Abstract
In higher eukaryotes, the expression of about 1 gene in 10 is strongly regulated at the level of messenger RNA (mRNA) translation into protein. Negative regulatory effects are often mediated by the 5′-untranslated region (5′-UTR) and rely on the fact that the 40S ribosomal subunit first binds to the cap structure at the 5′-end of mRNA and then scans for the first AUG codon. Self-complementary sequences can form stable stem-loop structures that interfere with the assembly of the preinitiation complex and/or ribosomal scanning. These stem loops can be further stabilized by the interaction with RNA-binding proteins, as in the case of ferritin. The presence of AUG codons located upstream of the physiological start site can inhibit translation by causing premature initiation and thereby preventing the ribosome from reaching the physiological start codon, as in the case of thrombopoietin (TPO). Recently, mutations that cause disease through increased or decreased efficiency of mRNA translation have been discovered, defining translational pathophysiology as a novel mechanism of human disease. Hereditary hyperferritinemia/cataract syndrome arises from various point mutations or deletions within a protein-binding sequence in the 5′-UTR of the L-ferritin mRNA. Each unique mutation confers a characteristic degree of hyperferritinemia and severity of cataract in affected individuals. Hereditary thrombocythemia (sometimes called familial essential thrombocythemia or familial thrombocytosis) can be caused by mutations in upstream AUG codons in the 5′-UTR of the TPO mRNA that normally function as translational repressors. Their inactivation leads to excessive production of TPO and elevated platelet counts. Finally, predisposition to melanoma may originate from mutations that create translational repressors in the 5′-UTR of the cyclin-dependent kinase inhibitor–2A gene.
Most human genetic disorders are caused by mutations affecting the protein-coding regions of genes, eg, missense or frame-shift mutations within exons or mutations within introns that disrupt pre–messenger RNA (pre-mRNA) processing.1Dysregulated gene expression at the level of transcription is another well-known, although less frequently encountered, mechanism in human disease.2 Recently, mutations that cause disease through increased or decreased efficiency of mRNA translation have been discovered, defining translational pathophysiology as a novel mechanism of human disease.
Translation of mRNA into protein was known to be a necessary and important step in gene expression, but until recently was largely considered to be a constitutive process. In the last few years, however, a variety of regulatory mechanisms acting at the level of translation have been discovered.3 In higher eukaryotes, the expression of about 1 gene in 10 is strongly regulated at this level. Translational control enables a cell to increase the concentration of a protein very rapidly and therefore appears to be especially suited to regulate genes implicated in cell proliferation and damage prevention.3 This article will review some of the basic molecular mechanisms involved in the regulation of translational efficiency and will illustrate the emerging principles of translational pathophysiology by describing the first examples of human disease attributable to mutations in mRNA regulatory sites.
Translational control of gene expression
The presence of equal amounts of different mRNA species does not necessarily ensure synthesis of equivalent quantities of the corresponding proteins. In fact, some mRNAs that reach the cytoplasm are not translated at all. Thus, negative or positive controls can influence the rate of translation.3 These controls can be exerted either by a global machinery that simultaneously affects all cellular mRNAs or by more specific mechanisms that influence individual mRNAs or subsets of mRNAs.4 5
Global control of translation is mediated by posttranslational modifications of initiation factors and ribosomal proteins. This type of control is especially important in early embryogenesis and during specific stages of the cell cycle. In contrast, the specific mechanisms operate through sequence motifs (cis-acting elements) in the mRNA untranslated regions (UTRs). The best-studied examples of translational regulation are mediated by the 5′-UTR and involve the control of translation initiation. The 3′-UTR most often affects translation indirectly by determining mRNA stability (eg, transferrin receptor; see “Translational control by proteins that bind stem-loop structures”). However, direct effects of the 3′-UTR on translation initiation or polyadenylation have also been described.6
Negative regulatory elements in the 5′-UTR can interfere with initiation of translation
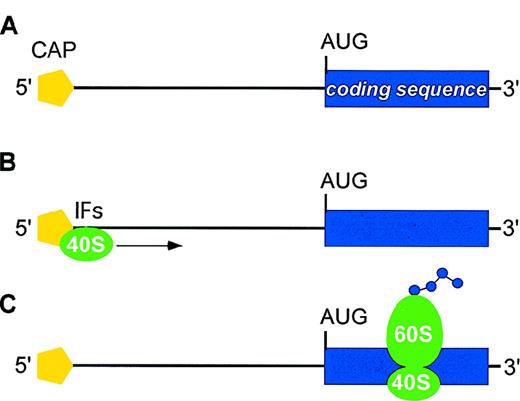
The majority of eukaryotic mRNAs are translated according to the scanning model of translation (Figure1). A preinitiation complex consisting of the ribosomal 40S subunit and several initiation factors (IFs) is assembled at the 5′-cap structure of mRNA (cap-dependent mechanism of initiation). This initiation complex then migrates along the 5′-UTR in search of an appropriate initiator AUG codon (scanning process). Recognition of an AUG leads to assembly of the 60S ribosomal subunit and initiation of protein synthesis. Sequence-specific inhibition of translation is mediated by negative regulatory elements present in the 5′-UTR of individual mRNAs that interfere with efficient translation initiation at the physiological start site. Two main categories of negative regulatory elements are known: secondary structure of mRNA and upstream open reading frames (uORFs) in the 5′-UTR (Figure2; Table1).7-41
Main steps in translation of eukaryotic mRNA.
(A) mRNAs display two structural features that are important for translation initiation: the methylated cap at the 5′ end (yellow pentagon) and the AUG initiation codon at the start of the coding region (blue box). The distance between the 5′-cap and the initiator AUG may range from about 40 to more than 1000 bases, although it usually is less than 100 bases. (B) A preinitiation complex consisting of the ribosomal 40S subunit (green) and several initiation factors (IFs) is assembled at the the methylated 5′-cap. This initiation complex then migrates along the 5′-UTR in search of an AUG initiation codon (scanning process). (C) Recognition of an AUG leads to assembly of the 60S ribosomal subunit and initiation of protein synthesis.
Main steps in translation of eukaryotic mRNA.
(A) mRNAs display two structural features that are important for translation initiation: the methylated cap at the 5′ end (yellow pentagon) and the AUG initiation codon at the start of the coding region (blue box). The distance between the 5′-cap and the initiator AUG may range from about 40 to more than 1000 bases, although it usually is less than 100 bases. (B) A preinitiation complex consisting of the ribosomal 40S subunit (green) and several initiation factors (IFs) is assembled at the the methylated 5′-cap. This initiation complex then migrates along the 5′-UTR in search of an AUG initiation codon (scanning process). (C) Recognition of an AUG leads to assembly of the 60S ribosomal subunit and initiation of protein synthesis.
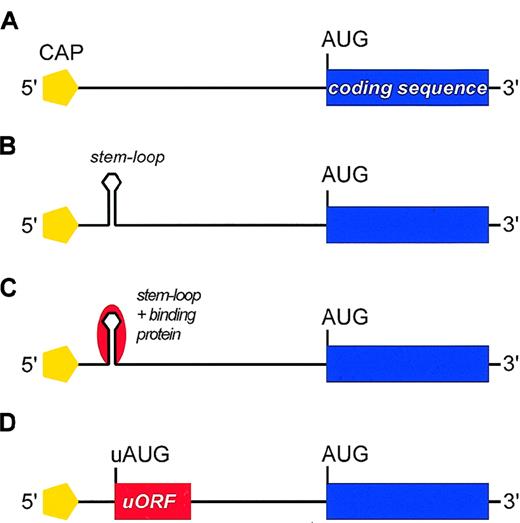
Negative regulatory elements in the 5′-UTR of mRNA that may repress translation.
(A) Schematic representation of mRNA with no negative regulatory element. (B) Self-complementary sequences within the 5′-UTR can form stable stem-loop structures (hairpins) located between the 5′-cap and the first AUG codon that interfere with assembling of the preinitiation complex and/or with ribosomal scanning. The stem-loop structure may be stable enough to resist the unwinding activity of the associated helicase, thus imposing a major barrier to conventional ribosomal scanning. (C) In some mRNAs, the stem loop can be further stabilized by the interaction with RNA-binding proteins (red). (D) The 5′-UTR may contain AUG codons upstream of the physiological start site (uAUG) that define short upstream ORFs (uORFs). These uAUGs can cause premature initiation and inhibit translation by preventing the ribosome from reaching the physiological start codon.
Negative regulatory elements in the 5′-UTR of mRNA that may repress translation.
(A) Schematic representation of mRNA with no negative regulatory element. (B) Self-complementary sequences within the 5′-UTR can form stable stem-loop structures (hairpins) located between the 5′-cap and the first AUG codon that interfere with assembling of the preinitiation complex and/or with ribosomal scanning. The stem-loop structure may be stable enough to resist the unwinding activity of the associated helicase, thus imposing a major barrier to conventional ribosomal scanning. (C) In some mRNAs, the stem loop can be further stabilized by the interaction with RNA-binding proteins (red). (D) The 5′-UTR may contain AUG codons upstream of the physiological start site (uAUG) that define short upstream ORFs (uORFs). These uAUGs can cause premature initiation and inhibit translation by preventing the ribosome from reaching the physiological start codon.
Proteins regulated at the translational level by negative regulatory elements in the 5′-UTR of mRNA
| Translational control mechanism . | Specific regulation . | Reference . |
|---|---|---|
| Stable stem-loop structures that interfere with initiation of translation | ||
| PDGF2 | IRES | 7, 8 |
| TGF-β1 | 9 | |
| Ornithine decarboxylase | 10, 11 | |
| Stem-loop structures that may be stabilized by mRNA-binding proteins | ||
| Ferritin (H- and L-subunit) | IRPs | 12, 13 |
| Transferrin receptor | IRPs | |
| Erythroid-specific δ-aminolevulinate synthase (ALAS2) | IRPs | |
| DMT1 (or Nramp 2) | IRPs | 14 |
| IGF-II | IMPs and IRES | 15, 16 |
| APP | 17 | |
| uORFs that repress translation | ||
| TPO | 18 | |
| IL-7 | 19 | |
| IL-15 | 20, 21 | |
| TGF-β3 | 22, 23 | |
| VEGF | IRES | 24 |
| Bcl-2 | 25 | |
| p18(INK4c) | 26 | |
| uORF-encoded peptides that inhibit translation | ||
| S-adenosylmethionine decarboxylase | 27 | |
| β2-adrenergic receptor | 28, 29 | |
| Alternative in frame initiation codons | ||
| C/EBPα | 30 | |
| C/EBPβ (LIP and LAP) | 31 | |
| int-2 | 32 | |
| bFGF | IRES | 33, 34, 35 |
| c-myc | IRES | 36, 37, 38 |
| pim-1 | 39 | |
| BAG-1 | 40, 41 |
| Translational control mechanism . | Specific regulation . | Reference . |
|---|---|---|
| Stable stem-loop structures that interfere with initiation of translation | ||
| PDGF2 | IRES | 7, 8 |
| TGF-β1 | 9 | |
| Ornithine decarboxylase | 10, 11 | |
| Stem-loop structures that may be stabilized by mRNA-binding proteins | ||
| Ferritin (H- and L-subunit) | IRPs | 12, 13 |
| Transferrin receptor | IRPs | |
| Erythroid-specific δ-aminolevulinate synthase (ALAS2) | IRPs | |
| DMT1 (or Nramp 2) | IRPs | 14 |
| IGF-II | IMPs and IRES | 15, 16 |
| APP | 17 | |
| uORFs that repress translation | ||
| TPO | 18 | |
| IL-7 | 19 | |
| IL-15 | 20, 21 | |
| TGF-β3 | 22, 23 | |
| VEGF | IRES | 24 |
| Bcl-2 | 25 | |
| p18(INK4c) | 26 | |
| uORF-encoded peptides that inhibit translation | ||
| S-adenosylmethionine decarboxylase | 27 | |
| β2-adrenergic receptor | 28, 29 | |
| Alternative in frame initiation codons | ||
| C/EBPα | 30 | |
| C/EBPβ (LIP and LAP) | 31 | |
| int-2 | 32 | |
| bFGF | IRES | 33, 34, 35 |
| c-myc | IRES | 36, 37, 38 |
| pim-1 | 39 | |
| BAG-1 | 40, 41 |
UTR indicates untranslated region; mRNA, messenger RNA; PDGF, platelet-derived growth factor; TGF, transforming growth factor; ALAS2, δ-aminolevulinate synthase; DMT, dimethyltryptamine; IGF, insulinlike growth factor; APP, amyloid precursor protein; uORF, upstream open reading frame; TPO, thrombopoietin; IL, interleukin; VEGF, vascular endothelial growth factor; bFGF, basic fibroblast growth factor; LIP, liver inhibitory protein; LAP, liver activator protein; IRES, internal ribosomal entry site; IRPs, iron regulatory proteins; IMPs, IGF II mRNA binding proteins.
Self-complementary sequences within the 5′-UTR can form stable stem-loop structures. When located near the 5′-cap, the stem loop may interfere with the assembly of the preinitiation complex. Stem loops located further downstream may be stable enough to resist the unwinding activity of the ribosome-associated helicase (Figure 2B) and thereby impose a major barrier to conventional ribosomal scanning (eg, mRNA for human platelet-derived growth factor 2 [PDGF2]) (Table1).8 Although less stable stem loops (free energies up to −126 kJ/mol) can be melted by the helicase,1 such hairpins can be further stabilized by the interaction with RNA-binding proteins (Figure 2C). In the case of mRNAs for ferritin and the erythroid-specific δ-aminolevulinate synthase (ALAS2), association with such RNA-binding proteins results in strong inhibition of translation (see “Translational control by proteins that bind stem-loop structures”).
The other major mechanism of translational inhibition involves recognition of AUG codons located upstream of the physiological start site (uAUG) by the scanning 40S subunit (Figure 2D).42uAUGs that are followed by stop codons can cause premature initiation and inhibit translation by preventing the ribosome from reaching the physiological start codon (eg, thrombopoietin [TPO], interleukin (IL)–7 and IL-15; Table 1). The sequences of these uORFs can be irrelevant, as demonstrated extensively for GCN4, the Jun homolog in yeast.43 By contrast, in the case of S-adenosylmethionine-decarboxylase,44 the β2-adrenergic receptor,28 and the human cytomegalovirus gpUL4 gene,45 an additional inhibitory effect on translation has been demonstrated for the uORF-encoded peptide. Finally, some genes are transcribed from alternative promoters resulting in mRNAs with variable 5′-ends that carry uAUGs in the same reading frame as the main translational product (eg, bFGF or c-myc). Use of such alternative initiation sites generates products with alternative N-termini that are either active or inactive, or that exert different functions. For instance, the transcription factor C/EBPβ translated from an upstream AUG functions as a transcriptional liver activator protein, whereas that translated from a downstream AUG behaves as a liver inhibitory protein. In some cases, inhibition of translation can be bypassed by initiation at internal ribosomal entry sites located downstream of stem loops or uORFs (eg, c-myc, PDGF, or VEGF; Table 1).
Translational control by proteins that bind stem-loop structures: the control of cellular iron metabolism as an example of highly integrated translational regulation
Iron, as an essential constituent of ribonucleotide reductase, a key enzyme in DNA synthesis, is required for growth and cell division. Physiologically, the transport, cellular uptake, and storage of iron are carried out by 3 proteins: transferrin, transferrin receptor, and ferritin. Ferritin is a protein shell with a molecular weight of about 500 kd made up of 24 subunits. The multiple forms, or isoferritins, that can be found in human tissues are composed of variable proportions of 2 subunits: L-ferritin (light, 19 kd, 174 residues) and H-ferritin (heavy, 21 kd, 182 aminoacids), encoded by genes located on chromosome 11 and 19, respectively. Two other proteins, divalent metal transporter 1 (DMT1) also called Nramp2,46 and HFE,47mediate and regulate iron absorption.
Cellular iron homeostasis in mammalian cells is maintained by the coordinated regulation of transferrin receptor and ferritin synthesis that occur at the translational level and is mediated by cytoplasmic mRNA-binding proteins, known as iron regulatory proteins (IRPs) (Figure 3).13 These proteins are capable of sensing cellular iron status and of interacting with mRNA stem-loop structures known as iron-responsive elements (IREs). IREs constitute the first well-characterized family of cis-acting noncoding regulatory sequences in eukaryotic mRNA.12 A single functional IRE is found in the 5′-UTR of mRNAs for ferritin H and L subunits and in ALAS2. In contrast, multiple IREs are present in the 3′-UTR of the mRNA for transferrin receptor, and a single (nonconsensus) IRE has been recently found in the 3′ UTR of the DMT1 gene.48
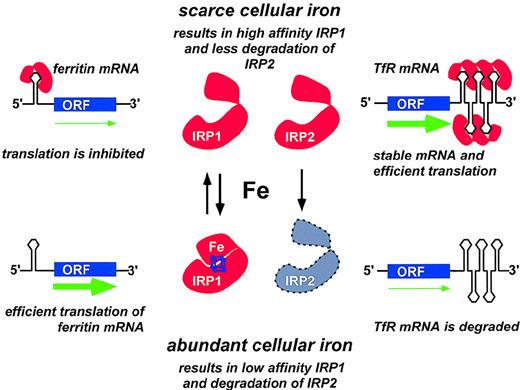
Coordinate regulation of transferrin receptor and ferritin synthesis through translational controls operated by the iron-responsive elements (IREs) and by the iron regulatory proteins IRP1 and IRP2.
Only one IRE is present in the 5′-UTR of ferritin mRNA. When cellular iron is scarce, IRP molecules are available for binding the 5′ IRE; initiation of translation is prevented; and ferritin synthesis is inhibited. By contrast, presence of abundant intracellular iron prevents binding of IRPs to the 5′ IRE and allows efficient mRNA translation to proceed (green arrow). Five IREs are present in the 3′-UTR of transferrin receptor (TfR) mRNA. When cellular iron is scarce, binding of one or more IRPs to the IREs in the 3′-UTR stabilizes TfR mRNA and increases TfR translation. Conversely, when iron is abundant, very few IREs are occupied by IRPs, and TfR mRNA is rapidly degraded. Adapted from Brittenham GM, Olivieri NF, Rouault TA. Iron physiology and iron overload. Hematology 1996. The American Society of Hematology, Orlando, FL, 1996, p. 177.
Coordinate regulation of transferrin receptor and ferritin synthesis through translational controls operated by the iron-responsive elements (IREs) and by the iron regulatory proteins IRP1 and IRP2.
Only one IRE is present in the 5′-UTR of ferritin mRNA. When cellular iron is scarce, IRP molecules are available for binding the 5′ IRE; initiation of translation is prevented; and ferritin synthesis is inhibited. By contrast, presence of abundant intracellular iron prevents binding of IRPs to the 5′ IRE and allows efficient mRNA translation to proceed (green arrow). Five IREs are present in the 3′-UTR of transferrin receptor (TfR) mRNA. When cellular iron is scarce, binding of one or more IRPs to the IREs in the 3′-UTR stabilizes TfR mRNA and increases TfR translation. Conversely, when iron is abundant, very few IREs are occupied by IRPs, and TfR mRNA is rapidly degraded. Adapted from Brittenham GM, Olivieri NF, Rouault TA. Iron physiology and iron overload. Hematology 1996. The American Society of Hematology, Orlando, FL, 1996, p. 177.
Two IRP family members, IRP1 and IRP2, have been identified in humans.49-51 Under conditions of intracellular iron depletion, both IRP1 and IRP2 function as RNA-binding proteins that bind IREs with high affinity (Figure 3). The binding of an IRP to the ferritin IRE prevents the association of the 43S translation preinitiation complex with the mRNA52 by precluding the recruitment of the small ribosomal subunit.53 Thereby, translation of the ferritin protein is repressed. Conversely, binding of IRPs to the IREs in the 3′-UTR of transferrin receptor increases the stability of mRNA and improves the efficiency of translation. When intracellular iron concentrations rise, the IRPs dissociate from the IREs. IRP1 may acquire a (4Fe-4S) cluster and behave as a cytosolic aconitase.54 IRP1 exhibits approximately 30% sequence homology to mitochondrial aconitase. IRP2 lacks aconitase activity and functions solely as an RNA-binding protein. At elevated intracellular iron concentrations, IRP2 is targeted for degradation by the proteasome.55
Thus, translational regulation by IRPs allows rapid and coordinated control of proteins that are crucial for cellular iron homeostasis. When cells have adequate iron, the expression of transferrin receptors decreases and the levels of ferritin rise to accommodate the excess iron. On the other hand, when cellular iron becomes scarce, the levels of ferritin fall while the expression of transferrin receptors increases to import more iron from the outside.
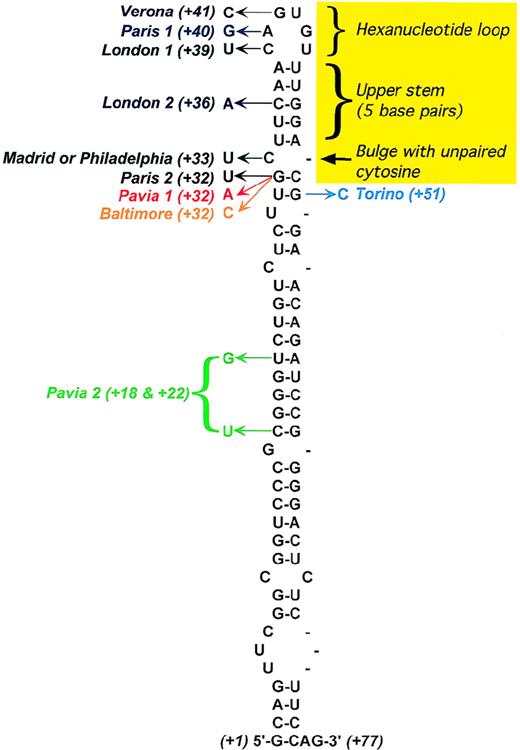
IREs can form a highly conserved stem-loop structure (Figure4). The conserved features include a hexanucleotide loop with the sequence CAGYCX, where Y = U or A, and X = U, C, or A (the first 5 bases are almost always CAGUG), and an upper stem—consisting of 5 base pairs—that is separated from a lower stem of variable length by an unpaired cytosine, which forms the bulge.
Secondary structure of the L-ferritin IRE.
The yellow area marks conserved features of the IRE consensus based on the IRE sequences of all known ferritin and transferrin receptor mRNAs. Positions of the point mutations responsible for hereditary hyperferritinemia/cataract syndrome are shown, with the arrow indicating the observed nucleotide substitution. In addition, the following deletions have been described: C10-A38,64A38-C39, and U42-G57.71 Adapted from Allerson et al.75
Secondary structure of the L-ferritin IRE.
The yellow area marks conserved features of the IRE consensus based on the IRE sequences of all known ferritin and transferrin receptor mRNAs. Positions of the point mutations responsible for hereditary hyperferritinemia/cataract syndrome are shown, with the arrow indicating the observed nucleotide substitution. In addition, the following deletions have been described: C10-A38,64A38-C39, and U42-G57.71 Adapted from Allerson et al.75
Additional translational controls are likely to exist, at least for ferritin. In fact, IL-1 and IL-6 also appear to elevate ferritin synthesis by translational mechanisms during inflammation, and a translational enhancer has been found in the L-ferritin mRNA 5′UTR.56 These sequences are distinct from the IRE and similar to a consensus reported for the 5′ leaders of other acute phase response mRNAs.
Hereditary hyperferritinemia/cataract syndrome as an established model of translational pathophysiology
Several reports in the last few years have described a new autosomal dominant disorder called hereditary hyperferritinemia/cataract syndrome (HHCS).57-74 This condition is characterized by a combination of elevated serum ferritin in the absence of iron overload and early-onset nuclear cataract.
With the use of monoclonal antibodies specific for the H- and L-ferritin subunits, the elevated serum ferritin in HHCS patients has been shown to be of the L-type.63 In addition, a close relationship has been established between mononuclear-cell L-type ferritin content and serum ferritin concentration, indicating that the excess production of ferritin in cells is directly responsible for the hyperferritinemia. This dysregulated L-subunit synthesis was found to result from different point mutations or deletions in the 5′-UTR IRE of the L-ferritin gene (Figure 4). The observed molecular lesions were shown to reduce the IRE affinity for the IRPs, which normally inhibit ferritin mRNA translation, thereby causing increased production of L-ferritin.
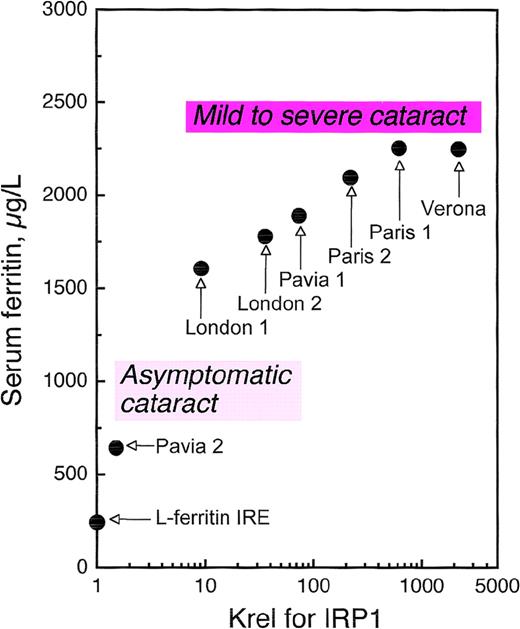
As several families with HHCS have been characterized, it has become clear that not all patients are equally affected. Some individuals have only slightly elevated serum ferritin and are asymptomatic for cataract, while others have serum ferritin levels more than 10-fold higher than normal and a history of severe cataract.63Allerson et al75 measured the in vitro affinity of the IRPs for the mutant IREs from HHCS patients and correlated decreases in binding affinity with clinical severity, showing close relationships with both degree of hyperferritinemia and cataract severity (Figure5). These investigators also used thermal denaturation methods to determine the effects of each HHCS mutation on the thermodynamic stability of the IRE. This analysis revealed that while some HHCS mutations lead to changes in the stability and secondary structure of the IRE, others appear to disrupt IRP-IRE recognition with minimal effect on IRE stability. The pathogenesis of cataract is not yet clear. It is likely that lens opacities are not congenital but rather develop during the first few years of life69,74 and appear to be a consequence of the elevated ferritin concentrations in the lens itself. In fact, Levi et al76 have found that a large proportion of L-ferritin accumulates as nonfunctional L-chain 24 homopolymers in HHCS cells, and that L-chain accumulation occurs also in the lens, where it probably induces cataract formation.
Correlation of serum ferritin with relative dissociation constants (Krel) of IRP1 complexes with wild-type IRE and IREs containing HHCS mutations.
Serum ferritin levels are maximal values observed in normal individuals and families with HHCS. A significant relationship was found between ferritin levels and dissociation constants: the higher the impairment in IRE-IRP1 binding affinity, the higher the serum ferritin.75 The relationship between degree of hyperferritinemia and severity of cataract75 is illustrated schematically for the reader's convenience. Modified from Allerson et al.75
Correlation of serum ferritin with relative dissociation constants (Krel) of IRP1 complexes with wild-type IRE and IREs containing HHCS mutations.
Serum ferritin levels are maximal values observed in normal individuals and families with HHCS. A significant relationship was found between ferritin levels and dissociation constants: the higher the impairment in IRE-IRP1 binding affinity, the higher the serum ferritin.75 The relationship between degree of hyperferritinemia and severity of cataract75 is illustrated schematically for the reader's convenience. Modified from Allerson et al.75
Thus, HHCS stands as a noteworthy example of translational pathophysiology. This human genetic disorder originates from RNA mutations within a protein-binding site, and its severity is determined by the energetics of the binding interaction, which varies considerably among different mutations (Figures 4, 5). A recent report of a de novo mutation in HHCS indicates that this disease should be searched for even in sporadic cases of early-onset cataract formation.77
uAUG codons as translational repressors: thrombopoietin as an example of an mRNA physiologically repressed by a translational mechanism
The production of several proto-oncogenes, cytokines, and other tightly regulated genes is in part controlled by the presence of uORFs in the 5′-UTR (Table 1 and references therein). The extent of uAUG-mediated inhibition of translation depends mainly on 2 factors. The first prerequisite for a strong inhibitory effect is efficient translational initiation at the uAUGs, which is strongly influenced by the sequence context of each individual uAUG (Kozak consensus). The rate of initiation at uAUGs rarely reaches 100%; therefore, some leaky scanning usually occurs, maintaining a basal level of translation from the physiological start site. The second factor determining the efficiency of translational inhibition is the rate of ribosomal reinitiation. After translation of the uORF and the encounter with the uORF stop codon, the 40S ribosomal subunit may remain associated with the mRNA and resume scanning for the next AUG codon. The rate of reinitiation depends on the distance between the stop codon and the next available AUG. A minimal intercistronic gap of 16 nucleotides was shown to be required to allow ribosomal reinitiation.78
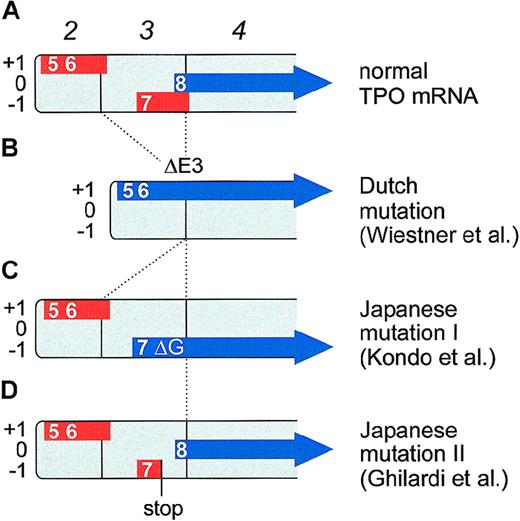
These principles are illustrated by the example of TPO mRNA (Figure6A). Translation of TPO mRNA is strongly inhibited by the presence of several uAUG codons in the 5′-UTR.18 Directed mutagenesis of all uAUGs in the TPO mRNA restored full translational efficiency, demonstrating that translational inhibition of TPO biosynthesis is entirely mediated by uORFs. The uORF defined by the seventh uAUG was shown to exert the strongest negative effect on translation.18 This uAUG is in a good Kozak consensus and the uORF extends beyond the physiological start site, thus preventing reinitiation (Figure 6A). The presence of this uAUG and the length of the resulting uORF are conserved between human, mouse, and rat TPO mRNAs.
Effect of TPO gene mutations on the composition of uORFs in TPO mRNA.
TPO transcripts starting from the main promoter in exon 2 are shown. Boxes numbered in italics represent exons. The uORFs are drawn as thick red lines and are placed into one of the three reading frames (+1, 0, and −1). The TPO coding region is shown as a thick blue arrow. Numbers indicate the order in which the uAUGs appear in the full-length TPO mRNA (therefore, uAUGs 1 through 4 are not shown). The eighth AUG is the physiological initiation codon. (A) Translation of normal TPO mRNA is physiologically almost completely inhibited by the presence of uORFs in the 5′-UTR. In particular, the uORF 7 is a potent inhibitor of translation, most likely because of its extension beyond the physiological start site. (B) A splice donor mutation in the Dutch HT family causes exon 3 skipping (ΔE3) that deletes uORF7 and shifts the TPO coding sequence into reading frame +1. TPO translation now initiates from the fifth and sixth AUGs. (C) The Japanese mutation I consists of a single G nucleotide deletion (ΔG) that shifts the TPO coding sequence into reading frame −1. TPO translation now initiates from the seventh AUG. Note that both the Dutch and the Japanese mutation I create altered TPO signal peptides, but do not alter the sequence of the mature TPO protein. Both signal peptides remain functionally active and promote secretion of a biologically active TPO protein. (D) The Japanese mutation II creates a premature stop codon in uORF7. This allows reinitiation of translation at the physiological start site (the eighth AUG).
Effect of TPO gene mutations on the composition of uORFs in TPO mRNA.
TPO transcripts starting from the main promoter in exon 2 are shown. Boxes numbered in italics represent exons. The uORFs are drawn as thick red lines and are placed into one of the three reading frames (+1, 0, and −1). The TPO coding region is shown as a thick blue arrow. Numbers indicate the order in which the uAUGs appear in the full-length TPO mRNA (therefore, uAUGs 1 through 4 are not shown). The eighth AUG is the physiological initiation codon. (A) Translation of normal TPO mRNA is physiologically almost completely inhibited by the presence of uORFs in the 5′-UTR. In particular, the uORF 7 is a potent inhibitor of translation, most likely because of its extension beyond the physiological start site. (B) A splice donor mutation in the Dutch HT family causes exon 3 skipping (ΔE3) that deletes uORF7 and shifts the TPO coding sequence into reading frame +1. TPO translation now initiates from the fifth and sixth AUGs. (C) The Japanese mutation I consists of a single G nucleotide deletion (ΔG) that shifts the TPO coding sequence into reading frame −1. TPO translation now initiates from the seventh AUG. Note that both the Dutch and the Japanese mutation I create altered TPO signal peptides, but do not alter the sequence of the mature TPO protein. Both signal peptides remain functionally active and promote secretion of a biologically active TPO protein. (D) The Japanese mutation II creates a premature stop codon in uORF7. This allows reinitiation of translation at the physiological start site (the eighth AUG).
TPO is the most potent humoral regulator of platelet formation.79 Physiologically, TPO serum concentrations are very low, ranging between 0.5 and 2 pmol/L. Circulating TPO is mainly produced by the liver and the kidneys, and TPO mRNA levels in these organs remain unchanged during thrombocytopenia.80 81However, it remains an open question whether TPO translation might be up-regulated in response to increased platelet demand. Irrespective of this possibility, translational repression is important to prevent overproduction of TPO, because the loss of this repression mechanism through germ-line mutations affecting the TPO 5′-UTR results in elevated TPO serum levels and thrombocythemia (see “Hereditary thrombocythemia as an example of uAUG-mediated translational pathophysiology”).
Hereditary thrombocythemia as an example of uAUG-mediated translational pathophysiology
Hereditary thrombocythemia (HT) (sometimes called familial essential thrombocythemia or familial thrombocytosis) is characterized by sustained proliferation of megakaryocytes, resulting in elevated platelet counts and thrombotic or hemorrhagic complications. In most kindreds, HT is inherited as an autosomal dominant trait with high penetrance and early age of onset. The clinical features of HT are indistinguishable from those of sporadic essential thrombocythemia (ET), a chronic myeloproliferative disorder. Hereditary syndromes resembling ET have been described in a number of families.82-99 In 4 of these families, thrombocythemia was found to be caused by gain of function mutations in the TPO gene, which result in systemic TPO overproduction (Table2). Interestingly, all 4 mutations affect the 5′-UTR of TPO mRNA.
Summary of gene mutations causing hereditary thrombocythemia
| Initial case report . | Mode of inheritance . | Plt count . | Complications . | Gene mutation . | Mechanism . | Remarks . |
|---|---|---|---|---|---|---|
| Schlemper et al82 | AD, 11 affected in 4 generations | 533-1516 | Vaso-occlusive | G > C in TPO intron 3, position +183 | Loss of uORFs through exon skipping83 | Leukemoid reaction in 1 child |
| Kondo et al84 | AD, 5 affected in 3 generations | 847-1600 | ΔG in TPO 5′-UTR84 | Frame shift in uORF785 | ||
| Kikuchi et al86 | AD, 4 affected in 3 generations | 833-1986 | G > T in TPO 5′-UTR87 | Premature stop codon in uORF787 | ||
| Jorgensen et al88 | AD | 700-1000 | Vaso-occlusive | A > G in TPO intron 3, position +588 | Polyclonal hematopoiesis |
| Initial case report . | Mode of inheritance . | Plt count . | Complications . | Gene mutation . | Mechanism . | Remarks . |
|---|---|---|---|---|---|---|
| Schlemper et al82 | AD, 11 affected in 4 generations | 533-1516 | Vaso-occlusive | G > C in TPO intron 3, position +183 | Loss of uORFs through exon skipping83 | Leukemoid reaction in 1 child |
| Kondo et al84 | AD, 5 affected in 3 generations | 847-1600 | ΔG in TPO 5′-UTR84 | Frame shift in uORF785 | ||
| Kikuchi et al86 | AD, 4 affected in 3 generations | 833-1986 | G > T in TPO 5′-UTR87 | Premature stop codon in uORF787 | ||
| Jorgensen et al88 | AD | 700-1000 | Vaso-occlusive | A > G in TPO intron 3, position +588 | Polyclonal hematopoiesis |
AD indicates autosomal dominant; TPO, thrombopoietin uORF, upstream open reading frame.
The question of how these mutations cause TPO overproduction was first elucidated in a Dutch HT family (Figure 6B).82,83 Affected family members carry a point mutation in the +1 position of the splice donor of intron 3. This G→C transversion causes exon skipping and results in loss of exon 3 that normally encodes a large part of the 5′-UTR. As a consequence, the mutant TPO mRNA lacks uORF7, which normally inhibits translation.18 Furthermore, a novel N-terminus is created by fusion of uORF5 with the TPO coding sequence. The resulting extended N-terminus was shown to be a functional signal peptide.83 Interestingly, an A→G substitution 4 bases downstream of the Dutch mutation in splice donor 3 has been reported in another HT-family.88 Although this mutation does not affect the most conserved residues of the splice donor, it is expected to cause the same aberrant splicing as the Dutch mutation.
A completely different mutation was recently found in a Japanese family with HT.84 Affected family members carry a single G-nucleotide deletion (ΔG) in the 5′-UTR of the TPO gene (Figure 6C). This ΔG causes a frameshift in the 5′-UTR of TPO mRNA, which places uORF7 in frame with the TPO coding sequence, neutralizing the strong inhibitory effect of uORF7 and again creating a novel N-terminus for the TPO signal peptide.85 Similar to the Dutch mutation, this novel N-terminus still functions as a signal peptide that can assure secretion of correctly processed, biologically active TPO protein.85 Thus, through a completely different mutation, translational repression is lost, resulting in TPO overproduction and thrombocythemia. Finally, in an unrelated Japanese HT family,86 a G→T mutation in the TPO 5′-UTR creates a novel stop codon, which shortens uORF7 by 42 nucleotides (Figure 6D).87 This generates a gap of 31 nucleotides between uORF7 and the physiological TPO start codon, which allows translational reinitiation, resulting in enhanced translational efficiency and overproduction of TPO.87
These 4 independent TPO gene mutations illustrate the physiological importance of the translational repression for TPO regulation. These examples also suggest that there are no platelet-sensing mechanisms that, under steady state conditions, are able to detect thrombocytosis and down-regulate TPO production in patients carrying the mutations.
Translational pathophysiology as a molecular basis of hereditary predisposition to melanoma
Several authors have documented familial inheritance in malignant melanoma.100 Approximately 10% of malignant neoplasms of melanocytes are inherited in an autosomal dominant fashion with variable penetrance, and in about two thirds of affected families, a chromosome 9p21 locus has been linked to this condition. Cyclin-dependent kinase inhibitor–2A (CDKN2A) (also known as p16, INK4, p16INK4A, and MTS1) maps to chromosome 9p21.101 This gene encodes a cdk4/cdk6 kinase inhibitor that constrains cells from progressing through the G1 restriction point. In the last few years, germ-line mutations in the CDKN2A coding sequence that result in loss of CDKN2A function have been described in families predisposed to melanoma. In a recent study, the incidence of CDKN2A mutations in families with 3 or more cases of melanoma and at least 1 member with multiple primary melanomas was found to be about 30%.102 A considerable proportion of mutation-negative families nevertheless demonstrate linkage of inherited melanoma to 9p21 markers, suggesting either that the majority of mutations in the CDKN2A gene causing malignant melanoma fall outside the CDKN2A coding sequence or that CDKN2A is not the only chromosome 9p melanoma-susceptibility locus.103
A subset of these kindreds with no coding-region mutation have recently been shown to carry a G→T transversion at position −34 in the 5′-UTR of CDKN2A mRNA.104 This mutation creates a novel uAUG and a novel uORF that inhibits translation from the physiological start site, thus leading to the loss of function of this allele. The G→34T mutation was not seen in controls while segregated with melanoma in families. Individuals carrying this germ-line mutation in CDKN2A are predisposed to melanoma through loss of heterozygosity. This study illustrates how characterization of noncoding mutations in genes controlling cell proliferation and differentiation may have an impact on current efforts to identify genetic susceptibility to cancer.
Other translational disorders and future perspectives
In addition to the ferritin L-subunit, several other genes encoding proteins involved in iron metabolism carry IREs. For instance, the mRNA for ferritin H-subunit has a consensus IRE in the 5′-UTR. Mutations in the gene encoding the H-subunit have not yet been described, but one might expect that they would cause overproduction of H-ferritin in a manner similar to IRE mutations in the L-subunit. The resulting disorder might be incompatible with life according to the studies by Picard et al,105 who showed that overexpression of the ferritin H-subunit in cultured erythroid cells markedly changed the intracellular iron distribution. Mutations in the 5′-UTR IRE of ALAS2 are also expected to cause ALAS2 protein overexpression, but it is unclear whether this may have clinical relevance. The transferrin receptor gene has 5 IREs in the 3′-UTR and a point mutation in 1 of the five 3′-UTR IREs is expected to have no impact on transferrin receptor production. The presence of 5 IREs might itself be the result of selection aimed at preventing defective production of transferrin receptor.
In mice, there are 2 classes of duodenal −DMT1 transcripts: 1 containing a 3′-UTR IRE, called DMT1(IRE), and 1 containing no IRE, called DMT1(non-IRE).48 Mutations in the 3′-UTR IRE are predicted to decrease the affinity for IRPs and result in reduced DMT1 protein production, causing deficient iron absorption. A large Sardinian family has been described in which hypochromic microcytic anemia with hypoferremia is inherited as a recessive characteristic.106 The disorder observed in this family closely resembles the anemia found in both homozygous mk/mk mice and Belgrade rats, which carry a glycine-to-arginine missense mutation (G185R) in DMT1.46 However, no linkage between anemia and highly polymorphic markers for the human DMT1 gene was found in the Sardinian family.
The 20210 G-to-A transition in the prothrombin gene was first described by Poort et al107 and found to be associated with both elevated plasma prothrombin levels and a moderately increased risk for first venous thrombotic events. This point mutation in the 3′-UTR of the prothrombin gene may simply represent a polymorphism associated with the increased risk for venous thrombosis. However, the possibility that the mutation occurred in a sequence participating in the control of mRNA translation should be investigated.
Translational mechanisms may also be involved in malignant disorders through somatic mutations in regulatory regions of genes controlling cell proliferation and differentiation. Thus, increased translation of IL-15 mRNA secondary to the production of a human T-lymphotropic virus–1 R-element fusion message that lacks many uAUGs has been shown in a human adult T-cell leukemia line.20,108 Moreover, enhanced translational efficiency of a novel transforming growth factor–β3 mRNA has been found in several human breast cancer cell lines.23
Regulation of translation may also contribute to the pathogenesis of Alzheimer disease. The amyloid precursor protein (APP) has been associated with Alzheimer disease because it is processed into the beta-peptide that accumulates in amyloid plaques, and also on the basis of the observation that APP gene mutations can cause early-onset disease. The 5′-UTR of APP mRNA contains a translational enhancer that responds to IL-1109 and is homologous to IL-6–responsive elements in the 5′-UTR of the L- and H- ferritin genes.56
In conclusion, the recent molecular characterization of HHCS and HT has opened a new chapter of translational pathophysiology. Although the 3 conditions described so far (HHCS, HT, and familial melanoma) are caused by germ-line mutations, somatic mutations may be responsible for translational pathophysiology as well. HHCS and HT mutations result in overexpression of the gene products, whereas the mutation CDKN2A causes loss of expression from the mutated allele. Translational pathophysiology can generate considerable diversity in disease states, as illustrated by the different HHCS-causing mutations (Figure 4). Finally, the possibility exists that naturally occurring polymorphisms within mRNA regulatory regions might contribute to phenotypic diversity in normal individuals.
Supported in part by grants from the Italian Association for Cancer Research (AIRC) Milan, Italy; IRCCS Policlinico S. Matteo, Pavia, Italy, Ministero dell'Università e della Ricerca Scientifica e Tecnologica (MURST), Rome, Italy; and Ferrata Storti Foundation, Pavia, Italy (to M.C.); and by grants from the Swiss National Science Foundation and the Swiss Cancer League (to R.C.S.).
Reprints:Mario Cazzola, Division of Hematology, IRCCS Policlinico S. Matteo, 27100 Pavia, Italy; e-mail: m.cazzola@iol.it; or Radek C. Skoda, Clinical Cooperation Unit for Molecular Hematology-Oncology, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; e-mail:r.skoda@dkfz.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal