Abstract
The activity of phosphodiesterase (PDE)3A requires divalent cations. Putative metal-binding sites are expected at 2 highly conserved metal-binding motifs, HXXXH(X)25E. A functional truncated recombinant PDE3A containing the catalytic domain (PDE3A▵1) and mutant proteins were expressed in a baculovirus/Sf9 cell system. All the mutant proteins had decreased catalytic efficiency (kcat/Km). Mutants H752A, H756A, and E825A had kcat of less than 0.0008 s−1 to 0.0475 s−1 compared to PDE3A▵1, with 1.86 second−1, with unchanged Km. Although E866A had a kcat of 0.235 s−1, the Kmfor cyclic adenosine monophosphate (cAMP) was increased 11-fold and the Ki for cyclic guanosine monophosphate (cGMP) was 27-fold higher than PDE3A▵1. The Ki of H836A for cGMP was 177-fold higher than that of PDE3A▵1. The Km for E971A was 5-fold higher than PDE3A▵1. These results suggest that the cAMP and cGMP binding sites are overlapping, but not identical, involving both common and different amino acids. Mutants E825A, H836A, and E866A showed low activity in a metal ion-free assay; however, their enzymatic activities were increased 4- to 10-fold in buffers containing Mn2+, Mg2+, or Co2+. This observation indicates that conserved amino acids in the second metal-binding motif might not be involved in binding divalent cations but may serve other functions such as substrate or inhibitor binding in PDE3A.
An increase of intracellular cyclic adenosine monophosphate (cAMP) produces potent inhibition of all platelet functions.1 Cyclic guanosine monophosphate (cGMP)-inhibited cAMP phosphodiesterase (PDE)3A, the most abundant cyclic nucleotide phosphodiesterase in platelets,2 hydrolyzes cAMP, lowering its intracellular concentration. Inhibitors function as potent antiplatelet agents. Currently, the 2 oral antiplatelet agents with proven efficacy are aspirin, which inhibits cyclooxygenase-dependent synthesis of thromboxane A2 (TXA2), and clopidogrel or ticlopidine, which blocks the ability of adenosine diphosphate (ADP) to inhibit stimulated adenyl cyclase.3Controlled trials show that both aspirin and ticlopidine are indicated in the secondary prevention of stroke, myocardial infarction, and peripheral vascular occlusion. However, standard antiplatelet drugs do not alter the impairment of hemostasis seen in cardiopulmonary bypass, and neither drug appears to prevent reocclusion of coronary arteries after either thrombolytic therapy or angioplasty. The failure of aspirin and ticlopidine in these situations largely results from their inability to inhibit thrombin-induced platelet activation. Although, at low concentrations of thrombin, platelet aggregation is dependent on ADP and TXA2, high concentrations of thrombin aggregate and activate platelets by independent pathways. In contrast, the elevation of cAMP blocks all activating pathways in platelets—among them increases in intracellular Ca++, shape change, aggregation, secretion, and the effects of phospholipases A2 and C—including the responses to thrombin. The potential for a PDE3A inhibitor is to modulate coronary artery reocclusion, which occurs in up to 30% of patients who undergo angioplasty or thrombolytic therapy.
The action of naturally occurring inhibitory prostaglandins—PGD2 from platelets and PGI2 (prostacyclin) from endothelial cells—is markedly enhanced by the inhibition of cGI–PDE. The validity of the strategy has been confirmed using a PDE3A inhibitor in a model of peripheral vascular thrombosis in rabbits,4 thrombolytic therapy of coronary artery occlusion in dogs,5 and coronary stents in humans.6 Nitric oxide (NO) is a potent platelet inhibitor that stimulates guanyl cyclase, which elevates cGMP. Because cGMP inhibits PDE3A,7 an inhibitor of PDE3A could synergize with NO. Nevertheless, current inhibitors have side effects,8-10and new potent, effective inhibitors are desirable. The potential of specific inhibitors is illustrated by the ability of the PDE5 inhibitor, sildenafil (Viagra; Pfizer, Groton, CT), to treat erectile dysfunction.11
The existence of distinct families of mammalian cyclic nucleotide PDEs, which are expressed in a cell-specific manner, has encouraged the further development of drugs that selectively inhibit specific cAMP–PDEs. The mammalian PDEs are currently classified into 10 distinct families based on a combination of amino acid sequence homology and a variety of biochemical properties, including substrate specificity, response to selective inhibitors, mode of regulation, and kinetic properties.12 The sequencing of the cDNA from at least 1 member of each PDE family suggests that the 10 families are coded for by related but distinct genes. They share a conserved region of approximately 250 amino acids at the C-terminal end that contains the catalytic domain, which is enzymatically competent by itself13-15; however, the extent of homology is only 28% to 40%.16 In contrast, the N-terminal region of each class is distinct and, in certain cases, appears to contain the regulatory domain(s).
Native PDE3A purified from human erythroleukemia (HEL) cells in the presence of chymostatin has a molecular mass of 110 kd17and hydrolyzes cAMP with a Km of 0.5 μmol/L. The enzyme is competitively inhibited by cGMP (Ki = 0.06 μmol/L). When the enzyme is purified in the absence of an inhibitor, the molecular mass is 61 kd with identical kinetic constants. PDE3A purified from outdated platelets is a 61-kd catalytically active hydrolase.18 We cloned HEL cell PDE3A,19obtained its cDNA sequence, which was identical to a cloned enzyme from heart,20 and coded for an amino acid sequence obtained from platelets. We expressed a deletion mutant, PDE3AΔ1, coding for amino acids 679 to 1141 in yeast with similar kinetics to the full-length molecule and a specific activity of 844 nmol cAMP hydrolyzed per minute per milligram protein. Further deletions led to a complete loss of activity.19
To elucidate the amino acids important in the active site, we showed that a pH profile of PDE3A yielded pKa values of 6.5 and 9.0, consistent with histidine and cysteine.21 We then performed group-specific modification on histidines with diethyl pyrocarbonate and cysteines with N-ethylmaleimide and 5,5′-dithiobis-(2-nitrobenzoic acid). From difference spectra and protection studies, 2 histidines were responsible for hydrolysis of cAMP. However, if cGMP was used to protect against modification, 2 different histidines were protected because the combination of cGMP and cAMP protected 4 histidine residues.21 A single cysteine was required for cAMP hydrolysis, but none was needed for cGMP binding. This study allowed us to formulate the hypothesis that cAMP and cGMP binding sites are overlapping but not identical.
PDE3A activity is dependent on the presence of a divalent cation, Mn++ > Mg++ > Co++. Zn++ is the most potent cation but is inhibitory.22 The sequence of the enzyme displayed 2 metal-binding motifs, HXXXH (X)25E, which are candidates for coordinating Mn++, Co++, and Mg++.23 These motifs are part of 26 perfectly conserved amino acids in PDE3A, PDE2, PDE4A, and PDE5A.24Twenty-three of these have been mutated in PDE5. We have previously mutated 6 of the 26 conserved amino acids. The protein containing C816A is poorly expressed. The mutation lies in a 44-amino acid insert unique for PDE3. The protein containing H840A is well expressed, and the mutation lies in the second metal-binding motif but displays no catalytic activity. The enzyme containing H869A, outside the 2 motifs, has kinetics consistent with defective binding for both cAMP and cGMP. Mutation of amino acids C942A, C945A, and C1013A resulted in enzymes indistinguishable from PDE3AΔ1 in their kinetic parameters.
We have now made 6 new mutations, 5 in the 2 metal-binding motifs, to test whether these motifs bind divalent cations. The results of our studies show that both histidines and the glutamate in the first motif have exceedingly low activity, consistent with impairment of either catalytic or metal binding. However, E866A has impaired binding for both cAMP and cGMP. H836 had only decreased cGMP binding, and E971A had decreased affinity only for cGMP. Neither mutant nor E825A showed loss of metal-induced activity. We conclude that the second metal-binding motif functions as a substrate binding region and that cAMP and cGMP binding involve common and distinct amino acids.
Materials and methods
Materials
3[H]-cAMP was purchased from Amersham Pharmacia Biotech (Piscataway, NJ). cAMP, cGMP, milrinone, rolipram, and high-quality chloride salts were from Sigma (St Louis, MO).
Expression of a recombinant PDE3A in baculovirus/Sf9 cell system
A vector pBSPDE3.319 containing a partial human platelet PDE3A DNA from nucleotide 1993 to 3841 of PDE3A (GenBank accession number U36 798) was digested by XhoI and SacI. The PDE3A DNA fragment was subcloned to a PCR.2.1. vector (Invitrogen) and then cut by XhoI and KpnI to add a KpnI site at the 3′ end of PDE3A for cloning purposes. The 2-kbXhoI–KpnI DNA fragment, which covers the entire catalytic region of PDE3A, was then cloned to the baculovirus expression vector pBlueBacHis 2 (Invitrogen, Carlsbad, CA) to generate pBBH3031. This vector contained 31 amino acids, including 6 histidines and an enterokinase cleaving site. When expressed, the specific activity was similar to that of the enzyme expressed in yeast and purified from platelets (see below). Sf9 cells were cultured in complete Grace's insect medium with 10% fetal bovine serum (Life Technologies) at 27°C. Sf9 cells were cotransfected with Bac-N-Blue DNA (Invitrogen) and the expression vector pBBH3031 by the lipofection method (insect transfection kit from Invitrogen). Preparation and purification of the recombinant virus were performed according to the protocol from Invitrogen. For expression, 5 × 107 cells were infected at a multiplicity of infection of 6 in a T-75 flask. The cells were collected 96 hours after infection, and cell lysates and enzyme preparation were performed as described below.
Site-directed mutagenesis
Site-directed mutagenesis of the recombinant PDE3A was performed with a QuikChange kit (Stratagene). Pairs of complementary oligonucleotide primers that contain desired mutants were synthesized as follows: (1) H752A, 5′-GGGATATTCCTTATGCTAACTGAATCCATGCC-3′ and 5′-GGCATGGATTCTGTTAGCA-TAAGGAATATCCC-3′; (2) H756A, 5′-CATAACAGAATCGCTGCCACTGATCTTTTACAT-GC-3′ and 5′-GCATGTAAAACATCAGTGGCAGCGATTCTGTTATG-3′; (3) E825A, 5′-GGAGTATCCCTGCCTTGGCGTTGATGGCGCTG-3′ and 5′-CAGCGCCATCAACGCCAA-GGCAGGGATATTCC-3′; (4) H836, 5′-GGCTGCAGCCATGGCCGATTATGATCATCC-3′ and 5′-GGATGATCATAATCATAATCGGCCATGGCTGCAGCC-3′; (5) E866A, 5′-CGATCGTTCAGTTTTGGCGAATCATCACGC-3′ and 5′-GCGTGATGATTCGCCAAAACT-GAACGATCG-3′; (6) E971A, 5′-GGACAGATGGTATTGTCAATGCATTTTATGAACAGG-G-3′ and 5′-CCCTGTTCATAAAATGCATTGACAATACCATCTGTCC-3′. pBlueBacHis 3031 plasmid DNA was used as a template of polymerase chain reaction (PCR) with plaque-forming unit DNA polymerase. The PCR products were treated with DpnI to digest the parent double-strand DNA chains. The mutated plasmid DNA was transformed in TOP 10 Escherichia coli competent cell (Invitrogen). The sequences of mutants were confirmed by automated DNA sequencing.
Preparation of cell extract
The Sf9 cells were harvested 96 hours after infection by centrifugation at 3000g for 15 minutes at 4°C, washed by PBS buffer, pH 7.4, and resuspended in a lysis buffer (50 mmol/L Tris-HCl, pH 7.8, 10 mmol/L MgCl2 with 0.5 μg/mL pepstatin, 0.5 μg/mL leupeptin, 2 μmol/L benzamidine, 10 μg/mL soybean trypsin inhibitor, and 50 μmol/L Tosyl phenylalanyl chloromethylketone) at 5 × 107 cells/mL. The cells were disrupted with a sonicator probe at 30-spare pulse for a total time of 2 minutes in ice. Crude cell extracts were centrifuged at 15 000g for 30 minutes at 4°C. The supernatant was either stored at −80°C or further purified.
Purification of the recombinant PDE3A protein and mutant proteins
ProBond nickel-chelating resin (Invitrogen) was used for the protein purification; 1 mL resin was added to 2 mL supernatant in a total volume of 8 mL with a binding buffer (50 mmol/L Tris-HCl, pH 8.0, 0.5 mol/L NaCl, and 25 mmol/L imidazole) and rotated for 20 minutes. The resin was washed 3 times with the same binding buffer and packed in a 10-mL column. The purified protein was eluted by the eluted buffer (50 mmol/L Tris-HCl, pH 7.0, 0.5 mol/L NaCl, and 250 mmol/L imidazole), and aliquots were collected at 0.5 mL of each. The purified proteins were further dialyzed against 50 mmol/L Tris-HCl, pH 7.8, 10 mmol/L MgCl2, and 20% glycerol. All procedures were performed at 4°C.
Phosphodiesterase activity assay
Enzymatic activity was measured as described previously.18 Briefly, 100 μL total reaction volume containing 50 mmol/L Tris-HCl, pH 7.8, 10 mmol/L MgCl, and 1 μmol/L3H-cAMP (40 000 cpm/assay) was incubated at 24°C for 30 minutes. Reactions were stopped by the addition of 0.2 mL of 0.2 mol/L ZnSO4 and 0.2 mL of 0.2 mol/L Ba(OH)2. Samples were mixed and centrifuged at 10 000g for 3 minutes. Radioactivity in the supernatants was determined by liquid scintillation. Vmax and Km for cAMP were determined by the Lineweaver–Burk plot with various concentrations of cAMP from 0.04 μmol/L to 20 μmol/L by Microsoft Excel program. For studies of enzyme activity inhibition by related inhibitor, compounds were present in the reaction mixture at concentrations that covered 4 orders of magnitude. Reactions were conducted at 24°C for 30 minutes and terminated by the addition of ZnSO4. Enzymatic assays were conducted in the linear range of the reaction, where less than 30% of the initial substrate cAMP was hydrolyzed. For mutants with low intrinsic activity, the amount of protein was increased until the total activity was comparable to the wild type. All assays were repeated 3 times, each on an independent transfection. Data are expressed as mean ± SD. Percentage expression of each recombinant protein was calculated by the following formula:
Multiple transfection of truncated recombinant PDE3A yielded specific activities that agreed within 15%.
Western blot analysis
Expressed cell lysates or purified recombinant protein and mutant proteins were separated on SDS–polyacrylamide gel electrophoresis using 10% polyacrylamide gels purchased from Fisher Scientific (Houston, TX). Proteins were transferred electrophoretically to nitrocellulose membrane (Bio-Rad, Hercules, CA). The membrane was blocked with 5% nonfat dry milk and 0.05% Tween 20 and incubated for 1.5 hours at room temperature with rabbit antiplatelet PDE3A polyclonal antibody (1:1000 dilution).19 Immunoreactivity was detected with horseradish-conjugated antirabbit IgG. Bands were visualized with substrate system (Bio-Rad) according to the manufacturer's protocol.
Effects of divalent metal cations on the purified recombinant PDE3A and mutant proteins
Metal-free water and metal-free buffer were made by the method previously described.22 Recombinant and mutant PDE3A were purified by the same method as above except in the metal-free buffer. Purified proteins were dialyzed in metal-free 50 mmol/L Tris-HCl buffer. Enzyme activity in metal-free buffer was assayed as a baseline, and the activity was measured in the presence of 10 μm to 1 mmol/L Mn2+, Mg2+, and Co2+ in chloride salt form. The value taken as 100% activity is the activity of purified recombinant PDE3A before dialysis.
Protein concentration determination
Protein concentrations in cell lysates and purified proteins were determined by bicinchonic acid (BCA) protein assay reagent (Pierce, Rockford, IL), and bovine serum albumin was used as a standard.
Results
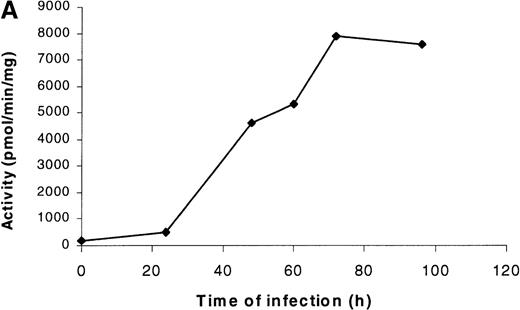
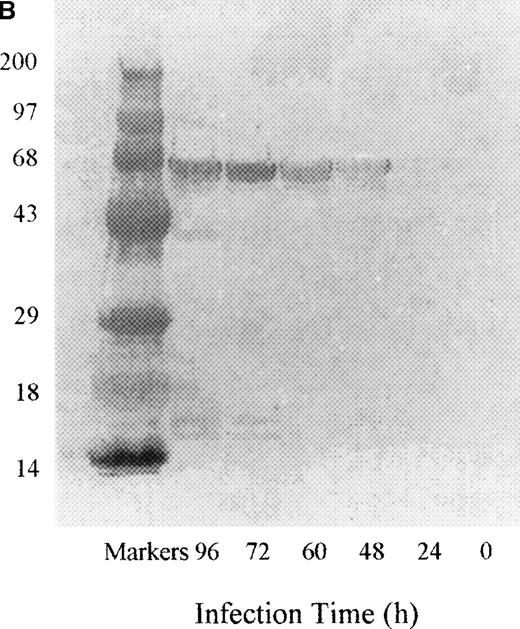
PDE3AΔ1 was expressed by infecting Sf9 insect cells with baculovirus containing the insert coding for the 66-kd C-terminal fragment of PDE3A, amino acids 679 to 1141. No PDE3A activity was detected at 24 hours, but the activity rapidly increased by 48 hours and reached a maximum value by 72 hours (Figure1A). The Western blot of the cell lysate shows a band at 66 kd starting at 48 hours (Figure 1B). When the gel was scanned, the reactive band followed the same pattern as the activity.
Time course of expression of recombinant PDE3A in Sf9 cells.
(A) At each time point, cells were collected, counted, and lysed, and the enzymatic activity and total protein were measured as described in “Materials and methods.” The left lane contains the molecular weight standards. (B) At each time point, 10 μg protein was applied to a 10% SDS gel, transferred electrophoretically, and analyzed by Western blotting as described in “Materials and methods.”
Time course of expression of recombinant PDE3A in Sf9 cells.
(A) At each time point, cells were collected, counted, and lysed, and the enzymatic activity and total protein were measured as described in “Materials and methods.” The left lane contains the molecular weight standards. (B) At each time point, 10 μg protein was applied to a 10% SDS gel, transferred electrophoretically, and analyzed by Western blotting as described in “Materials and methods.”
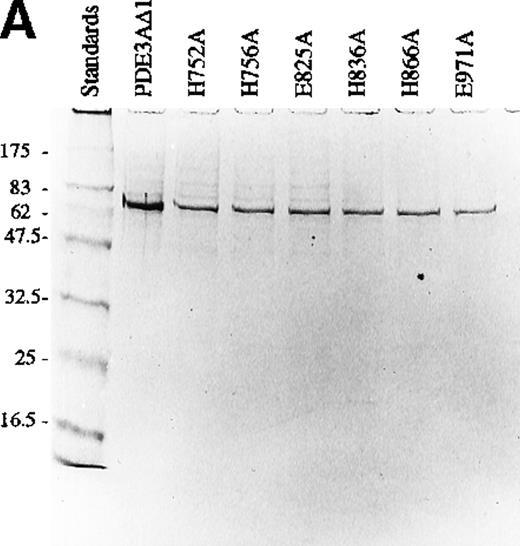
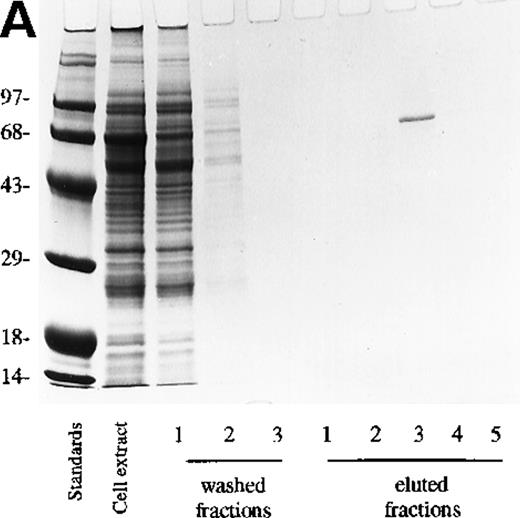
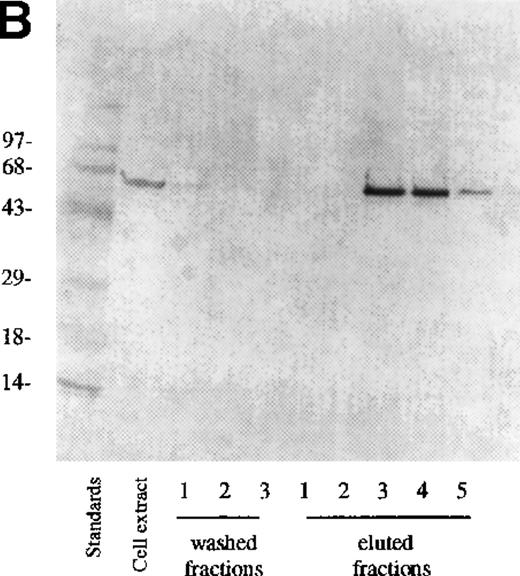
The cell lysate supernatant (cytosol) containing the recombinant protein was purified on Ni-column as described in “Materials and methods.” Coomassie blue staining of the lysate showed a large number of proteins (Figure 2A), but only a single protein was eluted from the Ni-column (Figure 2A, eluted fraction 3) because the construct contains a hexahistidine sequence. Western blot confirmed that in PDE3AΔ1, Mr = 66 kd (Figure 2B).
Analysis of mutant proteins of PDE3A with SDS-PAGE gel and Western blotting.
(A) SDS-PAGE gel of truncated recombinant PDE3A and 6 mutants. The left lane contains the molecular weight standards. (B) Western blot of truncated recombinant PDE3A and 6 mutants.
Analysis of mutant proteins of PDE3A with SDS-PAGE gel and Western blotting.
(A) SDS-PAGE gel of truncated recombinant PDE3A and 6 mutants. The left lane contains the molecular weight standards. (B) Western blot of truncated recombinant PDE3A and 6 mutants.
When 5 × 107 Sf9 cells were lysed, the PDE3AΔ1 showed a specific activity of 10.2 ± 3.0 nmol/min per milligram. The total protein was 7300 ± 754 μg. A pool of the eluted fractions showed a specific activity of 911 ± 67 nmol/min per milligram, which compares favorably with 844 nmol/min per milligram reported for the same construct in yeast19 and 855 nmol/min per milligram in platelets.18 A total protein of 56 ± 7.6 μg was recovered. The purification was 75-fold, and the yield was 46.2% ± 8.0%. After purification, only a single band appeared on Coomassie staining, exactly coinciding with the reactive band seen on Western blotting. Each mutant was purified similarly and showed a single band on both Coomassie (Figure 3A) and Western blot (Figure 3B), with an Mr = 66 kd. The expression of the mutants varied from 12% to 34% of PDE3AΔ1 (Table1).
Analysis of fractions from Ni-column chromatography.
Three fractions of wash buffer and 5 fractions of 250 mmol/L imidazole on 0.5 mol/L NaCl fractions of eluate were collected. (A) 10 μL were loaded onto a 10% SDS-PAGE gel and stained with Coomassie blue. (B) 10 μL was loaded on SDS-PAGE, transferred, and detected with anti-PDE3A polyclonal antibody as described in “Materials and methods.”
Analysis of fractions from Ni-column chromatography.
Three fractions of wash buffer and 5 fractions of 250 mmol/L imidazole on 0.5 mol/L NaCl fractions of eluate were collected. (A) 10 μL were loaded onto a 10% SDS-PAGE gel and stained with Coomassie blue. (B) 10 μL was loaded on SDS-PAGE, transferred, and detected with anti-PDE3A polyclonal antibody as described in “Materials and methods.”
Kinetic parameters of the PDE3A mutants
| . | Expression (%) . | Km μmol/L (mean ± SD) . | kcat s−1 (×100) (mean ± SD) . | kcat/Km μmol/L−1 s−1 . | ΔΔGT Kcal/mol . |
|---|---|---|---|---|---|
| PDE3AΔ1 | 100 | 0.21 ± 0.023 | 185.72 ± 23.55 | 8.759 | |
| H752A | 12 | 0.22 ± 0.029 | 0.14 ± 0.02* | 0.007 | 4.32 |
| H756A | 15 | 0.21 ± 0.034 | 0.08 ± 0.02* | 0.004 | 4.66 |
| E825A | 24 | 0.193 ± 0.015 | 4.75 ± 0.88* | 0.246 | 2.10 |
| H836A | 16 | 0.18 ± 0.185 | 0.97 ± 0.27* | 0.054 | 3.05 |
| E866A | 34 | 2.35 ± 0.308* | 23.58 ± 4.32* | 0.099 | 2.71 |
| E971A | 14 | 1.08 ± 0.253* | 2.06 ± 0.024* | 0.019 | 3.69 |
| . | Expression (%) . | Km μmol/L (mean ± SD) . | kcat s−1 (×100) (mean ± SD) . | kcat/Km μmol/L−1 s−1 . | ΔΔGT Kcal/mol . |
|---|---|---|---|---|---|
| PDE3AΔ1 | 100 | 0.21 ± 0.023 | 185.72 ± 23.55 | 8.759 | |
| H752A | 12 | 0.22 ± 0.029 | 0.14 ± 0.02* | 0.007 | 4.32 |
| H756A | 15 | 0.21 ± 0.034 | 0.08 ± 0.02* | 0.004 | 4.66 |
| E825A | 24 | 0.193 ± 0.015 | 4.75 ± 0.88* | 0.246 | 2.10 |
| H836A | 16 | 0.18 ± 0.185 | 0.97 ± 0.27* | 0.054 | 3.05 |
| E866A | 34 | 2.35 ± 0.308* | 23.58 ± 4.32* | 0.099 | 2.71 |
| E971A | 14 | 1.08 ± 0.253* | 2.06 ± 0.024* | 0.019 | 3.69 |
Mutants are listed in order of their position in the sequence. Results represent the mean of 3 determinations ± SD.
Statistical analyses were performed using Student t-test. Each mutant was compared to PDE3AΔ1.
P < .01.
Each mutant was then subjected to kinetic analysis. The enzyme catalytic efficiency was low in all cases. When compared with the truncated enzyme (8.759 μmol/L−1 second−1), the kcat/Km ranged from 0.007 to 0.25 μmol/L−1 second−1. The Km was virtually identical for all mutants compared with the truncated recombinant enzyme, except for E866A and E971A, which exhibited a 5- to 11-fold increase (Table 1). When cGMP was used as an inhibitor, the Ki for cGMP was similar for the first 2 mutants, making up the first metal-binding motif, but 5-fold elevated for E825A (Table 2). Both the first histidine of the second motif, H836A, and the glutamate residue, E866A, showed a 27- to 177-fold increase in the Ki of cGMP. The IC50 for milrinone was unaffected by any mutant except H836A, for which it was decreased. These data strongly suggested that the H836A and E866A mutants had defective cGMP binding and that only E866A had abnormal cAMP binding.
Studies of the effect of inhibitors of PDE3A and its mutants
| . | cGMP KI μmol/L (mean ± SD) . | Milrinone IC50 μmol/L (mean ± SD) . | Rolipram IC50 μmol/L . |
|---|---|---|---|
| PDE3AΔ1 | 0.76 ± 0.09 | 1.85 ± 0.26 | >100 |
| H752A | 0.84 ± 0.15 | 1.65 ± 0.23 | >100 |
| H756A | 1.01 ± 0.16 | 2.13 ± 0.41 | >100 |
| E825A | 4.03 ± 0.35* | 1.72 ± 0.17 | >100 |
| H836A | 135.3 ± 11.5* | 0.35 ± 0.10 | >100 |
| E866A | 20.6 ± 2.08* | 2.39 ± 0.37 | >100 |
| E971A | 1.06 ± 0.21 | 1.65 ± 0.24 | >100 |
| . | cGMP KI μmol/L (mean ± SD) . | Milrinone IC50 μmol/L (mean ± SD) . | Rolipram IC50 μmol/L . |
|---|---|---|---|
| PDE3AΔ1 | 0.76 ± 0.09 | 1.85 ± 0.26 | >100 |
| H752A | 0.84 ± 0.15 | 1.65 ± 0.23 | >100 |
| H756A | 1.01 ± 0.16 | 2.13 ± 0.41 | >100 |
| E825A | 4.03 ± 0.35* | 1.72 ± 0.17 | >100 |
| H836A | 135.3 ± 11.5* | 0.35 ± 0.10 | >100 |
| E866A | 20.6 ± 2.08* | 2.39 ± 0.37 | >100 |
| E971A | 1.06 ± 0.21 | 1.65 ± 0.24 | >100 |
Each value is the mean ± SD of three independent experiments. Each mutant was compared to PDE3AΔ1 by the Student t-test.
P < .01.
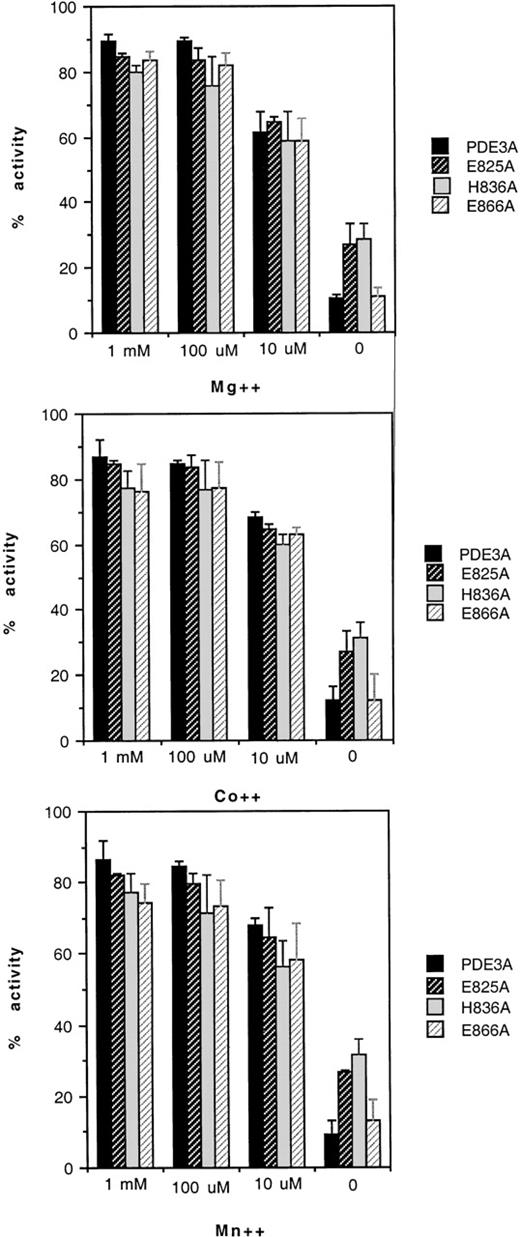
To test whether amino acids from the motifs were responsible for metal binding, the site-directed mutant enzymes were subjected to dialysis first against buffers devoid of divalent cations and assayed with buffer or with buffers containing concentrations of Mg++, Co++, and Mn++ (varying from 10 μmol/L to 1 mmol/L). The activities of H752A and H756A were too low to recover activity successfully in a control dialysis against assay buffer. However, the remainder of the mutants showed full activity, recovered in dialysis against the assay buffer (results not shown). When the truncated recombinant enzyme was dialyzed against metal-free buffer, more than 90% of the activity was lost compared with the activity before dialysis, assayed with Mg++.22 The activity with Mn++ and Co++ was 75% to 90%. E825A and H836A lost activity, but not as much as did the truncated recombinant enzyme. After dialysis against metal-free buffer, both regained activity similar to that in the assay of an undialyzed enzyme (Figure 4). The behavior of E866A with regard to metal depletion and repletion was identical to that of the unmutated recombinant enzyme.
Effects of metal ions on mutants of PDE3A.
Each enzyme was dialyzed against metal-free buffer, and the activity was measured and compared with the activity before dialysis after adding each cation at 10 μmol/L, 100 μmol/L, and 1 mmol/L, as indicated on the abscissa. Each mutant is represented by a bar, as indicated in the figures: (upper panel) MgCl2; (middle panel) CoCl2; (lower panel) MnCl2. The activity of PDE3A before dialysis is 100%.
Effects of metal ions on mutants of PDE3A.
Each enzyme was dialyzed against metal-free buffer, and the activity was measured and compared with the activity before dialysis after adding each cation at 10 μmol/L, 100 μmol/L, and 1 mmol/L, as indicated on the abscissa. Each mutant is represented by a bar, as indicated in the figures: (upper panel) MgCl2; (middle panel) CoCl2; (lower panel) MnCl2. The activity of PDE3A before dialysis is 100%.
Discussion
A limiting factor in the study of platelet PDE3A has been the quantity available. Conventional purification from outdated platelets requires starting material of 200 U outdated platelets derived from 100 L human blood. After purification of 2500-fold with a yield of approximately 20%, 100 to 200 μg enzyme is recovered.18,21 Overexpression of PDE3A in bacteria results in poor growth of the cells, presumably because of the effects of increased cAMP. Expression in yeast (S. cerevisiae) strain GL62, deficient in yeast PDE1 and PDE2, still required a purification of approximately500-fold with a yield of approximately 10%. The insect cell system first used for human myocardial PDE3A25 offers many advantages. With the use of a hexahistidine-containing construct, the purification of the recombinant enzyme on a Ni column is simple. A 1-step procedure results in a 75-fold purification, with a yield of almost 50% of an enzyme 90% to 95% homogeneous on SDS gel. The truncated recombinant enzyme PDE3AΔ1 has kcat and Km for cAMP similar to those of the platelet enzyme18 or those expressed in yeast.19 Moreover, the IC50 for cGMP and milrinone are similar, the immunologic reactivity is comparable, and the PDE3AΔ1 is cytosolic. The mutant enzymes show similar immunoreactivity when compared with PDE3AΔ1, indicating that their gross conformational structure is not altered.
As expected, the percentage expression for the mutants was reduced in comparison with the wild type by 12% to 34%. It is well-known that mutants are usually less efficiently synthesized than a wild-type enzyme because evolutionary selection optimizes the native form. Our method of calculation is only approximate and does not distinguish whether the decrease is caused by pre or posttranslational processes. Because the purpose of this study was to define the role of conserved amino acids in catalysts, our approach seemed reasonable.
Of the mutants chosen, 5 of 6 represent histidines and glutamates of the 2 canonic metal-binding motifs HXXXH (X)25E. The final histidine, H840, was mutated previously in our laboratory (Figure 5).26Histidines of the first motif substituted with alanine H752A and H756A have extremely low activities (less than 0.05% of PDE3AΔ1) with Km similar to the truncated recombinant enzyme. This result agrees with the results of Turko et al,24 who found that mutation of either of the homologous histidines in PDE5 led to a catalytic efficiency (kcat/Km) of less than 1.0% with an unchanged Km. We have shown that the mutation of H to S in the first motif of PDE4A results in a loss of most of the activity.23 Histidines in metal-dependent hydrolases can serve 2 functions. First, they can provide the protein ligands for the metal. In the case of zinc, the geometric association pattern provides for 4, 5, 6, and 8 amino-acid residues in the zinc–protein complexes27 in the structural database. Zinc is an appropriate metal for PDE3A. Even though it inhibits activity, we have shown that it competes for the activating metals Mn++, Co++, and Mg++.22 Unfortunately, the basal activity of PDE3AΔ1 in the metal-free assay is low, and, in the case of the mutants with the lowest activity (less than 0.05% of PDE3AΔ1), H752A and H756A, it was technically impossible to measure the enhancement due Mn++, Mg++ or Co++. Turko et al24 also experienced this problem in evaluating certain histidine-to-alanine mutants in PDE5. Therefore, we cannot determine whether the histidines in motif 1 are involved in metal binding. A second function for metals is catalysis, in which 1 or 2 histidines serve as general acids or bases. In such cases, 1 of the ligands is H20 because this is integral to all hydrolases. At the present time, we cannot decide between these possibilities, but formal metal-binding studies may in the future allow this distinction. These residues do not function in substrate binding because the Km is unchanged from PDE3AΔ1, nor are they at a site for cGMP or milrinone binding because the IC50 for these inhibitors is unchanged.
Alignment of catalytic region and site-directed mutants in 3 PDEs.
Alignment of part of the conserved domains of deduced amino acid sequences of members of 3 PDE families with the invariate histidine and glutamate residues that are likely to form ligands to the metal ion. The bold font indicates mutants of PDE3A from our laboratory (this report and Cheung et al26), mutants of PDE4A,23and mutants of PDE5.24
Alignment of catalytic region and site-directed mutants in 3 PDEs.
Alignment of part of the conserved domains of deduced amino acid sequences of members of 3 PDE families with the invariate histidine and glutamate residues that are likely to form ligands to the metal ion. The bold font indicates mutants of PDE3A from our laboratory (this report and Cheung et al26), mutants of PDE4A,23and mutants of PDE5.24
In many metal-binding motifs, the flanking amino acid is glutamic acid, which may be located 24 to 30 residues toward the C-terminal. In the first motif, this glutamate (E) is found in this position. If one omits the 44-amino acid insert unique for PDE3, E825 is located 25 amino acids from H756 in the first metal-binding motif. In the second motif, E866 is located 25 amino acids from H840. Despite their location in canonic binding sites for metals, alanine mutations of E825, H836, and E666 did not interfere with the enhancement of the activity of PDE3A by Mn++, Co++, and Mg++, indicating that their metal-binding activity was not impaired. In the case of E825, the 44-amino acid insert may have disrupted the proximity of this glutamate to the 2 histidines in the ternary structure.
Mutation of E825 to alanine results in an enzyme with a catalytic efficiency of 2.1% of PDEAδ1, 40 times higher than H752A or H756A. The decrease in specific activity is entirely attributable to a fall in kcat because Km is unchanged from the truncated recombinant enzyme. This decrease is therefore not caused by a change in cAMP binding. There is, however, a modest elevation (5-fold) in the binding site for cGMP as reflected by an increase in Ki. This change is specific because another inhibitor, milrinone, is not affected. Turko et al24 found that the homologous amino acid in PDE5, E632, when mutated to alanine, had an increase of the Km for cGMP of 2.5-fold.
Mutation of H836 in the second motif decreased the catalytic efficiency to approximately 0.5% of PDE3AΔ1. This decrease in activity is similar to the mutation of the homologous amino acid in PDE5, H643, to alanine, which decreases the activity to 0.4% of the unmutated enzyme,24 but it is markedly different from the H274S mutation in PDE4A that we found had 50% of the activity.23This latter difference could be caused by the mutation to serine instead of alanine, or it might be a change in function specific for PDE4. Thus, even perfectly conserved amino acids can exhibit quantitative differences in different PDEs.
Analysis of substrate binding for H836A indicates a normal Km for cAMP but a 177-fold increase in Ki for cGMP. This result supports our hypothesis, based on the results of chemical modification of histidines,21 that there are distinct amino acids in the active site that interact with cGMP to form a binding site different from that for cAMP. In contrast, the mutant E866A, located 25 amino acids C-terminal to H840, showed a Km for cAMP 11-fold elevated compared to PDE3AΔ1 and a Ki for cGMP 27-fold increased compared to the unmutated enzyme. Thus, E866 is required for cGMP and for cAMP and indicates that the cGMP and cAMP sites do share some, but not all, of their amino acid residues, accounting for the competitive inhibition shown by cAMP. A previous mutation from our laboratory, E869A, also elevates Km for cAMP and IC50 for cGMP and, thus, is important for binding to both cyclic nucleotide sites.26The homologous amino acid in PDE5 exhibits similar behavior, with an increase of the Km for cGMP of 14-fold compared to the wild-type enzyme. Finally, the mutant E971A showed a 5-fold increase in the Km for cAMP but no difference in the Ki for cGMP, indicating that this residue contributes to the cAMP site but not to the cGMP site, again supporting the existence of 2 separate sites that overlap.
In summary, we have constructed 6 new mutants in PDE3A in addition to 6 already published (Figure 5).26 The 2 histidines in the first metal-binding motif show the most profound loss of activity because of involvement in catalysis, metal binding, or both. Any interaction with these histidines by the conserved glutamate, which in the other PDE families follows the histidine by 24 to 30 amino acids, is disrupted by the unique 44-amino acid insert. Thus, it does not play a role in metal binding or catalysis but appears to interact with cGMP. The second motif is involved with substrate (cAMP) and inhibitor (cGMP) binding rather than metal binding. The occurrence of amino acids whose mutation disrupts only cAMP binding, only cGMP binding, or both further reinforces the suggestion of separate but overlapping binding sites. Finally, all the mutations involved alanine replacement, and further studies should involve more conservative replacements such as glutamine for histidine.
Acknowledgment
We thank Rita Stewart for skillful article preparation.
Supported in part by grants from the National Heart, Lung, and Blood Institute (NHLB I) (nos. P01-HL64943 and T32-HL07777).
Reprints:Robert W. Colman, Sol Sherry Thrombosis Research Center, Temple University School of Medicine, 3400 North Broad St, Philadelphia, PA 19140; e-mail: colmanr@astro.temple.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal