Abstract
Genes of the MAGE, BAGE, GAGE, and LAGE-1/NY-ESO-1families encode antigenic peptides that are presented by HLA class I molecules and that are recognized on human tumors by autologous cytolytic T lymphocytes. These genes are expressed in many solid tumor types but not in normal tissues, except male germline cells. Because the latter cells are devoid of HLA molecules, the derived antigens are strictly tumor-specific and should constitute safe immunogens for cancer immunotherapy. We detected a significant expression of these genes in a high proportion of bone marrow samples from patients with advanced multiple myeloma. This observation provides a basis for clinical trials aimed at inducing a cellular immune response directed at malignant plasma cells in advanced myeloma patients.
CONVENTIONAL chemotherapy or high-dose chemotherapy followed by autologous hematopoietic stem cell transplantation has improved the overall response rate and survival of myeloma patients. However, most, if not all of the patients who achieve a complete remission after such treatment will ultimately relapse, showing that chemotherapy alone is unable to cure multiple myeloma.1 Obviously, new approaches to treatment are needed.
There is increasing evidence that the immune system, and particularly T lymphocytes, can target malignant plasma cells. Clinical responses in myeloma patients treated with interleukin-2 (IL-2) have been reported.2 After allogeneic bone marrow transplantation, T lymphocytes from the donor can reject myeloma cells from the recipient, eventually leading to long-term complete remissions. This graft-versus-myeloma effect can also be induced by the reinfusion of donor lymphocytes in myeloma patients who are in relapse after bone marrow allografting.3 4 However, immune responses induced by IL-2 treatment or derived from the allogeneic graft are not myeloma-specific, and both are associated with significant toxicity. The efficiency and safety of immunotherapy against myeloma could therefore benefit from the identification of antigens present on myeloma cells and absent on nonmalignant cells.
The monoclonal Ig produced by a B-cell–derived malignancy constitutes a tumor antigen that is specific for this individual malignant clone. Promising immunological and clinical responses were obtained by using this immunogen in patients with B-cell lymphomas.5 In a case of myeloma, the monoclonal Ig produced by the recipient’s malignant plasma cells was used to immunize the donor before allogeneic bone marrow transplantation. This resulted in a detectable anti-idiotype cellular immune response in both donor and allografted recipient.6 In a recent report, 5 myeloma patients were immunized with their respective purified monoclonal Ig in combination with granulocyte-macrophage colony-stimulating factor (GM-CSF). An idiotype-specific cellular immune response was reported in all patients. One patient showed a decrease in the serum concentration of the monoclonal component after immunization, suggesting a tumor response to the vaccine.7
Another possible T-lymphocyte target present on myeloma cells is MUC1, an immunogenic epithelial mucin present in an underglycosylated form on breast, pancreatic, and ovarian carcinomas.8Underglycosylated MUC1 was also detected on malignant plasma cells,9 and anti-MUC1 cytolytic T lymphocytes (CTL) could be obtained from the bone marrow of myeloma patients.10
Another category of antigens is encoded by genes of the MAGEfamily. MAGE-A1 was initially isolated from a human melanoma cell line as a gene encoding an antigenic peptide presented to an autologous CTL clone by HLA-A1 molecules.11 This gene is 1 of 12 closely homologous members of the MAGE-A family, all located near the telomeric end of the long arm of chromosome X.12 Genes MAGE-A1, A2, A3, A4, A6, A10, andA12 are frequently expressed in many tumor types, such as melanoma, bladder carcinoma, non-small cell lung carcinoma, head and neck carcinoma, and esophagus squamous cell carcinoma, but are silent in normal tissues except testis and, in some cases, placental trophoblast cells. Immunohistochemistry studies have shown that, in testis, MAGE proteins are present in spermatogonia.13 These cells are known to lack expression of HLA molecules and are therefore unable to present peptides to CTL. Because this is also true for the trophoblast, the MAGE-derived antigens are strictly tumor-specific.
BAGE and GAGE-1 were also identified as genes encoding antigens recognized by autologous CTL on a melanoma cell line.14,15 They share the same pattern of expression asMAGE and are therefore referred to as MAGE-type genes. Another MAGE-type gene is LAGE-1. It was isolated by representational difference analysis as a gene expressed in a melanoma cell line but not in normal skin.16 It is homologous toNY-ESO-1, a gene that encodes an antigen recognized by autologous antibodies isolated from a patient with esophageal squamous cell carcinoma.17 Both are expressed in many solid tumors and in testis.
The MAGE-A1 gene becomes activated in tumor cells as a result of demethylation of CpG dinucleotides in its promoter. In nontumoral cells, the methylation of these sites inhibits the binding of activating transcription factors and results in gene silencing.18 Expression of the MAGE-type genes can be induced experimentally in nontumoral growing cells by incubation with the demethylating agent 5-aza-2′-deoxycytidine, which shows that DNA demethylation is the common mechanism that accounts for aberrant expression of these genes.
Antigens encoded by MAGE-type genes may be particularly suitable as targets for immunotherapy, because they are strictly tumor-specific and are shared by many different tumor types. In the present study, we investigated the expression of genes of theMAGE-A, GAGE, and LAGE-1/NY-ESO-1 families and of geneBAGE in bone marrow or blood samples from patients with multiple myeloma or monoclonal gammopathy of undetermined significance (MGUS). We also tested the expression of gene PRAME, which encodes an antigen recognized by a CTL clone on a melanoma cell line and is very frequently expressed in tumors of several types, as well as in testis. It is not as tumor-specific as the MAGE-type genes, because there is a low level of expression in the endometrium, ovary, and adrenals.19 Its mRNA has also been detected in acute leukemias and in a few samples of lymphoma and myeloma.20
MATERIALS AND METHODS
Tumor sample collection and processing.
Cells were collected from normal donors or from patients with MGUS or multiple myeloma by bone marrow aspiration. Samples from myeloma patients were collected before they received any treatment or at distance from previous chemotherapy. Cytologic examination confirmed that all myeloma samples contained malignant plasma cells. The mononuclear cells were purified by Lymphoprep (Nycomed, Oslo, Norway) density gradient centrifugation and were washed twice with Iscove’s medium (GIBCO Laboratories, Green Island, NY) containing 10% fetal calf serum (FCS; GIBCO) and twice with phosphate-buffered saline (PBS). After a final centrifugation, the dry cell pellet was stored at −80°C until needed for RNA extraction.
Reverse transcription-polymerase chain reaction (RT-PCR) assay.
RNA extraction and RT-PCR amplifications were performed as described previously,21 with slight modifications. Briefly, cDNA was synthesized from 2 μg of total RNA by extension with oligo(dT) primer and 200 U of Moloney murine leukemia virus (M-MLV) reverse transcriptase (GIBCO) at 42°C for 90 minutes. PCR amplification was performed on 1/40th of the cDNA solution with 0.625 U of Taq DNA polymerase (Takara, Shiga, Japan) in a final volume of 25 μL. The PCR conditions and primers were as follows: MAGE-A1: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, and 72°C for 2 minutes, primers 5′-CGGCCGAAGGAACCTGACCCAG-3′ and 5′-GCTGGAACCCTCACTGGGTTGCC-3′;MAGE-A2: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 67°C for 1 minute, and 72°C for 1 minute, primers 5′-AAGTAGGACCCGAGGCACTG-3′ and 5′-GAAGAGGAAGAAGCGGTCTG-3′;MAGE-A3: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute and 72°C for 2 minutes, primers 5′-TGGAGGACCAGAGGCCCCC-3′ and 5′-GGACGATTATCAGGAGGCCTGC-3′; MAGE-A4: 94°C for 4 minutes and 30 cycles of 94°C for 1minute, 68°C for 1 minute and 72°C for 1 minute, primers 5′-GAGCAGACAGGCCAACCG-3′ and 5′-AAGGACTCTGCGTCAGGC-3′;MAGE-A6: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 70°C for 2 minutes, and 72°C for 2 minutes, primers 5′-TGGAGGACCAGAGGCCCCC-3′ and 5′-CAGGATGATTATCAGGAAGCCTGT-3′;MAGE-A10: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 65°C for 1 minute, and 72°C for 1 minute, primers 5′-CACAGAGCAGCACTGAAGGAG-3′ and 5′-CTGGGTAAAGACTCACTGTCTGG-3′;MAGE-A12: 94°C for 4 minutes and 32 cycles of 94°C for 1 minute, 62°C for 2 minutes, and 72°C for 3 minutes, primers 5′-CGTTGGAGGTCAGAGAACAG-3′ and 5′-GCCCTCCACTGATCTTTAGCAA-3′;BAGE: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 62°C for 2 minutes, and 72°C for 2 minutes, primers 5′-TGGCTCGTCTCACTCTGG-3′ and 5′-CCTCCTATTGCTCCTGTTG-3′;GAGE-1/2: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 56°C for 2 minutes, and 72°C for 2 minutes, primers 5′-GACCAAGACGCTACGTAG-3′ and 5′-CCATCAGGACCATCTTCA-3′;GAGE-3/6: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 58°C for 2 minutes, and 72°C for 2 minutes, primers 5′-GACCAAGGCGCTATGTAC-3′ and 5′-CCATCAGGACCATCTTCA-3′; LAGE-1:94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 62°C for 1 minute, and 72°C for 1 minute, primers 5′-GCAGGATGGAAGGTGCCC-3′ and 5′-CTGGCCACTCGTGCTGGGA-3′; NY-ESO-1: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 62°C for 1 minute, and 72°C for 1 minute, primers 5′-CCCCACCGCTTCCCGTG-3′ and 5′-CTGGCCACTCGTGCTGGGA-3′;PRAME: 94°C for 4 minutes and 30 cycles of 94°C for 1 minute, 64°C for 1 minute, and 72°C for 1 minute, primers 5′-CTGTACTCATTTCCAGAGCCAGA-3′ and 5′-TATTGAGAGGGTTTCCAAGGGGTT-3′; β-ACTIN: 94°C for 4 minutes and 21 cycles of 94°C for 1 minute, 68°C for 1 minute, and 72°C for 1 minute, primers 5′-GGCATCGTGATGGACTCCG-3′ and 5′-GCTGGAAGGTGGACAGCGA-3′.
Cycling was concluded with a final extension step of 15 minutes at 72°C. Each primer was chosen in a different exon to avoid false-positives caused by DNA contamination of the RNA preparation. Assessment of the PCR product was performed visually on an ethidium bromide-stained agarose gel by comparing the intensity of the band with that resulting from RT-PCR performed on serial dilutions (1:1, 1:3, 1:9, and 1:27) of the RNA from 1 of 3 tumor cell lines (2 melanomas and 1 sarcoma) used as a positive control and reference for the level of expression. These cell lines were MZ2-MEL (all MAGE, BAGE, and GAGE PCR except MAGE-A4 and MAGE-A12), LB23-SARC (MAGE-A4 PCR), and LB373-MEL (MAGE-A12, LAGE-1, NY-ESO-1, and PRAME PCR). Samples were scored +++, ++, +, or ± if the amount of the amplified product was equal to or greater than that obtained with the 1:1, 1:3, 1:9, and 1:27 dilutions of the reference RNA, respectively. Lower levels of expression were scored negative. An expression level of the β-ACTIN gene comparable with that of the positive control was obtained with each sample.
Lysis assay.
All cell lines were grown in RPMI-1640 medium (GIBCO) supplemented with 5% FCS (EJM and U266) or 10% FCS (MZ2-MEL and SK23-MEL). The anti–MAGE-3.A1 CTL clone 434/1 was derived from the blood of an HLA-A1 patient with hemochromatosis after repetitive stimulation with autologous phytohemagglutinin (PHA)-stimulated T cells incubated with the MAGE-3.A1 peptide. The anti–MAGE-3.A2 CTL clone 297/22 was obtained similarly from an HLA-A2 patient.22 Chromium release assay was performed as described previously.23Briefly, target cells were labeled with 51Cr, washed, and dispensed into microwell plates at 1,000 cells per well in Iscove’s medium supplemented with 10% human serum and with L-arginine (116 mg/L), L-asparagine (36 mg/L), and L-glutamine (216 mg/L), which is further referred to as complete medium. CTL was added at increasing effector-to-target ratios. The cells were centrifuged and incubated at 37°C for 4 hours, and chromium release was determined by measuring the radioactivity in the supernatant. The myeloma cell lines were also tested for their ability to stimulate the production of tumor necrosis factor (TNF) by the CTL clones, as described previously.24Briefly, 5,000 CTL were added to microwells containing 10,000 target cells in 150 μL of complete medium supplemented with 25 U/mL of IL-2. After overnight incubation, the supernatant was collected and its TNF content was measured by testing its cytotoxic effect on WEHI-164 clone 13 cells in an MTT colorimetric assay.
Immunocytochemistry.
Myeloma cell lines EJM, U266, and Fravel and mononuclear cells isolated from the bone marrow of patient no. 43 were washed in Tris-buffered saline (TBS) and cytospun at 500 rpm for 4 minutes on microscope slides (105 cells per slide). Mononuclear cells isolated from the bone marrow of patient no. 21 were smeared on microscope slides. All of the slides were air-dried at room temperature, wrapped in aluminium foil, and stored at −80°C until needed. They were fixed in 10% buffered formalin at room temperature for 10 minutes and then washed in TBS for 2 minutes. The fixed slides were incubated with the anti-MAGE hybridoma supernatant 57B,25 either undiluted (patient no. 21) or diluted 1/10 (others) at 4°C for 18 hours, or with an isotype-matched irrelevant monoclonal antibody. They were further incubated with biotinylated antimouse Igs and alkaline phosphatase-conjugated streptavidin (LSAB+; Dako, Glostrup, Denmark), using New Fuchsin as a chromogen. Sections of formalin-fixed, paraffin-embedded gut biopsies from patient no. 29 were heated, after deparaffinization, in a 1,500 W microwave oven twice for 5 minutes in citrate buffer for antigen retrieval (Dako). They were washed in TBS containing 0.05% Tween 20 for 1 minute, incubated with antibody 57B diluted 1/10 or with the irrelevant antibody, at room temperature for 1 hour, and then incubated with a peroxydase-conjugated polymer backbone carrying antimouse Igs (En Vision; Dako) with AEC as chromogenic substrate.
RESULTS
Expression of MAGE-type genes in bone marrow samples from MGUS or myeloma patients.
We used RT-PCR to analyze 44 bone marrow samples from patients with MGUS (n = 6) or myeloma (n = 38) for the expression of 7 genes of the MAGE-A family, the gene BAGE, 2 subgroups of very closely related genes of the GAGE family, the geneLAGE-1, the gene NY-ESO-1, and the gene PRAME.Two samples were obtained from the peripheral blood of patients with plasma cell leukemia (Table 1 and Fig1). When considering only +++, ++, and + scores, all of the samples from patients with MGUS and with stage I and stage II myeloma were found negative, except for 1 MGUS sample that showed significant expression of gene LAGE-1. In contrast, a majority of the samples from the stage III myeloma patients expressed at least 1 of these genes. When considering stage III myelomas and plasma cell leukemias, the most frequently expressed genes wereLAGE-1 (52%), PRAME (48%), and GAGE (41%), followed by NY-ESO-1 and MAGE-A6 (31%),MAGE-A1 and MAGE-A3 (28%), MAGE-A2 andMAGE-A4 (17%), MAGE-A12 and BAGE (14%), and finally MAGE-A10 (7%). In total, 38% of these samples were positive for at least 1 of the MAGE-A genes, and 62% were positive for at least 1 of the MAGE-type genes. In addition, we tested 7 bone marrows from normal donors. All were negative (data not shown).
Detection of mRNA From the Indicated Genes by RT-PCR in Bone Marrow or Blood Samples From Patients with Monoclonal Gammopathy of Undetermined Significance (MGUS), Multiple Myeloma, and Plasma Cell Leukemia (PCL), Classified According to Durie and Salmon31
| Diagnosis . | Patient No. . | Sex . | Age . | Monocl.Ig . | Prev.Treat. . | % Tum.Cells . | RT-PCR Results . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAGE . | BAGE 1 . | GAGE . | LAGE 1 . | NY-ESO 1 . | PRAME . | ||||||||||||||
| A1 . | A2 . | A3 . | A4 . | A6 . | A10 . | A12 . | 1/2 . | 3-6 . | |||||||||||
| MGUS | 1 | M | 71 | IgG κ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2 | M | 62 | IgG λ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 3 | M | 62 | IgG κ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 4 | F | 39 | IgG κ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 5 | M | 51 | IgG λ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 6 | M | 64 | IgG λ | N | 0 | − | − | − | − | − | − | − | − | − | − | ++ | − | − | |
| Myeloma | 7 | F | 64 | IgG κ | N | 43 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| stage I | 8 | M | 71 | IgG λ | Y | 25 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 9 | M | 70 | IgA κ | N | 7 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 10 | M | 65 | IgA λ | N | 15 | − | − | − | − | − | − | − | − | − | − | − | − | ± | |
| 11 | M | 74 | IgG κ | N | 28 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 12 | F | 69 | IgA λ | N | 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 13 | M | 62 | IgG κ | N | 6 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 14 | F | 75 | IgG λ | N | 30 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 15 | F | 65 | IgG κ | N | 11 | − | − | − | − | − | − | − | − | − | ± | − | − | − | |
| Myeloma | 16 | F | 74 | IgA κ | N | 21 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| stage II | 17 | F | 65 | IgG κ | Y | 4 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Myeloma | 18 | M | 62 | IgG κ | Y | 89 | − | +++ | +++ | ++ | +++ | − | − | − | +++ | +++ | ++ | ++ | ++ |
| stage III | 19 | M | 87 | λ | N | 45 | +++ | ++ | ++ | ± | + | ± | ± | ± | +++ | +++ | +++ | +++ | +++ |
| 20 | F | 87 | IgG κ | N | 85 | + | − | ± | − | ++ | − | − | − | + | + | ± | − | +++ | |
| 21 | F | 81 | IgG κ | N | 17 | + | + | ++ | − | + | − | + | − | ± | ± | +++ | − | − | |
| 22 | M | 56 | κ | Y | 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 23 | M | 72 | IgA κ | Y | 32 | − | − | − | − | ± | − | − | − | +++ | +++ | + | − | ++ | |
| 24 | F | 84 | IgA λ | N | 90 | + | − | ± | − | − | − | − | ± | + | + | ++ | + | + | |
| 25 | M | 57 | IgG λ | Y | 81 | +++ | ± | ++ | − | − | − | ± | + | ++ | + | + | ++ | − | |
| 26 | F | 94 | IgG κ | Y | 33 | − | − | − | − | − | − | − | − | + | ++ | ++ | − | − | |
| 27 | M | 69 | IgG κ | Y | 10 | − | − | − | − | − | − | − | − | − | − | − | − | ++ | |
| 28 | M | 45 | IgG κ | N | 27 | − | − | − | − | − | − | − | − | − | ± | − | − | − | |
| 29 | M | 72 | IgA κ | Y | 6 | +++ | + | ++ | +++ | ++ | + | + | +++ | ++ | +++ | + | ++ | + | |
| 30 | F | 53 | IgG κ | Y | 9 | − | − | − | − | − | − | − | − | − | − | + | − | − | |
| 31 | M | 59 | IgA κ | N | 25 | +++ | − | +++ | ++ | +++ | − | ++ | +++ | +++ | +++ | +++ | +++ | ± | |
| 32 | M | 59 | IgG κ | Y | 7 | − | − | − | − | − | − | − | − | − | ± | +++ | − | − | |
| 33 | M | 61 | IgG κ | Y | 24 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 34 | F | 66 | λ | Y | 68 | ± | − | ± | − | − | − | − | ± | + | +++ | + | + | ++ | |
| 35 | F | 84 | IgA κ | N | 42 | ± | ± | − | − | ++ | − | − | − | +++ | +++ | ± | ± | − | |
| 36 | F | 62 | IgA κ | N | 100 | ± | − | − | − | − | − | − | − | − | − | ± | ± | + | |
| 37 | F | 71 | κ | Y | 47 | +++ | +++ | +++ | + | +++ | + | ++ | +++ | +++ | +++ | +++ | +++ | +++ | |
| 38 | F | 64 | IgA κ | Y | 33 | − | − | − | − | − | − | − | − | − | − | − | − | +++ | |
| 39 | M | 63 | κ | Y | 14 | − | − | − | − | − | − | − | − | − | − | + | − | − | |
| 40 | M | 79 | IgG κ | N | 32 | − | − | − | − | − | − | − | − | − | − | ± | − | +++ | |
| 41 | M | 77 | IgA κ | N | 46 | − | − | − | − | − | − | − | − | − | − | − | − | ± | |
| 42 | F | 56 | IgG κ | Y | 100 | − | − | − | − | − | − | − | − | − | − | − | − | ++ | |
| 43 | M | 53 | IgG κ | Y | 4 | − | − | + | + | + | − | − | − | − | − | − | − | + | |
| 44 | M | 61 | IgA κ | N | 23 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Plasma cell | 45 | F | 44 | IgA κ | N | 43 | ± | − | − | − | − | − | − | − | − | − | ++ | + | ± |
| leukemia | 46 | F | 38 | IgG κ | N | 10 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Total (stage III and PCL) = 29 positives: | 8 | 5 | 8 | 5 | 9 | 2 | 4 | 4 | 12 | 12 | 15 | 9 | 14 | ||||||
| Diagnosis . | Patient No. . | Sex . | Age . | Monocl.Ig . | Prev.Treat. . | % Tum.Cells . | RT-PCR Results . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAGE . | BAGE 1 . | GAGE . | LAGE 1 . | NY-ESO 1 . | PRAME . | ||||||||||||||
| A1 . | A2 . | A3 . | A4 . | A6 . | A10 . | A12 . | 1/2 . | 3-6 . | |||||||||||
| MGUS | 1 | M | 71 | IgG κ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2 | M | 62 | IgG λ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 3 | M | 62 | IgG κ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 4 | F | 39 | IgG κ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 5 | M | 51 | IgG λ | N | 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 6 | M | 64 | IgG λ | N | 0 | − | − | − | − | − | − | − | − | − | − | ++ | − | − | |
| Myeloma | 7 | F | 64 | IgG κ | N | 43 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| stage I | 8 | M | 71 | IgG λ | Y | 25 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 9 | M | 70 | IgA κ | N | 7 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 10 | M | 65 | IgA λ | N | 15 | − | − | − | − | − | − | − | − | − | − | − | − | ± | |
| 11 | M | 74 | IgG κ | N | 28 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 12 | F | 69 | IgA λ | N | 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 13 | M | 62 | IgG κ | N | 6 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 14 | F | 75 | IgG λ | N | 30 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 15 | F | 65 | IgG κ | N | 11 | − | − | − | − | − | − | − | − | − | ± | − | − | − | |
| Myeloma | 16 | F | 74 | IgA κ | N | 21 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| stage II | 17 | F | 65 | IgG κ | Y | 4 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Myeloma | 18 | M | 62 | IgG κ | Y | 89 | − | +++ | +++ | ++ | +++ | − | − | − | +++ | +++ | ++ | ++ | ++ |
| stage III | 19 | M | 87 | λ | N | 45 | +++ | ++ | ++ | ± | + | ± | ± | ± | +++ | +++ | +++ | +++ | +++ |
| 20 | F | 87 | IgG κ | N | 85 | + | − | ± | − | ++ | − | − | − | + | + | ± | − | +++ | |
| 21 | F | 81 | IgG κ | N | 17 | + | + | ++ | − | + | − | + | − | ± | ± | +++ | − | − | |
| 22 | M | 56 | κ | Y | 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 23 | M | 72 | IgA κ | Y | 32 | − | − | − | − | ± | − | − | − | +++ | +++ | + | − | ++ | |
| 24 | F | 84 | IgA λ | N | 90 | + | − | ± | − | − | − | − | ± | + | + | ++ | + | + | |
| 25 | M | 57 | IgG λ | Y | 81 | +++ | ± | ++ | − | − | − | ± | + | ++ | + | + | ++ | − | |
| 26 | F | 94 | IgG κ | Y | 33 | − | − | − | − | − | − | − | − | + | ++ | ++ | − | − | |
| 27 | M | 69 | IgG κ | Y | 10 | − | − | − | − | − | − | − | − | − | − | − | − | ++ | |
| 28 | M | 45 | IgG κ | N | 27 | − | − | − | − | − | − | − | − | − | ± | − | − | − | |
| 29 | M | 72 | IgA κ | Y | 6 | +++ | + | ++ | +++ | ++ | + | + | +++ | ++ | +++ | + | ++ | + | |
| 30 | F | 53 | IgG κ | Y | 9 | − | − | − | − | − | − | − | − | − | − | + | − | − | |
| 31 | M | 59 | IgA κ | N | 25 | +++ | − | +++ | ++ | +++ | − | ++ | +++ | +++ | +++ | +++ | +++ | ± | |
| 32 | M | 59 | IgG κ | Y | 7 | − | − | − | − | − | − | − | − | − | ± | +++ | − | − | |
| 33 | M | 61 | IgG κ | Y | 24 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 34 | F | 66 | λ | Y | 68 | ± | − | ± | − | − | − | − | ± | + | +++ | + | + | ++ | |
| 35 | F | 84 | IgA κ | N | 42 | ± | ± | − | − | ++ | − | − | − | +++ | +++ | ± | ± | − | |
| 36 | F | 62 | IgA κ | N | 100 | ± | − | − | − | − | − | − | − | − | − | ± | ± | + | |
| 37 | F | 71 | κ | Y | 47 | +++ | +++ | +++ | + | +++ | + | ++ | +++ | +++ | +++ | +++ | +++ | +++ | |
| 38 | F | 64 | IgA κ | Y | 33 | − | − | − | − | − | − | − | − | − | − | − | − | +++ | |
| 39 | M | 63 | κ | Y | 14 | − | − | − | − | − | − | − | − | − | − | + | − | − | |
| 40 | M | 79 | IgG κ | N | 32 | − | − | − | − | − | − | − | − | − | − | ± | − | +++ | |
| 41 | M | 77 | IgA κ | N | 46 | − | − | − | − | − | − | − | − | − | − | − | − | ± | |
| 42 | F | 56 | IgG κ | Y | 100 | − | − | − | − | − | − | − | − | − | − | − | − | ++ | |
| 43 | M | 53 | IgG κ | Y | 4 | − | − | + | + | + | − | − | − | − | − | − | − | + | |
| 44 | M | 61 | IgA κ | N | 23 | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Plasma cell | 45 | F | 44 | IgA κ | N | 43 | ± | − | − | − | − | − | − | − | − | − | ++ | + | ± |
| leukemia | 46 | F | 38 | IgG κ | N | 10 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Total (stage III and PCL) = 29 positives: | 8 | 5 | 8 | 5 | 9 | 2 | 4 | 4 | 12 | 12 | 15 | 9 | 14 | ||||||
Samples are scored +++, ++, +, or ± if the amount of the amplified product is equal to or greater than that obtained with the 1:1, 1:3, 1:9, and 1:27 dilutions of the reference RNA used for each gene, respectively. Lower levels of expression are scored negative. RT-PCR amplification of β-actin mRNA was obtained in each sample.
Abbreviations: Monocl. Ig, monoclonal immunoglobulin associated with the gammopathy; Prev. Treat., previous treatment was either chemotherapy, or intensive chemotherapy followed by autologous hematopoietic cell transplantation, or both; % Tum. Cells, proportion of malignant plasma cells in the sample tested by RT-PCR, assessed by cytological examination.
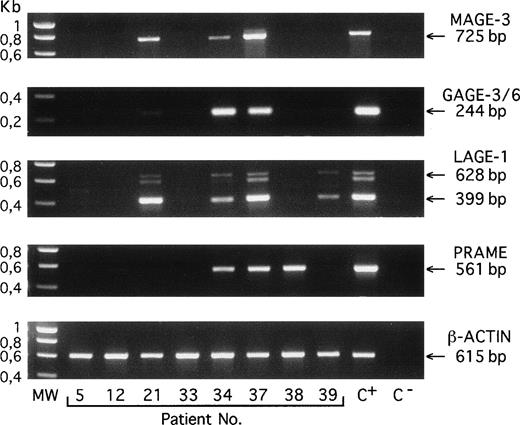
RT-PCR amplification products of the indicated genes obtained with bone marrow samples from 8 different patients with MGUS (no. 5), myeloma stage I (no. 12), or myeloma stage III (nos. 21 through 39). C+, positive control melanoma line (MZ2-MEL for MAGE-A3, GAGE-3/6, and β-ACTIN; LB373-MEL for LAGE-1 and PRAME). C−, negative control (no RNA present in the RT reaction). MW, molecular weight marker is SmartLadder (Eurogentec, Seraing, Belgium). Amplifications of MAGE-A3, GAGE-3/6, PRAME, and β-ACTIN transcripts give unique bands of 725, 244, 561, and 615 bp, respectively. Amplification of LAGE-1 transcript gives 2 bands that correspond to fully spliced (399 bp) and partially spliced (628 bp) mRNA, respectively, and a band of approximately 600 bp representing heteroduplexes formed during PCR between the products amplified from the partially and the fully spliced cDNAs.
RT-PCR amplification products of the indicated genes obtained with bone marrow samples from 8 different patients with MGUS (no. 5), myeloma stage I (no. 12), or myeloma stage III (nos. 21 through 39). C+, positive control melanoma line (MZ2-MEL for MAGE-A3, GAGE-3/6, and β-ACTIN; LB373-MEL for LAGE-1 and PRAME). C−, negative control (no RNA present in the RT reaction). MW, molecular weight marker is SmartLadder (Eurogentec, Seraing, Belgium). Amplifications of MAGE-A3, GAGE-3/6, PRAME, and β-ACTIN transcripts give unique bands of 725, 244, 561, and 615 bp, respectively. Amplification of LAGE-1 transcript gives 2 bands that correspond to fully spliced (399 bp) and partially spliced (628 bp) mRNA, respectively, and a band of approximately 600 bp representing heteroduplexes formed during PCR between the products amplified from the partially and the fully spliced cDNAs.
Recognition of myeloma cells by anti-MAGE CTL.
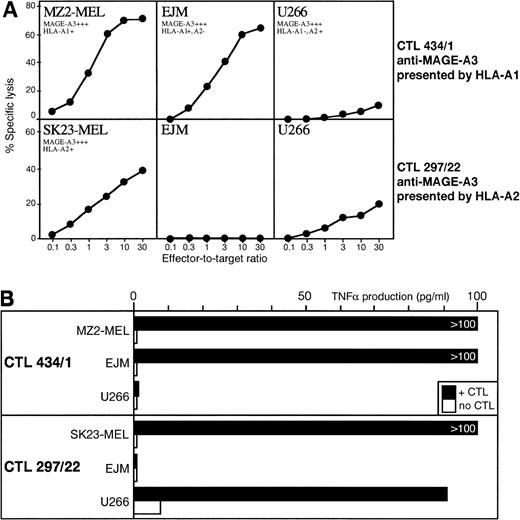
We used 2 anti–MAGE-A3 CTL clones, 1 restricted by HLA-A1 and the other by HLA-A2. Only 2 myeloma cell lines with proper HLA type and MAGE expression were available. The anti–MAGE-A3.A1 CTL lysed myeloma EJM as well as control melanoma target cells. The anti–MAGE-A3.A2 CTL showed weak but significant lysis against myeloma U266, but this CTL appears to recognize a poorly processed peptide and lyses weakly most melanoma cell lines (Fig 2A). A more sensitive TNF release assay demonstrated recognition of the 2 myeloma cell lines by the appropriate anti-MAGE CTL (Fig 2B).
(A) Lytic activity of anti–MAGE-A3.A1 CTL clone 434/1 and anti–MAGE-A3.A2 CTL clone 297/22 against myeloma cell lines EJM and U266. Expression of gene MAGE-A3 was assessed by RT-PCR by using the same procedure as for the primary myeloma samples. (B) TNF- release by the same CTL clones after incubation with the same myeloma cell lines.
(A) Lytic activity of anti–MAGE-A3.A1 CTL clone 434/1 and anti–MAGE-A3.A2 CTL clone 297/22 against myeloma cell lines EJM and U266. Expression of gene MAGE-A3 was assessed by RT-PCR by using the same procedure as for the primary myeloma samples. (B) TNF- release by the same CTL clones after incubation with the same myeloma cell lines.
Staining of myeloma cells with a MAGE-specific monoclonal antibody.
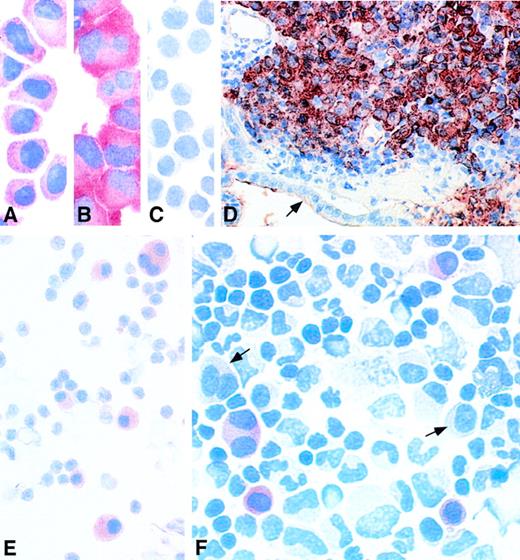
The monoclonal antibody 57B was initially reported to recognize the MAGE-A3 protein.25 We have observed that this antibody recognizes not only the MAGE-A3 protein, but also the MAGE-A1, A2, A4, A6, and A12 proteins. It stained the MAGE-expressing human myeloma cell lines EJM and U266 (Fig 3A and B) and did not stain theMAGE-negative myeloma cell line Fravel (Fig 3C). We tested samples of a few patients with a MAGE-expressing myeloma, recorded in Table 1. Patient no. 29 developed skin and gut plasmacytomas during the course of a stage III myeloma treated with chemotherapy. His bone marrow was found to be positive for expression of all the genes tested in this report. A gut biopsy section was stained with antibody 57B. The gut wall showed a massive infiltration by malignant plasma cells (Fig 3D). A similar result was obtained with a cutaneous plasmacytoma from the same patient (data not shown). In both samples, all the malignant plasma cells were homogeneously stained, whereas the normal cells were negative. A bone marrow smear from patient no. 21, who had an untreated MAGE+ stage III myeloma, was stained with the same antibody. Here also, all of the malignant plasma cells were positive (Fig 3E). A cytospin performed with bone marrow mononuclear cells from patient no. 43, who had aMAGE+ myeloma in relapse after 2 autologous bone marrow transplantations, was stained with 57B. Approximately 30% of all the malignant plasma cells were positive, whereas normal bone marrow cells and the remaining myeloma cells were negative (Fig 3F).
Immunocytochemistry performed on myeloma samples with anti-MAGE monoclonal antibody 57B. (A) MAGE+ myeloma cell line EJM. (B) MAGE+ myeloma cell line U266. (C) MAGE− myeloma cell line Fravel. (D) Gut biopsy from patient no. 29, a patient with MAGE+ stage III myeloma treated by chemotherapy, who developed skin and gut plasmacytomas. The cells stained in brownish red are malignant plasma cells infiltrating the gut mucosa. The epithelium is indicated by an arrow. (E) Bone marrow smear from patient no. 21, a patient with MAGE+ stage III myeloma. All the plasma cells are stained in red. (F) Bone marrow cytospin from patient no. 43, who had a myeloma in relapse after 2 autografts. Some malignant plasma cells are stained in red. The arrows indicate 2 unstained myeloma cells. The slides incubated with the isotype-matched irrelevant monoclonal antibody remained negative, as did cytospins of MAGE-negative bone marrows from stage III myeloma patients incubated with antibody 57B (data not shown).
Immunocytochemistry performed on myeloma samples with anti-MAGE monoclonal antibody 57B. (A) MAGE+ myeloma cell line EJM. (B) MAGE+ myeloma cell line U266. (C) MAGE− myeloma cell line Fravel. (D) Gut biopsy from patient no. 29, a patient with MAGE+ stage III myeloma treated by chemotherapy, who developed skin and gut plasmacytomas. The cells stained in brownish red are malignant plasma cells infiltrating the gut mucosa. The epithelium is indicated by an arrow. (E) Bone marrow smear from patient no. 21, a patient with MAGE+ stage III myeloma. All the plasma cells are stained in red. (F) Bone marrow cytospin from patient no. 43, who had a myeloma in relapse after 2 autografts. Some malignant plasma cells are stained in red. The arrows indicate 2 unstained myeloma cells. The slides incubated with the isotype-matched irrelevant monoclonal antibody remained negative, as did cytospins of MAGE-negative bone marrows from stage III myeloma patients incubated with antibody 57B (data not shown).
DISCUSSION
Our results show that MAGE-type genes are expressed in the bone marrow of myeloma patients. There is an obvious correlation between expression of these genes and the stage of the disease, because they are almost always silent in MGUS and stage I and II myelomas and are frequently activated in stage III myelomas. A similar correlation has already been observed in melanoma, in which metastases were found to be more frequently positive for MAGE expression than primary tumors26 and in bladder cancer, in which MAGE was expressed more frequently in infiltrative tumors than in local tumors.21 In addition, as in many solid tumors, several of these genes are frequently coexpressed in individual myeloma samples.
Our observations indicate that myeloma cells may frequently present tumor-specific shared antigens. It has been shown previously that myelomas have a high level of expression of HLA class I molecules and that they have the capacity to present antigenic peptides to T lymphocytes and to activate these cells. In addition, they often carry functional HLA class II molecules.27 Our immunocytochemistry data confirm the presence of MAGE proteins in the cytoplasm of malignant plasma cells from MAGE-expressing myeloma bone marrows. Our lysis assays show that aMAGE-expressing myeloma cell line can be killed by a CTL clone recognizing a MAGE epitope. Immunotherapy with tumor-specific shared antigens therefore constitutes a possible new treatment modality against stage III myeloma.
Several antigenic peptides that are derived from the MAGE, BAGE, GAGE, NY-ESO-1, and PRAME proteins and that are presented to CTL by HLA class I molecules have been described. Table 2gives a prediction of their frequency in advanced myeloma. It is quite probable that many additional antigens derived from the same proteins and from LAGE-1 still have to be identified. Thus, even if the probability of having a given antigen on an individual myeloma is low, many different antigens should be present simultaneously. For immunotherapy, the use of multiple immunogens is important, because it increases the probability of inducing a specific immune response and reduces the risk of tumor escape by selection of antigen-loss variants. Multiple immunogens can be delivered as a combination of peptides or as a recombinant protein. They can also be provided as a defective virus containing a gene or several minigenes coding for selected epitopes.
Currently Identified Antigenic Peptides Expressed by MAGE-type Genes
| Gene . | HLA . | Antigenic Peptide . | Position in Protein . | Predicted Frequency (%) . | Reference . |
|---|---|---|---|---|---|
| MAGE-A1 | A1 | EADPTGHSY | 161-169 | 8 | 32 |
| Cw16 | SAYGEPRKL | 230-238 | 2 | 32 | |
| MAGE-A2 | A2 | KMVELVHFL | 112-120 | 10 | 33 |
| A2 | YLQLVFGIEV | 157-166 | 10 | 33 | |
| MAGE-A3 | A1 | EVDPIGHLY | 168-176 | 8 | 32 |
| A2 | KVAELVHFL | 112-120 | 16 | 34 | |
| A2 | FLWGPRALV | 271-279 | 16 | 32 | |
| A24 | IMPKAGLLI | 195-203 | 6 | 35 | |
| B44 | MEVDPIGHLY | 167-176 | 7 | 32 | |
| MAGE-A12 | A2 | FLWGPRALV | 271-279 | 8 | * |
| BAGE | Cw16 | AARAVFLAL | 2-10 | 1 | 32 |
| GAGE-1/2 | Cw6 | YRPRPRRY | 9-16 | 7 | 32 |
| PRAME | A24 | LYVDSLFFL | 301-309 | 10 | 19 |
| NY-ESO-1 | A2 | QLSLLMWITQCFL† | 155-167 | 18 | 36 |
| A31 | ASGPGGGAPR | 53-62 | 2 | 37 | |
| LAAQERRVPR | 18-27‡ | 2 | 37 |
| Gene . | HLA . | Antigenic Peptide . | Position in Protein . | Predicted Frequency (%) . | Reference . |
|---|---|---|---|---|---|
| MAGE-A1 | A1 | EADPTGHSY | 161-169 | 8 | 32 |
| Cw16 | SAYGEPRKL | 230-238 | 2 | 32 | |
| MAGE-A2 | A2 | KMVELVHFL | 112-120 | 10 | 33 |
| A2 | YLQLVFGIEV | 157-166 | 10 | 33 | |
| MAGE-A3 | A1 | EVDPIGHLY | 168-176 | 8 | 32 |
| A2 | KVAELVHFL | 112-120 | 16 | 34 | |
| A2 | FLWGPRALV | 271-279 | 16 | 32 | |
| A24 | IMPKAGLLI | 195-203 | 6 | 35 | |
| B44 | MEVDPIGHLY | 167-176 | 7 | 32 | |
| MAGE-A12 | A2 | FLWGPRALV | 271-279 | 8 | * |
| BAGE | Cw16 | AARAVFLAL | 2-10 | 1 | 32 |
| GAGE-1/2 | Cw6 | YRPRPRRY | 9-16 | 7 | 32 |
| PRAME | A24 | LYVDSLFFL | 301-309 | 10 | 19 |
| NY-ESO-1 | A2 | QLSLLMWITQCFL† | 155-167 | 18 | 36 |
| A31 | ASGPGGGAPR | 53-62 | 2 | 37 | |
| LAAQERRVPR | 18-27‡ | 2 | 37 |
The predicted frequency of these antigens in advanced myelomas (stage III or plasma cell leukemia) is obtained by multiplying the probability of having the corresponding gene expressed by the myeloma and the prevalence of the HLA allele in the Caucasian population.
Identical to MAGE-A3(271-279) peptide.
Optimal antigenic sequence remains to be determined.
Alternative open reading frame.
Clinical trials aimed at evaluating MAGE-derived immunogens against solid tumors, mainly melanomas, are ongoing. In a recent report, immunization of advanced melanoma patients with the MAGE-3.A1 peptide led to objective regressions of metastases in 7 of 25 patients with measurable disease who completed the treatment. No toxicity was observed. However, no specific CTL response could be detected in the blood of a subset of patients, including 2 who showed tumor regressions.28 The encouraging clinical observation deserves further investigation. It is hoped that increased frequency of tumor regressions and correlation with measurable immune response will be achieved by using better immunogens, such as recombinant proteins or viruses, by associating these immunogens with immunological adjuvants or immunostimulatory cytokines, and by developing improved assays for measuring antigen-specific CTL responses.
Myeloma might represent a valuable model to study the immunological and clinical responses to vaccination with MAGE-type immunogens. It is a chronic, incurable disease whose evolution can be monitored easily. Moreover, myeloma bone marrows may constitute a reproducible source of tumor-infiltrating lymphocytes, allowing for a more precise assessment of CTL responses.
Our observations also have an implication with respect to the follow-up of advanced myeloma patients treated by chemotherapy. Given that genesMAGE, BAGE, GAGE, LAGE-1, and NY-ESO-1 are transcribed in myeloma cells but not in normal nongerminal tissues, including hematopoietic cells, the presence of the corresponding mRNA can be considered as a tumor-specific marker. The same applies toPRAME, because it is not expressed in normal bone marrow or peripheral blood leukocytes. Thus, the high sensitivity and specificity of RT-PCR could allow the detection of a very low proportion of myeloma cells in the bone marrow or in the blood. This would be particularly useful to assess the response to high-dose chemotherapy, to detect residual disease in patients with cytological remission, and to detect early relapse in these patients. Moreover, the detection of low expression levels of MAGE-type mRNA in bone marrow from patients with MGUS and stage I and stage II myeloma may predict the evolution towards more advanced disease. It can be deduced from Table 1 that the detection of all the samples that express at least 1 of the tumor-specific genes analyzed in this report can be performed by RT-PCR using only primers specific for GAGE, LAGE-1, andPRAME.
The expression of the MAGE-type genes in tumors has been demonstrated to be linked to overall DNA demethylation. Because almost 80% of stage III myelomas tested in our series expressed at least 1 of these genes or PRAME, we consider it likely that overall DNA demethylation occurs in many advanced myelomas. Genome-wide demethylation acquired by a few tumor cells may result in transcription of oncogenes that would otherwise remain silent. Their aberrant activation could confer a growth advantage to these cells as compared with normally methylated cells. It is unclear which oncogenes could become activated in myeloma as a result of demethylation. Among possible candidates, c-MYC is known to be frequently overexpressed in malignant plasma cells. In some myelomas, its overexpression seems to be a consequence of a fusion with the IgH locus in the t(8;14) translocation.29 In 1 report, a CpG site in the third exon of the c-MYC gene was found to be demethylated in 5 myeloma cell lines that had a significant overexpression ofc-MYC as compared with normally methylated cells.30However, a link between demethylation and c-MYC overexpression remains to be demonstrated in fresh myeloma cells. It should be mentioned that the function of the MAGE, BAGE, GAGE, LAGE-1, NY-ESO-1, and PRAME proteins is unknown, and their involvement in tumor progression remains unproven. The activation of these genes may be a neutral side effect of a global demethylation process that activates other genes involved in tumor progression.
It is important to note that, in 1 of the 3 samples analyzed by immunocytochemistry, only a subset of the myeloma cells were stained by the anti-MAGE antibody. This observation is consistent with the acquisition of DNA demethylation by a few cells that will grow faster and then progressively overwhelm the others. Targeting these more tumorigenic cells by specific CTL might remain a valuable aim, because the remaining myeloma cells would probably show less aggressiveness. Immunization of stage I and II myeloma patients to prevent the emergence of demethylated myeloma cells and the resulting progression to more aggressive disease should also be considered.
ACKNOWLEDGMENT
The authors are grateful to M. Hérin and J.-M. Scheiff for reviewing the immunohistochemistry data. We thank C. Mondovits, B. Tollet, and M. Swinarska for excellent technical assistance; S. Mapp for his help in the preparation of the manuscript; and P. Coulie for helpful discussions.
Supported by the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming, and by grants from the Association contre le Cancer (Brussels, Belgium), from the BIOMED2 programme of the European Community, from the Fonds J. Maisin (Belgium), from CGER-Assurances and VIVA (Brussels, Belgium), from the Fonds National de la Recherche Scientifique (TELEVIE grants) (Brussels, Belgium), and from the Swiss National Fonds Grant No. 31-45560.95.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Nicolas van Baren, MD, Ludwig Institute for Cancer Research, 74 avenue Hippocrate, UCL 74.59, B-1200 Brussels, Belgium.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal