Abstract
Neutrophil elastase, proteinase-3, and azurocidin are primary components of neutrophil azurophilic granules and are encoded by closely linked genes (gene symbols ELA2, PRTN3, and AZU1, respectively) in a region of approximately 50 kb. These genes are coordinately expressed in a granulocyte-specific fashion, but the mechanisms defining this pattern of expression are unknown. To understand the role of chromatin organization in governing the expression of ELA2, PRTN3, and AZU1, we mapped this region of chromosome 19 and identified the adipsin (complement factor D) gene in proximity to the 3′ end of ELA2. We then examined the changes in chromatin structure at the locus which accompany myeloid cell differentiation and identified 17 DNase I hypersensitive sites (DHS 1 to 17) in U-937 cells, an early myelomonocytic cell line expressing high levels of neutrophil elastase. Chemically induced differentiation and concomitant downregulation of AZU1, PRTN3, and ELA2 transcription in U-937 cells is not accompanied by changes in the DHS-pattern. Mature neutrophils, however, do not carry any of these hypersensitive sites, indicating a large degree of chromatin remodeling at this locus accompanying terminal granulocytic differentiation. Sixteen of the 17 DHS identified in U-937 cells are also present in the HL-60 myelomonocytic cell line. Hematopoietic cell lines representing the early erythroid and lymphocyte lineages, and a nonhematopoietic cell line display a subset of the hypersensitive sites. The altered chromatin structure specific to cells that actively transcribe the AZU1-PRTN3-ELA2 genes suggests that chromatin reorganization is an important mechanism regulating the myeloid-specific transcription of this gene cluster.
HUMAN NEUTROPHIL elastase (also known as leukocyte elastase and medullasin), proteinase 3 (also known as myeloblastin and AGP-7), and azurocidin (also known as CAP37) are major components of the azurophilic (primary) granules of neutrophils. Neutrophil elastase possesses a broad proteolytic spectrum, functioning primarily as an antimicrobial agent against gram-negative bacteria1; it is also capable of degrading many extracellular matrix components and has thus been extensively studied as a potential agent of tissue destruction in a number of diseases, particularly those accompanied by evidence of inflammation and connective tissue damage such as pulmonary emphysema,2-4rheumatoid arthritis,5 cystic fibrosis,6 and glomerulonephritis.7 Proteinase 3 has proteolytic properties similar to neutrophil elastase and is believed to affect myeloid cell differentiation8-10; it has been most widely studied for its role as the autoantigen in Wegener’s granulomatosis.11 Azurocidin is a serine protease homolog with no proteolytic activity, but possesses bactericidal and chemotactic properties.12
Transcription of the human neutrophil elastase, proteinase 3, and azurocidin genes (gene symbols ELA2, PRTN3, and AZU1, respectively) is restricted to the promyelocyte stage of granulocytic differentiation; expression ceases with differentiation to mature granulocytes, and is coordinately downregulated on phorbol ester induction of U-937 myelomonocytic cells.13,14 AZU1, PRTN3, and ELA2 are structurally related, belonging to class 6 of the trypsin superfamily of serine protease genes, and are closely linked in a region of approximately 50 kb (the ELA2 locus).15 The mechanisms governing the high-level of transcription of AZU1, PRTN3, and ELA2 specifically in myeloid cells that are committed to granulocytic differentiation are poorly understood. Previous studies of ELA2 and the mouse neutrophil elastase homolog have shown minimal 5′ proximal promoter regions, which appear to have myeloid specific, but not necessarily stage-restricted activity in transient transfection assays.16,17 The ELA2 promoter and mouse neutrophil elastase promoter share a notable degree of homology, containing consensus sequences for c-myb, C/EBP, and PU.1/GABP, which are prevalent in many myeloid-specific promoters.18,19 An enhancer of transcription has also been identified approximately 1 kb upstream of the transcription initiation site of ELA2 with activity in both myeloid and heterologous cell types.20Cis-acting elements important in regulation of expression of PRTN3 and AZU1 are less well characterized than those of ELA2; however, the physical proximity of the 3 genes, their similar gene structure, and coordinate pattern of expression suggest that the transcription of these genes may be similarly regulated and perhaps under the influence of common regulatory elements.
We have previously produced transgenic mice using a transgene whose expression is driven by the ELA2 promoter and −1 kb enhancer region. There was no detectable expression of the transgene in any of the lines established, indicating that despite the apparent activity of the proximal ELA2 promoter region in transient transfection assays, it is not sufficient to reproduce the expression pattern of human neutrophil elastase when stably integrated in vivo (E.W., unpublished observations, July 1996). This implied that additionalcis-regulatory elements are necessary for directing the high levels of promyelocyte-specific expression of neutrophil elastase that are normally observed in vivo, when ELA2 is in the context of chromatin.
Chromatin structure has been shown to play an important role in the transcriptional regulation of eukaryotic genes.21,22 DNase I hypersensitive sites (DHS) in particular, indicate regions of chromatin that are preferentially devoid of regular nucleosomal arrangement, and in many cases have been shown to have functional significance, often coinciding with active cis-transcriptional regulatory elements, origins of replication, matrix attachment sites, and recombinational hot-spots.23 In the present study, we characterize the genomic regions flanking the AZU1-PRTN3-ELA2 gene cluster and identify a fourth member of the serine protease gene family, adipsin (complement factor D, properdin factor D; gene symbol, ADN) in close proximity to the 3′ end of ELA2. We then identify regions of open chromatin structure within and surrounding the AZU1-PRTN3-ELA2-ADN (APEA) cluster of genes that are specific to cell types that actively transcribe ELA2, PRTN3, and AZU1, indicating a potentially complex mode of gene control, and identifying regions of putative transcriptional regulatory activity.
MATERIALS AND METHODS
Cells.
U-937 (American Type Culture Collection [ATCC] CRL1593), HL-60 (ATCC CCL240), HUT-78 (ATCC TIB161), Jurkat (ATCC TIB152), and K-562 (ATCC CCL243) cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (GIBCO-BRL, Gaithersburg, MD) and maintained at 37°C and 5% CO2; COLO 201 cells (ATCC CCL224) were grown in RPMI 1640 medium supplemented with 20% fetal calf serum and maintained under the same conditions. For induction of differentiation, U-937 cells were cultured for 72 hours in the presence of 25 ng/mL TPA (12-O-tetradecanoyl-phorbol-13-acetate, also phorbol 12-myristate 13-acetate) (GIBCO-BRL). Polymorphonuclear granulocytes were isolated from heparinized donor blood from a healthy donor using Histopaque-1119 and -1077 (Sigma Diagnostics, St Louis, MO) double density gradient centrifugation according to the manufacturer’s directions.
DNase I hypersensitive site mapping.
Near-confluent cultures were obtained, and nuclei were isolated and subjected to incremental DNase I digestion as previously described.24
Southern blotting.
Restriction-digested genomic DNA was separated using electrophoresis in 0.65% to 0.85% agarose, blotted onto Nytran Plus membrane (Schleicher and Schuell, Keene, NH) and hybridized with [α-32P]dCTP random primed labeled probes: The probes are derived from AZU1 and PRTN3 human cDNA clones,13 from the human ELA2 cDNA clone, a gift from M. Naruto (Kamakura, Japan),25 and through standard polymerase chain reaction (PCR) amplification of human cDNA fragments as indicated in Fig 3. Hybridized blots were washed at a final stringency of 0.1 × SSC/ 0.1% SDS for 2 hours and exposed to Kodak X-AR film.
Isolation of RNA and Northern blot analysis.
Total RNA was isolated from cells with Trizol Reagent (GIBCO-BRL) according to the manufacturer’s directions. For Northern analysis, 10 μg of total RNA was denatured, separated on a 1% agarose-formaldehyde gel, transferred to Nytran Plus membrane, UV cross-linked and hybridized as above. The same blots were then stripped and hybridized with a human β-actin cDNA radiolabeled probe as described.
Isolation of genomic clones and restriction site mapping.
Because we suggested the location of ADN on chromosome 19p13.3 in close vicinity to the AZU1-PRTN3-ELA2 gene cluster, a partial cDNA probe for human adipsin (pDJ3) was generated by PCR using the primers DJ21 (5′-CTGGGGCATAGTCAACCA-3′) and DJ34 (5′-TGTCGATCCAGGCCGCATAG-3) with cDNA from U937 cells and was used to screen a chromosome 19–specific cosmid library. Three positive clones, R27353, R32285, and R27805, were identified and further characterized by restriction site mapping and Southern blotting using BamHI, EcoRI, Hind III, and Sac I (Fig1). The latter restriction sites were mapped after linearization of cosmid DNA with lambda terminase and partial enzyme digestion. On the basis of cDNA sequence, oligonuceotides were designed to amplify genomic fragments from cosmid R27805 using Taq polymerase and Elongase (GIBCO-BRL) to determine the size and location of the introns. Exon-intron boundaries were identified by directing sequencing of purified cosmid DNA using the same primers.
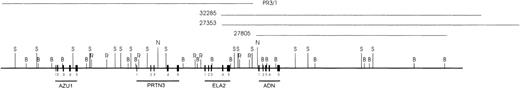
Restriction enzyme map of the AZU1-PRTN3-ELA2-ADN (APEA) locus. Cosmids used to identify the relative position of the ADN gene to the known genes in the locus are shown as thin horizontal lines above the restriction map. Restriction sites mapped are indicated. (B, BamHI; N, Not I; R, EcoRI; S, Sal I). Relative positions of the identified exons are shown for ADN (thick lines), and also for AZU1, PRTN3, and ELA2.
Restriction enzyme map of the AZU1-PRTN3-ELA2-ADN (APEA) locus. Cosmids used to identify the relative position of the ADN gene to the known genes in the locus are shown as thin horizontal lines above the restriction map. Restriction sites mapped are indicated. (B, BamHI; N, Not I; R, EcoRI; S, Sal I). Relative positions of the identified exons are shown for ADN (thick lines), and also for AZU1, PRTN3, and ELA2.
RESULTS
Physical relationship between the ELA2 and ADN genes.
Adipsin and complement factor D were shown to represent orthologous gene products with identical functions in humans and mice.26 The structure of the mouse adipsin gene has been previously reported.27 Examining these exon-intron data we noticed that the location and phase type of introns in mouse adipsin was completely identical to that of other neutrophil serine protease homologs found on human chromosome 19p13.3, but differed in a subtle manner from other serine protease homologs clustered on 5q11-12 and 14q11-12.15 Moreover, many structural features like the length of the putative propeptide and the number of cysteine bonds in complement factor D were shared with known members of the 19p13.3 serine protease gene cluster. Thus we wished to prove our hypothesis that complement factor D gene is a fourth member of the neutrophil enzyme cluster on 19p13.3. As expected, screening of a human cosmid library for chromosome 19 with the pDJ3 partial cDNA probe was successful. The three cosmids R27805, R32285, and R27353 identified were shown to contain identical restriction site fragments and hybridized to the ADN cDNA probe in Southern blots.
Figure 1 shows the composite restriction site map for the 3 ADN-positive cosmids and the PRTN3-positive cosmid, PR3/1, previously reported.13 The latter cosmid appeared to overlap cosmid R32285 and R27353 by a few kb (Fig 1). To confirm this overlap, both ends of the R32285 insert were sequenced. One end was found to match the sequence of intron 3 of ELA2. Figure 1 also shows the location of the 5 ADN exons. Exon-intron junctions of the ADN gene and transcriptional orientation were the same as for AZU1, PRTN3, and ELA2. To confirm the 5′ end of the ADN cDNA and translational start site as predicted from genomic sequences, we performed 5′ RACE and compared these cDNA sequences with those obtained from cosmid clones and an ADN cDNA sequence previously reported (data not shown). Indeed, 7 additional codons including the first methionine codon were found at the 5′ end which, together with the following 12 amino acid residues, make up the signal peptide of human ADN.
Models for analysis of chromatin structure at the AZU1/PRTN3/ELA2/ADN (APEA) locus.
We chose to use the U-937 cell line as a model to study the chromatin structure of the APEA gene locus because they are arrested at an early stage of myeloid differentiation and express neutrophil elastase at high levels.28 To determine whether chromatin structure at the APEA locus is specific to cell lines that actively transcribe ELA2 (the human neutrophil elastase gene), we also studied nonmyeloid hematopoietic cell lines and a representative nonhematopoietic cell line. HUT-78 is a mature T-cell line and Jurkat cells are lymphoid cells derived from an acute T-cell leukemia. K-562 cells are derived from erythromyeloblastic cells arrested at an early stage of differentiation. COLO 201 cells are a nonhematopoietic, colonic epithelial cell line. Among these cell lines, ELA2 transcription is restricted to the myeloid cell type (Fig2). Treatment of U-937 cells with the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) results in downregulation of AZU1, PRTN3, and ELA transcription. Although downregulation of ELA2 and PRTN3 in U-937 is complete in 5 days, we observed that the same cells still produce detectable though greatly reduced levels of AZU1 transcripts. A previous study did not detect differences in the levels of downregulation between the 3 genes13 and may be because of differences between U-937 sublines, to prolonged TPA exposure, or to differences in culture conditions. ADN exhibits a less restricted pattern of expression than ELA2, PRTN3, and AZU1. ADN transcripts are present in all the hematopoietic cell lines examined and are also downregulated to low levels in TPA activated U-937 cells like AZU1 transcripts, but are not detectable in the nonhematopoietic cell line COLO 201. The level of ADN transcription in TPA-treated U-937 cells is similar to that of other cultured hematopoietic cell lines. HL-60 cells were examined to confirm the presence of the DHS identified in U-937 cells. HL-60 cells are also myelomonocytic cells, which express high levels of NE.28Mature peripheral blood granulocytes do not express any of the 4 genes of this cluster (not shown).
Expression of the AZU1-PRTN3-ELA2-ADN cluster of genes in cell lines studied. Northern analysis was performed on U-937 cells, U-937 cells treated with 25 ng/mL TPA for 3 days (TPA), COLO 201 (COLO), HUT 78 (HUT), Jurkat, and K-562 cell lines and hybridized to32P-labeled human ELA2, AZU1, PRTN3, ADN, and human β-actin cDNA probes as indicated for each blot.
Expression of the AZU1-PRTN3-ELA2-ADN cluster of genes in cell lines studied. Northern analysis was performed on U-937 cells, U-937 cells treated with 25 ng/mL TPA for 3 days (TPA), COLO 201 (COLO), HUT 78 (HUT), Jurkat, and K-562 cell lines and hybridized to32P-labeled human ELA2, AZU1, PRTN3, ADN, and human β-actin cDNA probes as indicated for each blot.
DNase I hypersensitive mapping of the APEA gene cluster in neutrophil elastase expressing cells.
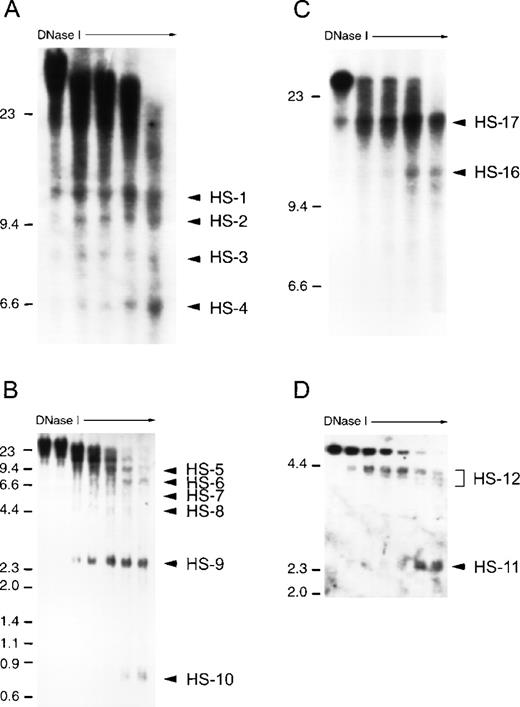
We performed DHS analyses on a greater than 50 kb region surrounding the APEA gene cluster. DHS analysis of more distant DNA was not performed because no serine protease gene sequences were detected beyond this gene cluster within 50 kb in either direction using PCR primer pairs for conserved serine protease gene sequences. During the process of locating genes and ESTs on our cosmid contig, we identified the gene for polypyrimidine tract binding protein (PTB) on the telomeric side of AZU1 and the human transcript Hs.24441 (Unigene designation) on the centromeric side of ADN (data not shown), indicating that the region encompassing the 4 serine protease genes represents a complete multigene locus. An array of distinct DHS are detected at the APEA locus in U-937 cells, within a region from 15 kb upstream of the AZU1 gene to approximately 15 kb centromeric to ADN. Multiple overlapping restriction enzyme series were used to confirm the location of each DHS (Fig 3). Typical blots are shown, showing each of the 17 DHS observed (Figs 4 and5). All DHS present in U-937 cells were present in HL-60 cells, with the exception of DHS-16. DHS-12 (Fig 4A) and 14 (Fig 5) appear as doublets or triplets on some Southern blots; on others they appear as a single intense band and they have each been considered to be a single DHS. Although analysis of nuclease hypersensitivity is not a quantitative technique, differing intensities of each DHS relative to adjacent sites on the same blot can be recognized. DHS-4, 5, 10, and 12 coincide with the promoter regions of AZU1, PRTN3, ELA2, and ADN, respectively (Figs2, 4A, 4B, and 5). Two sets of hypersensitive sites, DHS-1 to -4 and DHS-16 and -17, flank the locus (Figs 2, 4A and 4C). DHS-6 is located within the PRTN3 gene (Figs 2 and 4B), and DHS-11 appears in the 3.5 kb region between ELA2 and ADN (Figs 2, 4D, and 5). DHS-9 is a particularly prominent site in U-937 and HL-60 cells, appearing approximately 2 kb upstream of ELA2 (Fig 4B). DHS-13 and -15 are very weak sites in U-937 cells, appearing stronger in nonexpressing cells such as COLO 201 (Fig 5).
Scale map of DHS in U-937 cells (vertical arrows) relative to the AZU1, PRTN3, ELA2, and ADN genes and to restriction fragments used for DHS mapping (short vertical lines), (A, Hpa I; B, BamHI; C, Sca I; G, Bgl II; L, Bcl I; M, Mlul; R, EcoRI; X, Xbal, and Y, BssHII). Horizontal arrows depict orientation of end-labeled detection of DHS. Exons from the 4 genes used as probes for DHS analysis are shown (half-boxes) for each restriction fragment used for DHS mapping. The EcoRI fragment encompassing AZU1 extends 24 kb beyond the AZU1 gene and the EcoRI fragment at the ADN end of the locus is 31 kb. Multiple analyses using different restriction fragments were used to confirm the location of the DHS in U-937 and in other cell lines.
Scale map of DHS in U-937 cells (vertical arrows) relative to the AZU1, PRTN3, ELA2, and ADN genes and to restriction fragments used for DHS mapping (short vertical lines), (A, Hpa I; B, BamHI; C, Sca I; G, Bgl II; L, Bcl I; M, Mlul; R, EcoRI; X, Xbal, and Y, BssHII). Horizontal arrows depict orientation of end-labeled detection of DHS. Exons from the 4 genes used as probes for DHS analysis are shown (half-boxes) for each restriction fragment used for DHS mapping. The EcoRI fragment encompassing AZU1 extends 24 kb beyond the AZU1 gene and the EcoRI fragment at the ADN end of the locus is 31 kb. Multiple analyses using different restriction fragments were used to confirm the location of the DHS in U-937 and in other cell lines.
Analysis of chromatin structure at the AZU1-PRTN3-ELA2-ADN (APEA) locus in untreated U-937 cells. Nuclei were prepared and treated with increasing concentrations of DNase I (0 to 5 μg/mL, increasing from left to right in each case). The location of molecular weight DNA markers are shown on the left, in kilobases. DHS are indicated by solid arrowheads, with DHS-1 lying toward the AZU1 portion of the locus and DHS-15 at the ADN end of the locus. (A) DHS-1 to -4. DNase I–treated DNA was digested to completion with EcoRI and hybridized with a PCR amplified, 32P-labeled AZU1 exon 4/5 cDNA probe. (B) DHS-5 to -10. DNase I–treated DNA was digested to completion with Mlu I and hybridized with a PCR-amplified ELA2 exon I/II probe. (C) DHS-16 and -17. DNase I–treated DNA was digested to completion with Eco RI and hybridized with a 32P-labeled ADN cDNA probe. (D) DHS-11 and -12. DNase I–treated DNA was digested to completion with Bam HI and hybridized with a 32P-labeled ELA2 exon IV/V probe.
Analysis of chromatin structure at the AZU1-PRTN3-ELA2-ADN (APEA) locus in untreated U-937 cells. Nuclei were prepared and treated with increasing concentrations of DNase I (0 to 5 μg/mL, increasing from left to right in each case). The location of molecular weight DNA markers are shown on the left, in kilobases. DHS are indicated by solid arrowheads, with DHS-1 lying toward the AZU1 portion of the locus and DHS-15 at the ADN end of the locus. (A) DHS-1 to -4. DNase I–treated DNA was digested to completion with EcoRI and hybridized with a PCR amplified, 32P-labeled AZU1 exon 4/5 cDNA probe. (B) DHS-5 to -10. DNase I–treated DNA was digested to completion with Mlu I and hybridized with a PCR-amplified ELA2 exon I/II probe. (C) DHS-16 and -17. DNase I–treated DNA was digested to completion with Eco RI and hybridized with a 32P-labeled ADN cDNA probe. (D) DHS-11 and -12. DNase I–treated DNA was digested to completion with Bam HI and hybridized with a 32P-labeled ELA2 exon IV/V probe.
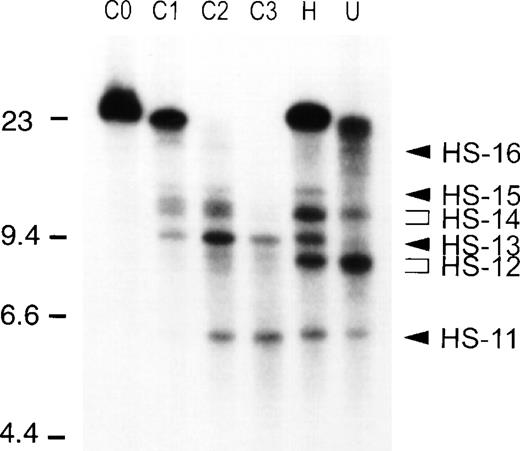
Analysis of DHS-11 to -16 in COLO 201, U-937, and HL-60 cells. All lanes are DNase I–treated DNA digested to completion with Bgl II and hybridized with a 32P-labeled ELA2 exon IV/V cDNA probe. From left to right: C0, COLO 201 DNA not treated with DNase I; C1 to C3, COLO 201 DNA from nuclei treated with increasing concentrations of DNase I; H, HL-60 DNase I treated time-point; U, U-937 DNase I treated time-point. The locations of the DNA molecular weight markers used are shown on the left, and the location of DHS-11 to -16 are indicated by arrowheads on the right.
Analysis of DHS-11 to -16 in COLO 201, U-937, and HL-60 cells. All lanes are DNase I–treated DNA digested to completion with Bgl II and hybridized with a 32P-labeled ELA2 exon IV/V cDNA probe. From left to right: C0, COLO 201 DNA not treated with DNase I; C1 to C3, COLO 201 DNA from nuclei treated with increasing concentrations of DNase I; H, HL-60 DNase I treated time-point; U, U-937 DNase I treated time-point. The locations of the DNA molecular weight markers used are shown on the left, and the location of DHS-11 to -16 are indicated by arrowheads on the right.
Differential chromatin structure at the APEA cluster of genes in cells that do not express neutrophil elastase.
Treatment of U-937 cells with TPA has previously been shown to induce partial differentiation of these cells.29 Although TPA-induced differentiation is accompanied by downregulation of ELA2, PRTN3, and AZU1 transcription, the DHS pattern at the APEA locus in these cells is identical to that of untreated, expressing U-937 cells in terms of position and relative intensity (Table1 and not shown), indicating that change in chromatin structure at this locus is not necessary for transcriptional downregulation of the ELA2, PRTN3, and AZU1 genes. The DHS observed in the early myeloid U-937 cell line, however, are not present in mature peripheral neutrophils (PMN), evidence that chromatin reorganization at this region accompanies terminal neutrophilic differentiation. In the nonmyeloid hematopoietic cell lines studied, only a subset of the DHS observed in U-937 cells exist (Table 1): DHS-1 appears only in the erythroid line, K-562. DHS-2 appears in both K-562 and COLO 201 cells. DHS-16 appears in COLO 201 and also in HUT-78 cells but is not present in HL-60 cells. The other sites, DHS-3 to -10, appear exclusively in the early myeloid cell type. DHS-17 is present in COLO 201 cells and in all the hematopoietic lineages but not in mature neutrophils and is the only site common to all the hematopoietic and nonhematopoietic lineages. The sites located within the ADN gene are present in all cell lines examined, with the exception of mature neutrophils.
Presence of DNase Hypersensitive Sites in Myeloid and Nonmyeloid Cell Lines
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U-937 | ↓ | ↓ | ↓ | ||||||||||||||
| +TPA | ↓ | ↓ | ↓ | ||||||||||||||
| HL-60 | |||||||||||||||||
| COLO | ↓ | ||||||||||||||||
| HUT | ↓ | ||||||||||||||||
| Jurkat | ↓ | ||||||||||||||||
| K562 | ↓ | ||||||||||||||||
| PMN |
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U-937 | ↓ | ↓ | ↓ | ||||||||||||||
| +TPA | ↓ | ↓ | ↓ | ||||||||||||||
| HL-60 | |||||||||||||||||
| COLO | ↓ | ||||||||||||||||
| HUT | ↓ | ||||||||||||||||
| Jurkat | ↓ | ||||||||||||||||
| K562 | ↓ | ||||||||||||||||
| PMN |
The presence of DHS-1 to -17 (columns) are indicated by open arrows or thin arrows (extremely weak or barely distinguishable site) in each of the cell lines studied (rows). The presence of each DHS was confirmed by overlapping DHS analyses as for Fig 3.
Abbreviations: +TPA, TPA-treated U-937 cells; PMN, peripheral blood granulocytes.
DISCUSSION
Studies to date on the regulation of transcription of the human neutrophil elastase and proteinase 3 genes (ELA2 and PRTN3, respectively) have focused only on the proximal promoter regions of these genes and the putative trans-acting factors that may interact with these regions. Although the ELA2 and PRTN3 promoters have functional activity in transient transfection assays, they are unable to direct expression of a transgene in vivo, indicating that additionalcis-regulatory elements are necessary to reproduce the endogenous pattern of expression of neutrophil elastase and proteinase 3 when stably integrated into chromatin. An “active” chromatin conformation is an important factor in permitting high-level, cell-type specific, and position-independent expression of stably integrated genes.30-32
To show changes in chromatin structure at the ELA2 locus with probes from the flanking regions, we extended the map beyond the 3′ UTR of the ELA2 gene and identified an additional member of the serine protease gene family, which encodes human adipsin (also complement factor D). In contrast to AZU1, PRTN3, and ELA2, adipsin is not found in granules of neutrophils, but is present in the circulation at relatively high levels of 50 μg/mL. The enzyme is mainly synthesized and secreted by adipocytes, but also by monocytes and macrophages and is already converted into the mature form before secretion.33Adipsin/factor D is a key regulator of the alternative pathway of the complement system with absolute substrate specificity for factor B as a component of the reversible C3bB complex. It only acquires its proteolytically active conformation during cleavage of the C3bB complex. Although adipsin has not been regarded as a genuine member of the hematopoietic serine protease gene family previously, exon-intron organization of the adipsin gene (ADN) is completely conserved, indicating that it is indeed derived from a common ancestor for hematopoietic serine proteases. In support of this view is the finding that adipsin is also expressed by monocytes and macrophages, but not by human or rodent hepatocytes, which synthesize all other components of the complement system. Furthermore, adipsin and the other neutrophil enzymes are still related in their biological functions in that all these serine protease homologs contribute to bactericidal defense mechanisms.9,12 34
In the present study, we observed an altered chromatin conformation at the APEA locus in cells that are actively transcribing the ELA2, PRTN3, and AZU1 set of genes. The specificity of DHS-3 to -10 to both U-937 and HL-60 cells, which express the ELA2, PRTN3, and AZU1 genes, indicates that they may be involved in transcriptional upregulation of these genes; however, TPA-induced downregulation of ELA2, PRTN3, and AZU1 transcription is not accompanied by a corresponding reorganization of the active chromatin conformation. These observations indicate that although active chromatin conformation appears to be an important event in permitting gene expression of ELA2, PRTN3, and AZU1, it is not sufficient in itself to result in normal levels of myeloid specific transcription. This observation contrasts with the myeloperoxidase (MPO) model, in which chemically induced granulocytic differentiation and downregulation of expression of MPO is associated with loss of 3 DHS in the region proximal to the 5′ end of the gene.35,36Although there are regions of extensive homology between the promoters of MPO and ELA2,37 the promoters of ELA2, PRTN3, and AZU1 possess a TATAA consensus sequence, unlike the promoters of MPO and the majority of other myeloid-specific gene promoters that have been studied. Furthermore, unlike expression of the AZU1, PRTN3, and ELA2 genes, MPO expression is not specific to the neutrophilic path of myeloid differentiation but is also expressed during monocytic differentiation. The significance of the differences between chromatin remodeling at the ELA2 and MPO loci in relation to potential similarities or differences between their mechanisms of transcriptional regulation is presently unknown. DHS-11 to -15 are present in both myeloid and nonmyeloid cell types, which, together with their location in or adjacent to the ADN gene, suggests a role in the positive regulation of this gene.
The absence of observable change in chromatin organization in TPA-treated U-937 cells suggests that downregulation of the AZU1, PRTN3, and ELA2 genes induced by TPA is likely to be a consequence of the effects of TPA on trans-acting factors involved in the transcription of these genes. C/EBPα and c-myb are candidate transcription factors that may be involved in TPA-mediated downregulation of these genes. Both C/EBPα and c-myb are expressed in early myeloid cells and their levels are decreased on differentiation. Consensus binding sites for C/EBP factors are present in the promoters of all 3 genes and c-myb binding sequences are present in the promoters of ELA2 and PRTN3; both C/EBP and c-myb consensus sequences are important in ELA2 and PRTN3 promoter activity, indicating a role for these factors in mediating transcription of these genes.38,39 In particular, TPA-induced differentiation in HL-60 cells has been shown to reduce c-myb levels.40 The ets factors PU.1 and GABP are also likely to be involved in transcriptional regulation at the ELA2 locus. Ets binding sites are present in the promoters of ELA2 and PRTN3, and mutation of this site decreases promoter function.41 TPA-induced differentiation of U-937 cells is accompanied by phosphorylation of PU.1, affecting PU.1 binding activity.42 Treatment with a protein kinase C inhibitor antagonizes TPA-induced differentiation, indicating that decrease in ELA2 and PRTN3 gene expression after TPA treatment in U-937 cells may be mediated in part by alteration of PU.1 activity. Hence, though transcription at the locus may require an active chromatin structure, regulation by trans-acting factors alone may be sufficient for mediating the initial downregulation of transcription in myeloid cells accompanying differentiation of the promyelocyte. The broad similarity in the pattern of nuclease hypersensitivity between HL-60 and U-937 cells in comparison to nonmyeloid cells, however, suggests that this pattern of chromatin organization is likely to be important in regulating expression of the genes at this locus.
Although the AZU1, PRTN3, and ELA2 genes can be downregulated in myeloid cells in the presence of an active chromatin conformation, it is possible that an “inactive” chromatin conformation characterized by the absence of DHS seen in nonmyeloid cell types may be important in preventing aberrant expression of these genes. This would allow transcriptional silencing of the AZU1, PRTN3, and ELA2 genes even in cell types where C/EBPα, c-myb, PU.1, or othertrans-acting factors involved in transcription of these genes are present. Because TPA-induced transcriptional downregulation in U-937 cells is not accompanied by change in the active chromatin conformation of the APEA locus, loss of the DHS accompanying terminal neutrophil differentiation occurs subsequent to transcriptional downregulation and may possibly serve as an additional barrier against aberrent expression of these genes. The biological significance of such a stringent control of transcription of these genes is substantiated by the destructive nature of neutrophil elastase and proteinase 3 and their potential contribution to inflammatory-related disease. Specific downregulation of expression at the APEA locus mediated by inactivation of transcriptionally permissive chromatin structure is a potential means through which this repression may be implemented.
The physical proximity, common spatial and temporal pattern of expression, homologous gene structure, and coordinate downregulation of AZU1, PRTN3, and ELA2 suggest that the mechanisms governing the transcriptional regulation of these genes are related and exist in a common transcriptional regulatory domain. If regulation of transcription of these genes is indeed governed by commoncis-acting elements, then it is conceivable that an insulatory element exists between the ELA2 and ADN gene, as ADN exhibits a pattern of expression that is distinct from the other genes in the region. Studies in other model systems have shown the existence of such insulatory elements that can serve to delimit transcriptional units.43 The most distal 3′ DHS, DHS-17, is present in all the cell types examined except mature neutrophils. It may perhaps serve an analogous role to the chicken β-globin constitutive DHS at the 5′ domain border, which insulates the domain from neighboring chromatin and transcriptional regulatory influences.44 Functional characterization of the DHS at this locus both in vitro and in transgenic animals will show their associated functions, some of which are likely to be involved in mediating the stringent, high-level expression of ELA2, PRTN3, and AZU1 in granulocytic precursors in vivo.
The cathepsin G cluster of genes on chromosome 14 shares structural similarities with the ELA2 group of genes, consisting of the granzyme B, granzyme H, cathepsin G, and mast cell chymase genes. Cathepsin G is expressed concomitantly with ELA2.45 A 6-kb genomic fragment encompassing the cathepsin G gene with 2.7 kb of 5′ flanking DNA has been shown to direct myeloid-specific expression when used as a transgene46 whereas a 1.2-kb 5′ flanking fragment of granzyme H was shown to direct expression to lymphokine-activated natural killer cells.47 Further functional analysis of the DHS at the ELA2 locus or chromatin structural analysis of the cathepsin G locus may show potential similarities or differences in how these 2 structurally similar loci are regulated.
ACKNOWLEDGMENT
We are grateful to Heike Reimann for excellent technical assistance and to Dr H. Mohrenweiser, Lawrence Livermore National Laboratory, Livermore, CA, for screening their chromosome 19 library with pDJ3.
Supported by grants from the British Columbia Health Research Foundation, Medical Research Council of Canada, and Sonderforschungsbereich 469.
REFERENCES
Author notes
Address reprint requests to C. Blake Gilks, MD, ACU Building, G227-2211 Wesbrook Mall, Vancouver, BC, Canada, V6T 2B5.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal