Abstract
-Thalassemia is very common throughout all tropical and subtropical regions of the world. In Southeast Asia and the Mediterranean regions, compound heterozygotes and homozygotes may have anemia that is mild to severe (hemoglobin [Hb] H disease) or lethal (Hb Bart's hydrops fetalis). We have developed a reliable, single-tube multiplex–polymerase chain reaction (PCR) assay for the 6 most frequently observed determinants of -thalassemia. The assay allows simple, high throughput genetic screening for these common hematological disorders. (Blood. 2000;95:360-362)
α-Thalassemia is the most common inherited disorder of hemoglobin (Hb) synthesis in the world, with gene frequencies varying between 1% and 98% throughout the tropics and subtropics.1 Unlike β-thalassemia, in which nondeletional mutations predominate, > 95% of recognized α-thalassemia involves deletion of 1 or both α-globin genes from chromosome 16p13.3.1,2 The most common of these are the -α.3.7 and -α.4.2 single gene deletions, the —SEA and —FIL Southeast Asian double gene deletions, and the —MED and -α.20.5 Mediterranean double gene deletions. Although a high incidence of the — THAI deletion has been reported in Taiwan,3,4 it was recently suggested that these patients have the —FIL mutation.5 With the exception of the original patient with this mutation,6 no others have been described, and the allele frequency is unknown.
Until several years ago, the standard procedure for molecular characterization of α-thalassemia was Southern blot analysis. DNA sequence analysis of each deletion breakpoint has now enabled PCR-based testing,4 7-10 which is faster, less expensive, safer (no radioactivity involved), and easier to interpret than Southern blot analysis. Currently, however, separate tests are required for the different mutations because of different reagent and thermocycling requirements. Additionally, reproducibility of some PCR-based tests have been problematic, particularly those involving the -α.37 allele. These problems stem in part from the differences in GC nucleotide content of the various deletion junctions and also from the considerable sequence homology within the α-globin cluster, especially at the α2 and α1 loci.
We have developed a single-tube multiplex-PCR assay capable of detecting any combination of these 6 common single and double gene deletions.
Study design
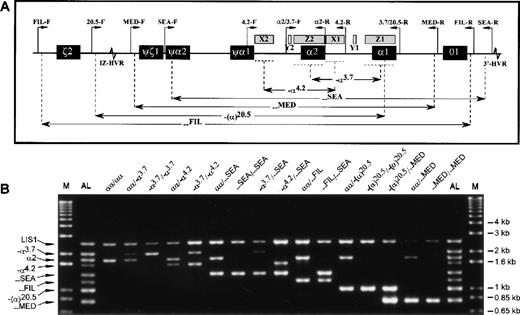
Sequence information on the deletion breakpoints and a control locus (the LIS1 gene at 17p13.3) has been published previously.4,5 11-15 Primers were designed to amplify the junction fragments of the α-thalassemia determinants that could be easily identified by size (Table). Since each of the 6 deletions either partially or completely removes the α2 gene (Figure 1A), its positive amplification was used to indicate heterozygosity when a deletion allele was also present. Additionally, amplification of a large (2.5 kilobase) segment of the LIS1 gene 3′UTR was included as a control for amplification success.
Multiplex-PCR genotype analysis of the -globin gene cluster.
(A) Schematic representation of the α-globin gene cluster. Figure indicates extents of each deletion represented in the multiplex-PCR assay and relative positions of the primers (except for the control LIS1-F and LIS1-R primers, which are located on a different chromosome). Locations of sequence homology (X, Y, Z boxes) and hypervariable regions (HVR) are also shown. (B) Representative results from DNA samples with various α-globin genotypes. The M indicates the 1 kb-Plus molecular weight marker (Life Technologies), and the AL, the allelic ladder comprising all 8 possible amplification fragments.
Multiplex-PCR genotype analysis of the -globin gene cluster.
(A) Schematic representation of the α-globin gene cluster. Figure indicates extents of each deletion represented in the multiplex-PCR assay and relative positions of the primers (except for the control LIS1-F and LIS1-R primers, which are located on a different chromosome). Locations of sequence homology (X, Y, Z boxes) and hypervariable regions (HVR) are also shown. (B) Representative results from DNA samples with various α-globin genotypes. The M indicates the 1 kb-Plus molecular weight marker (Life Technologies), and the AL, the allelic ladder comprising all 8 possible amplification fragments.
Each 50 μL reaction contained 20 mmol/L Tris-HCl pH 8.4, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 1 mol/L betaine (SIGMA, St. Louis, MO), 0.2 μL of each primer (Table 1), 0.2 mmol/L of each dNTP, 2.5 units of polymerase (Platinum Taq; Life Technologies, Gaithersburg, MD), and 100 ng of genomic DNA. Reactions were carried out on a thermal cycler (Genius; Techne, Cambridge, UK), with an initial 5-minute denaturation at 95°C, 30 cycles of 97°C for 45 seconds, 60°C for 1 minute 15 seconds, 72°C for 2 minutes 30 seconds, and a final extension at 72°C for 5 minutes. Following amplification, 10 μL of product was electrophoresed through a 1% agarose, 1 × TBE gel at 5-6 volts/cm for 1 hour, stained in ethidium bromide, and visualized on an ultraviolet transilluminator.
Table. Primers for single-tube multiplex-PCR analysis of common-thalassemia deletions
| Name . | 5′ → 3′ Sequence . | GenBank ID: Nucleotides . | Amplicon (Size) . |
|---|---|---|---|
| LIS1-F | GTCGTCACTGGCAGCGTAGATC | HSLIS10:407 → 428 | LIS1 3′UTR frag |
| LIS1-R | GATTCCAGGTTGTAGACGGACTG | HSLIS10:2909 → 2887 | (2503 bp) |
| α2/3.7-F | CCCCTCGCCAAGTCCACCC | HUMHBA4:5676 → 5694 | -α3.7jxn frag |
| 3.7/20.5-R | AAAGCACTCTAGGGTCCAGCG | HUMHBA4:11514 → 11494 | (2022/2029 bp) |
| α2/3.7-F | see above | α2 gene | |
| α2-R | AGACCAGGAAGGGCCGGTG | HUMHBA4:7475 → 7457 | (1800 bp) |
| 4.2-F | GGTTTACCCATGTGGTGCCTC | HUMHBA4:3064 → 3084 | -α4.2jxn frag |
| 4.2-R | CCCGTTGGATCTTCTCATTTCCC | HUMHBA4:8942 → 8920 | (1628 bp) |
| SEA-F | CGATCTGGGCTCTGTGTTCTC | HSGG1:26120 → 26140 | --SEAjxn frag |
| SEA-R | AGCCCACGTTGTGTTCATGGC | HSCOS12:3817 → 3797 | (1349 bp) |
| FIL-F | TGCAAATATGTTTCTCTCATTCTGTG | HSGG1:11684 → 11709 | --FILjxn frag |
| FIL-R | ATAACCTTTATCTGCCACATGTAGC | HSCOS12:570 → 546 | (1166 bp) |
| 20.5-F | GCCCAACATCCGGAGTACATG | HSGG1:17904 → 17924 | -(α)20.5jxn frag |
| 3.7/20.5-R | see above | (1007 bp) | |
| MED-F | TACCCTTTGCAAGCACACGTAC | HSGG1:23123 → 23144 | --MEDjxn frag |
| MED-R | TCAATCTCCGACAGCTCCGAC | HSGG1:41203 → 41183 | (807 bp) |
| Name . | 5′ → 3′ Sequence . | GenBank ID: Nucleotides . | Amplicon (Size) . |
|---|---|---|---|
| LIS1-F | GTCGTCACTGGCAGCGTAGATC | HSLIS10:407 → 428 | LIS1 3′UTR frag |
| LIS1-R | GATTCCAGGTTGTAGACGGACTG | HSLIS10:2909 → 2887 | (2503 bp) |
| α2/3.7-F | CCCCTCGCCAAGTCCACCC | HUMHBA4:5676 → 5694 | -α3.7jxn frag |
| 3.7/20.5-R | AAAGCACTCTAGGGTCCAGCG | HUMHBA4:11514 → 11494 | (2022/2029 bp) |
| α2/3.7-F | see above | α2 gene | |
| α2-R | AGACCAGGAAGGGCCGGTG | HUMHBA4:7475 → 7457 | (1800 bp) |
| 4.2-F | GGTTTACCCATGTGGTGCCTC | HUMHBA4:3064 → 3084 | -α4.2jxn frag |
| 4.2-R | CCCGTTGGATCTTCTCATTTCCC | HUMHBA4:8942 → 8920 | (1628 bp) |
| SEA-F | CGATCTGGGCTCTGTGTTCTC | HSGG1:26120 → 26140 | --SEAjxn frag |
| SEA-R | AGCCCACGTTGTGTTCATGGC | HSCOS12:3817 → 3797 | (1349 bp) |
| FIL-F | TGCAAATATGTTTCTCTCATTCTGTG | HSGG1:11684 → 11709 | --FILjxn frag |
| FIL-R | ATAACCTTTATCTGCCACATGTAGC | HSCOS12:570 → 546 | (1166 bp) |
| 20.5-F | GCCCAACATCCGGAGTACATG | HSGG1:17904 → 17924 | -(α)20.5jxn frag |
| 3.7/20.5-R | see above | (1007 bp) | |
| MED-F | TACCCTTTGCAAGCACACGTAC | HSGG1:23123 → 23144 | --MEDjxn frag |
| MED-R | TCAATCTCCGACAGCTCCGAC | HSGG1:41203 → 41183 | (807 bp) |
Results and discussion
The assay was tested on a total of 115 blood and prenatal DNA archival samples. The α-globin genotypes had been determined previously by Southern blot analysis. Samples were amplified, products were separated on agarose gels, and genotypes were scored before checking against the previously determined Southern results.
In 16 samples tested (number of samples noted in brackets), there were no amplification products observed: (—SEA/αα [9],-α.37/αα [6], —MED/αα [1]) (data not shown). Repeat reactions with a fresh multiplex-PCR mix or a previously tested primer pair specific to a different locus did not alter the outcome (data not shown). This suggests that rather than a specific failure of the multiplex-PCR system, these DNA samples were inadequate for any standard PCR reaction.
In 10 additional samples (—SEA/αα [5], -α.3.7/αα [3], —MED/αα [1], —MED/—MED [1]), 1 or 2 α-globin fragments successfully amplified, but the LIS1 positive control fragment was absent (data not shown). In accordance with the intended function of the LIS1 positive control, these samples were excluded from further analysis, since situations where smaller fragments amplify but larger ones do not can occur with partially degraded or impure DNA template or with incorrect PCR conditions. In the case of heterozygous or compound heterozygous samples, failure to detect the second α-globin fragment would lead to misdiagnosis.
In the remaining 89 samples, the LIS1 positive control fragment amplified successfully: (αα/αα [10], α.3.7/αα [21],—SEA/αα [24], -α.4.2/αα [2], —FIL/αα [7], —MED/αα [2], -(α).20.5/αα [3], -α.3.7/—SEA [3], -α.3.7/ -α.4.2 [2], -α.4.2/—SEA [2], —FIL/—SEA [2], -(α).20.5/—MED [1], -α.3.7/-α.3.7 [5], —SEA/—SEA [3], —MED/—MED [1], -(α).20.5/-(α).20.5 [1]). For each of these samples, co-amplification of the expected α-globin fragment or fragments was also observed, and the assigned α-globin genotypes were confirmed for all samples (Figure 1B).
The intensities of the α2 and -α.3.7 fragments were often weaker than for the other fragments, probably due to a combination of their higher GC content and large size. Nevertheless, allele dropout (amplification failure) of either of these fragments was not observed. Unexpected or inappropriate fragments were not observed in any of the samples tested, confirming the specificity of this assay. These results demonstrate the successful development of a single-tube multiplex-PCR assay for common deletional determinants of α-thalassemia.
The simplicity and universality of this multi-ethnic multiplex-PCR assay should significantly reduce the cost and complexity of screening for these common α-thalassemia deletions. The assay is especially useful in cosmopolitan populations, such as in the United States and the United Kingdom, where in the majority of cases and irrespective of patient ethnicity, only a single diagnostic test needs to be performed.
Acknowledgment
We thank P. Sanders for technical assistance.
Supported by research funds from the Johns Hopkins University and by a grant from the Academic Research Fund of the National University of Singapore (NUS/ARF 3 690 044).
Reprints:Samuel S. Chong, Department of Pediatrics, National University of Singapore, Level 4, Main Building, National University Hospital, 5 Lower Kent Ridge Road, Singapore 119074, Singapore; e-mail:paecs@nus.edu.sg.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal