript served as a reference for comparison of the gp91-phox hybridization signals from the CGD and normal EBV-transformed B cell lines.
The transcription rates of genes encoding gp91-phox, and p47-phox were assessed by nuclear run-on assays, with minor modifications of previously published procedures.33Briefly, EBV-transformed B-lymphocyte nuclei were isolated by cell lysis in 0.05% Nonidet P-40. Freshly prepared nuclei were incubated 30 minutes at 30°C in a reaction mixture that contained [32P]UTP (9.25 MBq [250 μCi], 1.11 × 1014 Bq/mmol [3000 Ci/mmol]) in buffer modified from Greenberg et al33 by the addition of 0.8 mmol/L MnCl2. Newly synthesized RNA was prepared by guanidine extraction and ethanol precipitation.26 Equal amounts of incorporated label from each group (1-2 × 107 cpm) were then hybridized to saturating amounts of cDNA probes, immobilized on filters by slot blotting. The probes used in these experiments included cDNAs for the genes encoding gp91-phox6 and p47-phox,28 a hybridization negative control (plasmid without insert), and a constitutively expressed gene (β-actin or α-tubulin).22,32 34 We calculated relative rates of transcription by a computer analysis of phosphorimager data.
Reverse transcription–polymerase chain reaction
The cDNA from the CYBB gene encoding gp91-phox was produced by RT of total and nuclear RNA, isolated by standard procedures from the EBV-transformed patient or normal B-cell lines. Total monocyte RNA (kindly provided by Dr R. A. B. Ezekowitz) was similarly isolated from the patient's peripheral blood monocytes, both before and during therapy, with recombinant human IFN-γ, 50 μg/m2 injected subcutaneously 3 times per week.25 RT was performed with SuperScript II RT (GIBCO BRL) from random hexamer primers. The cDNA was amplified by a single-step PCR with the following primers: CYBB cDNA nucleotides 32→56 (5′→3′) and 634→600 (3′→5′); also 913→932 (5′→3′) and 1163→1144 (3′→5′); and β-actin (GenBank accession number NM001101) cDNA nucleotides 920→943 (5′→3′) and 1494→1471 (3′→5′).
For CYBB gene splicing studies, RT-PCR analysis compared the products of amplification from transcripts in which the first intron had been spliced versus those in which it had remained unspliced. The spliced transcript was amplified using primers corresponding toCYBB cDNA nucleotides 28→52 (5′→3′) (exon 1) and 407→381 (3′→5′) (exon 5). The unspliced transcript was amplified using primers corresponding to CYBB cDNA nucleotides 28→52 (5′→3′) (exon 1), and intron 1 nucleotides 226→200 (3′→5′) (CCTCCTAGGAATTCAGAAGTAAGCTAG; intron sequence kindly provided by Dr S. H. Orkin).
The semiquantitative PCR was performed by amplifying serial dilutions of cDNA preparations at successive cycle numbers to obtain conditions for linear amplification of target cDNAs. Such conditions forCYBB transcripts were generally 35 cycles (monocyte cDNA) to 40 cycles (B cell cDNA), with normal cDNA diluted 1:20 and CGD cDNA undiluted or 1:2; for β-actin transcripts, 25 cycles for all cDNAs, with no dilution necessary. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining, and quantitated by digital photography and computer analysis with Molecular Analyst software (Bio-Rad). Levels of CYBB transcripts were adjusted for cDNA dilutions and normalized to β-actin signals from the same cDNA preparations.
Statistics
Results are represented as mean ± SD. The Wilcoxon rank sum test (Mann-Whitney U test) or the paired Student t test for matched pairs were used for comparisons between groups35; P < .05 was considered significant.
Results
Molecular analysis of the genomic DNA of the X-linked CGD patient has previously revealed a single base substitution, ATT/gtaagt→ ATT/gtaagc,9,14 at the sixth position of first intron in the CYBB gene, which encodes the gp91-phox component of the phagocyte NADPH oxidase.6 Our first hypotheses predicted that this mutation would produce abnormal splicing with both exon skipping and normal product, as previously noted with abnormalities of intron sequences distal to the splice site, both in CYBB and other genes.9 36-41 To investigate the nature of the mutantCYBB transcripts, we amplified the cDNA of the X-linked CGD patient by RT-PCR, followed by subcloning and sequencing. No structural changes were detected in or near the first or second exons of the gp91-phox transcript of the X-linked CGD patient (results not shown).
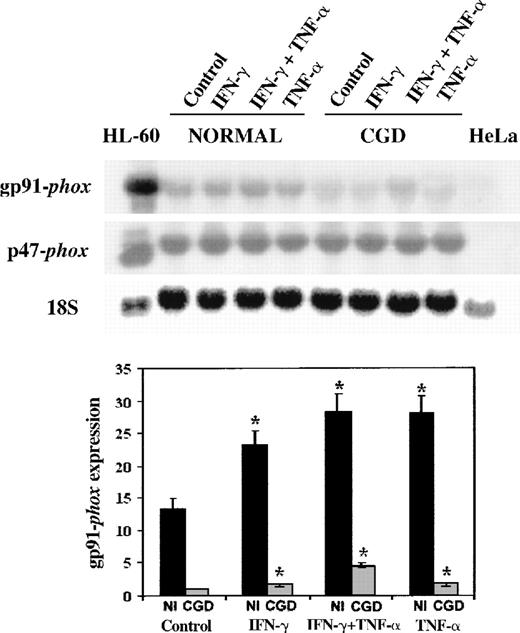
We next examined CYBB gene expression, using patient and normal EBV-transformed B-cell lines as a model system.19 42 Figure1 shows a representative Northern blot (above) and a graphical analysis of 4 such experiments (below). These data demonstrate that IFN-γ (100 U/mL) and TNF-α (1000 U/mL) increased the expression of gp91-phox gene 1.7- to 4.5-fold in both the patient's and the normal cells (P < .05 in all situations, n = 4, Mann-Whitney U test). The cytokines also increased the expression of p47-phox gene, an unaffected component of the NADPH oxidase system, but did not reach statistical significance (P > .05 in all situations, n = 4, Mann-Whitney U test). The expression of gp91-phoxwas 6- to 13-fold higher in the normal cells compared with the patient's cells in basal conditions or after cytokine stimulation (P < .05 in all situations, n = 4, Mann-WhitneyU test). A synergistic response occurred when the patient's cells were cultured with IFN-γ and TNF-α in combination.
Northern blot of mRNA transcripts encoding gp91-phox and p47-phox.
Each lane contains 10 μg total RNA from IFN-γ–differentiated HL-60 cells, normal, and CGD B-cell lines, or HeLa cells, as indicated above. The autoradiograph shows a representative blot successively probed with32P-labeled cDNAs encoding gp91-phox, p47-phox, and (as a control for equal loading of lanes) 18S rRNA, as indicated in the left margin. The EBV-transformed B-cell lines were cultured in control conditions or in the presence of IFN-γ (100 U/mL), TNF-α (1000 U/mL), alone or in combination for 7 days, as indicated in the top margin.
Northern blot of mRNA transcripts encoding gp91-phox and p47-phox.
Each lane contains 10 μg total RNA from IFN-γ–differentiated HL-60 cells, normal, and CGD B-cell lines, or HeLa cells, as indicated above. The autoradiograph shows a representative blot successively probed with32P-labeled cDNAs encoding gp91-phox, p47-phox, and (as a control for equal loading of lanes) 18S rRNA, as indicated in the left margin. The EBV-transformed B-cell lines were cultured in control conditions or in the presence of IFN-γ (100 U/mL), TNF-α (1000 U/mL), alone or in combination for 7 days, as indicated in the top margin.
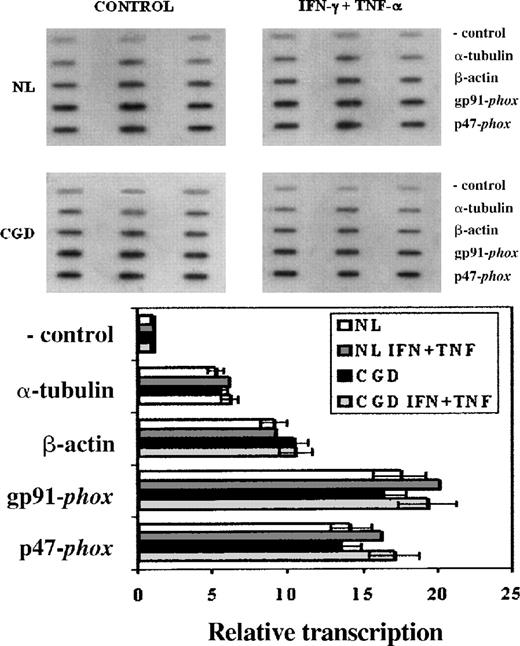
Nuclear run-on assays (Figure 2) were performed to investigate any impairment in the transcription of the patient's CYBB gene that could account for its low expression. These experiments showed similar transcription rates of theCYBB and NCF1 genes—encoding, respectively, gp91-phox and p47-phox—in both the CGD patient's and the normal cell lines (Figure 2, P > .05 in all situations, n = 3, Mann-Whitney U test). IFN-γ (100 U/mL) and TNF-α (1000 U/mL) enhanced the transcription rates of these genes by 10% to 15% in both the CGD patient's and the normal cell lines, but the changes did not reach statistical significance (P > .05 in all situations, n = 3, Mann-Whitney U test). α-Tubulin and β-actin transcription rates were not affected by these cytokines (P > .05 in all situations, n = 3, Mann-Whitney U test).
Transcription rates of genes encoding gp91-phox, and p47-phox in CGD patient's and normal B-cell lines.
Upper panel: Representative nuclear run-on assay showing transcription rates of genes encoding gp91-phox (CYBB), and p47-phox (NCF1) in CGD and normal B cell lines. Lower panel: Graph representing compiled data from 3 experiments; columns indicate means and error bars show SDs. EBV-transformed B cells were cultured under control conditions or with IFN-γ (100 U/mL) and TNF-α (1000 U/mL) for 7 days. The negative control lanes contain pBluescript plasmid alone. Genes encoding α-tubulin and β-actin were included as transcriptionally constitutive controls.
Transcription rates of genes encoding gp91-phox, and p47-phox in CGD patient's and normal B-cell lines.
Upper panel: Representative nuclear run-on assay showing transcription rates of genes encoding gp91-phox (CYBB), and p47-phox (NCF1) in CGD and normal B cell lines. Lower panel: Graph representing compiled data from 3 experiments; columns indicate means and error bars show SDs. EBV-transformed B cells were cultured under control conditions or with IFN-γ (100 U/mL) and TNF-α (1000 U/mL) for 7 days. The negative control lanes contain pBluescript plasmid alone. Genes encoding α-tubulin and β-actin were included as transcriptionally constitutive controls.
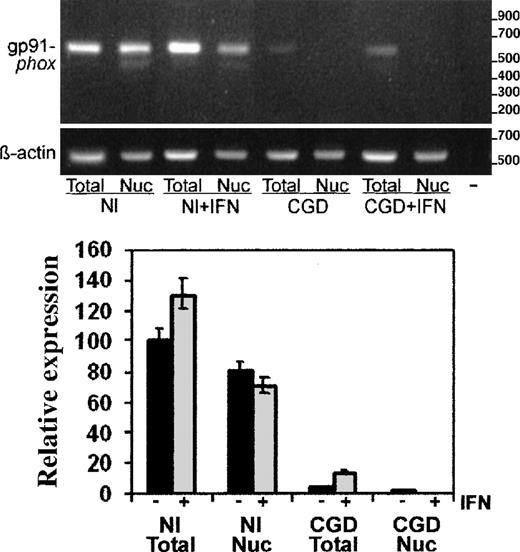
Using the semiquantitative RT-PCR techniques, we performed more detailed analyses of CYBB transcript abundance, distribution, and splicing. Figure 3 shows a representative image of the PCR products in an ethidium bromide-stained gel (above) and a graphical analysis of image intensity from 4 such experiments (below). The digital camera data showed markedly decreased levels of gp91-phox messenger RNA (mRNA), normalized to β-actin transcripts, in both total (5% of normal) and nuclear (1.4% of normal) RNA preparations from patient-derived cells compared with normal cells. In vitro treatment of the CGD cells with IFN-γ (Figure3) increased the level of gp91-phox transcripts 3-fold in total RNA (P < .05, n = 3, Mann-Whitney U test), while decreasing the amount of gp91-phox transcripts in the nuclear RNA preparations to undetectable levels.
RT-PCR analysis of gp91-phox transcripts in total cell and nuclear RNA preparations.
RT-PCR products of gp91-phox transcripts from the normal (Nl) or CGD patient's EBV-transformed B-cell lines, cultured with or without IFN-γ, as indicated (+IFN). Upper panel: Representative agarose electrophoresis gels, stained with ethidium bromide, of RT-PCR products from the RNA sources indicated below the image. Primers corresponded to cDNAs encoding gp91-phox or β-actin, as indicated in the left margin; size markers are indicated in the right margin. Lower panel: Graph representing compiled data from 3 experiments. Band intensity was quantitated by digital photography and computer analysis with Molecular Analyst software. Data are expressed as percentage expression relative to gp91-phox transcript levels in normal total cellular RNA, adjusted for β-actin signals; columns indicate means and error bars show SDs.
RT-PCR analysis of gp91-phox transcripts in total cell and nuclear RNA preparations.
RT-PCR products of gp91-phox transcripts from the normal (Nl) or CGD patient's EBV-transformed B-cell lines, cultured with or without IFN-γ, as indicated (+IFN). Upper panel: Representative agarose electrophoresis gels, stained with ethidium bromide, of RT-PCR products from the RNA sources indicated below the image. Primers corresponded to cDNAs encoding gp91-phox or β-actin, as indicated in the left margin; size markers are indicated in the right margin. Lower panel: Graph representing compiled data from 3 experiments. Band intensity was quantitated by digital photography and computer analysis with Molecular Analyst software. Data are expressed as percentage expression relative to gp91-phox transcript levels in normal total cellular RNA, adjusted for β-actin signals; columns indicate means and error bars show SDs.
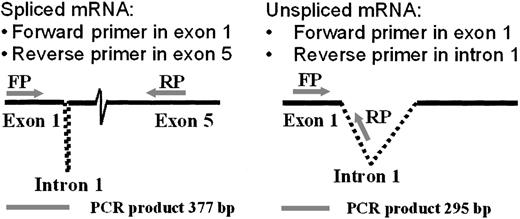
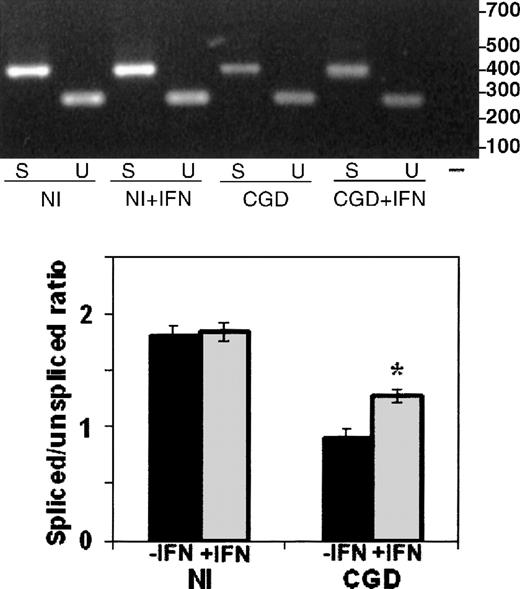
The RT-PCR analysis of RNA splicing used a forward primer in the first exon and alternative reverse primers in the fifth exon (to detect spiced mRNA) or the first intron (to detect unspliced mRNA), as illustrated in Figure 4. Comparison of the ratios of spliced to unspliced gp91-phox transcripts in the normal and the CGD patient-derived B-cell lines revealed slightly more unspliced than spliced transcripts in the patient's nuclear RNA (0.9 spliced/unspliced ratio), in contrast to a predominance of spliced transcripts in normal nuclear RNA (1.8 spliced/unspliced ratio), as shown in Figure 5. In vitro incubation with IFN-γ increased the amount of spliced relative to unspliced mRNA by 40% in the CGD B-cell line nuclei (P < .05, n = 4, Mann-Whitney U test).
Diagram of methodology for mRNA splicing analysis by RT-PCR.
A forward primer in the first exon and alternative reverse primers in the first intron or the fifth exon amplified products of different sizes, as indicated below the figure (bp = base pairs). The spliced transcript was amplified using a forward primer (FP) corresponding to gp91-phox cDNA exon 1 nucleotides 28→52 (5′→3′) and a reverse primer (RP) corresponding to exon 5 nucleotides 407→381 (3′→5′). The unspliced product was detected by the same forward primer, coupled with a reverse primer corresponding to intron 1 nucleotides 226→200 (3′→5′).
Diagram of methodology for mRNA splicing analysis by RT-PCR.
A forward primer in the first exon and alternative reverse primers in the first intron or the fifth exon amplified products of different sizes, as indicated below the figure (bp = base pairs). The spliced transcript was amplified using a forward primer (FP) corresponding to gp91-phox cDNA exon 1 nucleotides 28→52 (5′→3′) and a reverse primer (RP) corresponding to exon 5 nucleotides 407→381 (3′→5′). The unspliced product was detected by the same forward primer, coupled with a reverse primer corresponding to intron 1 nucleotides 226→200 (3′→5′).
The effect of IFN-γ on splicing of nuclear CYBBtranscripts in CGD and normal B-cell lines.
RT-PCR products of CYBB nuclear mRNA amplification performed with a forward primer in the first exon and alternative reverse primers in the fifth exon or the first intron (as diagramed in Figure 4) detect, respectively, spliced (S) and unspliced (U) transcripts from nuclei prepared from normal (Nl) or CGD B-cell lines, incubated with (+IFN) or without IFN-γ. Upper panel: Representative ethidium bromide-stained agarose gel of RT-PCR products from the RNA sources indicated below the image; size markers are shown in the right margin. Lower panel: Graph representing compiled data from 3 experiments. Band intensity was quantitated by digital photography and computer analysis with Molecular Analyst software. Data are expressed as the ratio of spliced to unspliced products; columns represent means and error bars show SDs.
The effect of IFN-γ on splicing of nuclear CYBBtranscripts in CGD and normal B-cell lines.
RT-PCR products of CYBB nuclear mRNA amplification performed with a forward primer in the first exon and alternative reverse primers in the fifth exon or the first intron (as diagramed in Figure 4) detect, respectively, spliced (S) and unspliced (U) transcripts from nuclei prepared from normal (Nl) or CGD B-cell lines, incubated with (+IFN) or without IFN-γ. Upper panel: Representative ethidium bromide-stained agarose gel of RT-PCR products from the RNA sources indicated below the image; size markers are shown in the right margin. Lower panel: Graph representing compiled data from 3 experiments. Band intensity was quantitated by digital photography and computer analysis with Molecular Analyst software. Data are expressed as the ratio of spliced to unspliced products; columns represent means and error bars show SDs.
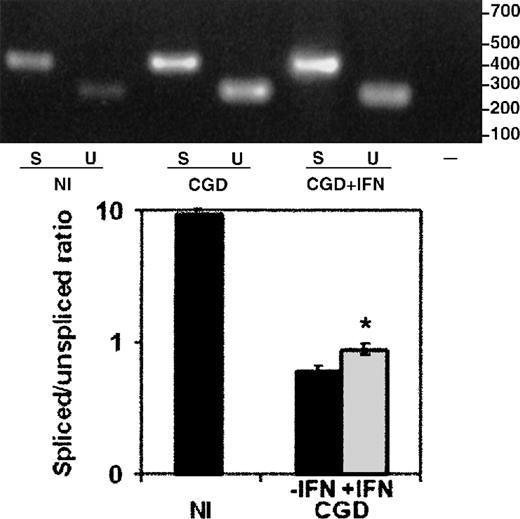
The total RNA from peripheral blood monocytes (Figure6), harvested from the same CGD patient, on and off therapy with IFN-γ, showed a similar improvement, with a 47% increase in the ratio of spliced to unspliced message (P < .05, n = 3, paired Student t test). The nuclear RNA preparations could not be performed because of limitations on the frequency and volume of blood collections from a patient.
The effect of IFN-γ on splicing of CYBBtranscripts in total cellular RNA from the CGD patient's peripheral blood monocytes.
RT-PCR products amplified from total RNA extracted from peripheral blood monocytes harvested from the same patient on and off therapy with IFN-γ. Upper panel: Representative ethidium bromide-stained agarose gel of RT-PCR products from the RNA sources indicated below the image; size markers are shown in the right margin. Lower panel: Graph representing compiled data from 3 experiments. Labeling as in Figure5.
The effect of IFN-γ on splicing of CYBBtranscripts in total cellular RNA from the CGD patient's peripheral blood monocytes.
RT-PCR products amplified from total RNA extracted from peripheral blood monocytes harvested from the same patient on and off therapy with IFN-γ. Upper panel: Representative ethidium bromide-stained agarose gel of RT-PCR products from the RNA sources indicated below the image; size markers are shown in the right margin. Lower panel: Graph representing compiled data from 3 experiments. Labeling as in Figure5.
Discussion
The X-linked CGD derives from defects in the CYBB gene, which encodes the gp91-phox component of the NADPH oxidase.1,2 The propositus has a variant form of X-linked CGD, due to a single base substitution in the first intron ofCYBB gene.9,14 His phagocytes show a dramatic increase in superoxide release and cytochrome b levels in response to IFN-γ in vitro or in vivo.15,16 Mutations at splice junctions interfere with RNA processing and usually result in multiple splicing products, including both normal transcripts and mRNA species that are nonfunctional because of exon skipping or diminished RNA stability.9 36-41
Our results in this CGD kindred show that the mutation, located 6 nucleotides downstream from the first splice site of the CYBBgene, did not cause any structural changes in the transcript, as demonstrated by the PCR analysis and the sequencing of the cDNA. However, the mutation caused an impairment in the CYBB gene expression, as demonstrated by the Northern blot and RT-PCR analysis. Transcription rates were equal in the nuclear run-on assays for both normal and CGD EBV-transformed B cells, indicating that the low levels of CYBB transcripts reflect instability rather than decreased synthesis. Both the nuclear and the cytoplasmic transcript levels are markedly diminished, indicating increased degradation in the nucleus; the relative preservation of CYBB transcript levels in the total RNA suggests that they are more stable once exported to the cytoplasm. The proportions of spliced and unspliced mRNA indicate a failure to process the nuclear pre-mRNA, most likely at the mutated splicing region in the first intron. Thus, the intronic mutation appears to interfere with the normal splicing process, and hence leads to low gp91-phox expression and the phenotype of variant X-linked CGD.
Most notably, in vitro incubation with IFN-γ increased the ratio of spliced relative to unspliced CYBB transcripts in nuclei from the B-cell line of the patient with CGD. A less dramatic but similar improvement in the splicing defect occurred in vivo, shown in the proportions of spliced and unspliced transcripts in the total RNA isolated from the patient's peripheral blood monocytes before and during therapy with IFN-γ. Thus, in this kindred, IFN-γ may improve cytochrome b558 levels and partially correct the CGD phenotype by increasing the rate of splicing at the CYBBfirst intron, which thus allows the nuclei to export greater quantities of the spliced mRNA to be translated into gp91-phox, the normal protein product.
These results provide a molecular basis for our previous observations of IFN-γ induction of the CYBB gene expression in this kindred.15,16,43 Previous studies of these patients with variant X-linked CGD have shown phagocyte superoxide production at 5% to 10% normal rates, with corresponding levels of cytochromeb13; they show an unusual, dramatic increase in these measures in response to IFN-γ, both in vitro and in vivo.15,16 This response derives at least in part from an enhancement on the expression of CYBB gene, as demonstrated in both previous15,16 and current experiments. In contrast, most patients with CGD show no biochemical or molecular response to IFN-γ,44 despite demonstrated clinical efficacy.25 45
Previous studies of the current CGD kindred demonstrated larger increases in the cytochrome b558 protein levels, and still greater rises in superoxide generation and bacterial killing, compared with the magnitude of change in mRNA encoding the gp91-phox component.15,16,43 As might be expected from a multicomponent system, NADPH oxidase activity does not strictly correlate with the amount of any single component.15,16,46In addition, further posttranslational mechanisms could contribute to the activation of the NADPH oxidase system and hence, the quantitative differences between the cytochrome b558 content and the NADPH oxidase activity results.47-50 Thus, the physiological responses observed in these patients probably derives in part from the change in splicing reported here, with a major additional contribution from the functional amplification provided by the partial restoration of a missing component to an otherwise competent, or even primed, system.
The model system of EBV-transformed B lymphocytes has some limitations because of the relatively low transcriptional expression of genes encoding components of the NADPH oxidase system, compared with monocytes or phagocytic cell lines.32 Studies of cytokine-mediated gene regulation in granulocytes and monocyte-derived macrophages,23,24 human monocytic THP-1 cells,22 and human myeloid leukemia cells51have demonstrated synergistic induction of CYBB expression by IFN-γ and TNF-α that was at least in part transcriptional. Thus, unlike the EBV-transformed B-cell lines, cells in the monocytic-myeloid lineage probably up-regulate gp91-phox expression in response to IFN-γ by both transcriptional and posttranscriptional mechanisms.
Interferon-γ has previously been reported to augment expression of several other genes by increasing both transcription rates and mRNA stability. Examples include human Clara cell secretory protein production by human epithelial cells,52 synthesis of complement components in monocytes and fibroblasts,53-55expression of intercellular adhesion molecule 1 in fibroblasts56 and monocytes,57 and TNF-α generation in macrophages.58 The effect of IFN-γ onCYBB splicing has some precedent in previously reported cytokine effects on alternative splicing of transcripts encoding Trp-tRNA synthetase.59
The effect of IFN-γ on expression of the gp91-phox gene in phagocytes from patients with X-linked CGD15 provided, in part, the basis for the clinical use of IFN-γ for prevention of infection in CGD.25 However, the large majority of patients appear to benefit clinically from other, unknown mechanisms of IFN-γ action, because most do not show a change in oxidase activity.44 The current studies demonstrate a new mechanism by which IFN-γ can up-regulate CYBB gene expression and may explain in part its unusually dramatic biochemical effect in this kindred.
Acknowledgments
We thank Drs. John Curnutte and Alan Ezekowitz for sharing data, materials, and helpful discussions; we thank Constance Whitney and Carolyn Padden for excellent technical assistance.
Supported by Brazil's Conselho Nacional de Desenvolvimento Cientı́fico e Tecnolólogico grant 200955/95-0, Fundação de Amparo à Pesquisa do Estado de São Paulo grant 96/11666-2, and State University of Campinas Medical School in house grant; by US National Institutes of Health grants DK54369 and TW00883; and by an award from the Howard Hughes Medical Institute to the University of Massachusetts Medical School under the Research Resources Program for Medical Schools.
Reprints:Peter E. Newburger, Department of Pediatrics, University of Massachusetts Medical School, 373 Plantation St, Worcester, MA 01605; e-mail: peter.newburger@umassmed.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal