Abstract
We determined the type and frequency of chromosomal aberrations in leukemic cells of 478 children diagnosed with acute myeloid leukemia and enrolled in the Pediatric Oncology Group study 8821. Of the 478 cases, 109 (22.8%) had normal karyotypes. Chromosomal abnormalities of 280 patients (58.6%) were classified into subgroups: 11q23 abnormalities (n = 88, 18.4%), t(8;21) (n = 56, 11.7%), t(15;17) (n = 55, 11.5%), inv(16)/t(16;16) (n = 28, 5.9%), trisomy 8 alone (n = 10, 2.1%), monosomy 7 (n = 9, 1.9%), non–Down-associated trisomy 21 alone (n = 7, 1.5%), and rare recurrent chromosomal translocations (n = 27, 5.6%). The remaining 89 patients (18.6%) had miscellaneous clonal abnormalities. Overall, 84.9% of the children achieved a complete remission; the 4-year event-free survival (EFS) estimate was 33.8% ± 2.4%. Remission rates were significantly higher (96.4%,P = .011) for patients with t(8;21) and inv(16)/t(16;16) but significantly lower (74.5%, P = .022) for those with t(15;17). The 4-year survival rate for all patients was 43.5% ± 2.4%; for those with an inv(16)/t(16;16), 75.0% ± 8.6%; a normal karyotype, 53.8% ± 4.9%; a t(8;21), 51.6% ± 7.3%; a t(15;17), 39.8% ± 6.9%; and an 11q23 abnormality, 32.9% ± 5.1%. Four-year EFS estimates for patients with inv(16)/t(16;16) (58.2% ± 10.9%,P = .007), t(8;21) (45.1% ± 7.7%,P = .014), or normal karyotypes (43.1% ± 5.0%,P = .012) were higher than the 4-year EFS estimate for all patients, but EFS estimates for patients with t(15;17) (19.6% ± 8.0%, P = .033) or 11q23 abnormalities (23.8% ± 4.8%, P = .0013) were lower. EFS estimates did not differ significantly among 11q23 subgroups. Limited analysis suggested that patients with inv(16) can be salvaged better following relapse than those with t(8;21). Thus, patients with an inv(16)/t(16;16) may have high survival rates when treated with chemotherapy alone.
IN ACUTE MYELOID LEUKEMIA (AML), various chromosomal abnormalities have been recognized as primary pathogenetic changes. Clonal chromosomal abnormalities are found in approximately 80% of children with AML.1-6 Data from International Workshops and large prospective studies of adults and children have shown correlations between specific recurrent chromosomal abnormalities and clinico-biological characteristics and outcome.7-9Favorable-prognosis karyotypes in patients with AML include constitutional trisomy 21 (Down syndrome), and t(8;21) associated with myelocytic leukemia (AML French-American-British [FAB]-M2) and inv(16)/t(16;16), which is primarily associated with myelomonocytic leukemia and increased abnormal eosinophils (AML FAB-M4eo). Poor-prognosis karyotypes include 11q23 abnormalities in young patients and monosomy 7. Recently, the use of chemotherapy and all-trans-retinoic acid (ATRA) has improved outcome for patients with t(15;17)-associated acute promyelocytic leukemia (APL).10 11
With current treatment protocols, only 30% to 50% of children with AML will have a successful outcome.3,5,7,12 Therefore, it is important to correlate characteristic cytogenetic subgroups with disease outcome to identify patients that may benefit from prospective individualized therapy.12-14 We report here the results of the cytogenetic analysis of a cooperative group study, the largest study of chromosomal abnormalities in children with AML published to date. The 478 cases were segregated into cytogenetic subgroups for comparison of presenting clinical and biological features, response to induction therapy, and long-term outcome.
MATERIALS AND METHODS
Patients
Between June 1988 and March 1993, 666 patients younger than 21 years with previously untreated AML were registered on the POG-8821 AML study. Informed consent was obtained from each patient or guardian at their local institution. The treatment regimen for the POG-8821 AML protocol was reported recently.15 Briefly, after remission induction, eligible patients were randomized to compare the efficacy of autologous transplantation and intensive consolidation chemotherapy. Patients with matched siblings could opt for allogeneic bone marrow transplantation. Of the 666 patients enrolled on the protocol, 649 were evaluable for treatment response. The 5 patients with secondary AML [(ie, patients previously treated for other types of malignant disease) had an inv(16), t(8;21), t(2;11)(q37;q23), t(11;16)(q23;p13), or t(11;22)(q23;q13)], and the 34 patients with Down syndrome were excluded from this analysis. Of the remaining 610 evaluable patients, 132 lacked chromosome studies and the 478 patients (78%) for whom cytogenetics data were available were the subjects of the present study.
Cytogenetic Studies
Bone marrow samples were collected at diagnosis and mailed to the reference cytogenetic laboratories at the University of Alabama, Birmingham, AL (1988-1991) or St Jude Children’s Research Hospital, Memphis, TN (1992-1993).16,17 After overnight culture, the samples were processed according to standard techniques. The cytogenetic analysis was performed according to the International System of Human Cytogenetic Nomenclature.18 19 A patient was classified as having a normal karyotype only after 20 normal metaphases were analyzed. FAB morphology was continually reviewed (by M.V.G.).
Statistical Analysis
Cases were segregated into commonly accepted cytogenetic subgroups for evaluation of clinical features at presentation, response to treatment, and outcome. Patients were each assigned to a single subgroup reflecting the primary change observed. The categories were normal karyotype, t(8;21), t(15;17), inv(16) or t(16;16), 11q23 abnormalities, monosomy 7, trisomy 8 or trisomy 21 alone, rare recurrent translocations [ie, t(10;11)(p13;q21), t(6;9)(p23;q34), t(3;5)(q25;q34), t(1;22)(p13;q13), and t(8;16)(p11;p13)], and miscellaneous (with single or multiple clonal abnormalities). A patient achieved remission if a marrow status of either M1 (<5% blast cells) or M2a (<15% blast cells) resulted after 2 courses of induction therapy. Event-free survival (EFS) was calculated from the date of registration until the earlier of the date of first relapse or the date of death. Overall survival was calculated from the date of resistance to DAT (daunomycin, cytarabine [ara-c], and 6-thioguanine), regardless of cause. The actuarial curves of EFS for various cytogenetic subgroups were generated according to the Kaplan-Meier method,20 and the log-rank test was used to compare survival rates.21 Median values were compared by the Wilcoxon rank sum test,22 and the homogeneity of proportions was tested by the chi-square test. The P values were computed from the 2-sided test by comparing the groups of patients with and without the characteristic of interest.
RESULTS
Of the 478 children diagnosed with de novo AML and an evaluable chromosome analysis, 109 (22.8%, 57 males and 52 females) had an apparently normal karyotype (Table 1). Of the remaining 369 cases with an abnormal karyotype (77.2% of the study population), 280 (75.9%) had consistent or recurrent abnormalities, and 89 (18.6%) had miscellaneous chromosomal changes (Fig1 and Table2).
Presenting Clinico-Biological Features of 478 Children With AML and Outcome
| Cytogenetic Subgroup . | No. of Cases (% of population) . | Median Age (yr) . | Sex (M/F) . | Median WBC Count (×109/L) . | Remission Rate (%) . | 4-yr EFS (% ± SE, P Value) . | |
|---|---|---|---|---|---|---|---|
| All patients | 478 (100.0) | 7.8 | 259/219 | 20.1 | 406 (84.9) | 33.8 ± 2.4 | |
| Normal karyotype | 109 (22.8) | 11.3 | 57/52 | 24.5 | 93 (85.3) | 43.1 ± 5.0 | .012 |
| t(8;21)(q22;q22) | 56 (11.7) | 10.7 | 29/27 | 15.9 | 54 (96.4) | 45.1 ± 7.7 | .014 |
| t(15;17)(q21;q12-21) | 55 (11.5) | 12.8 | 32/23 | 8.4 | 41 (74.5) | 19.6 ± 8.0 | .033 |
| inv(16)(p13q22)/t(16;16) | 28 (5.9) | 7.8 | 19/9 | 60.5 | 27 (96.4) | 58.2 ± 10.9 | .0071 |
| 11q23 Abnormalities | 88 (18.4) | 3.0 | 44/44 | 26.7 | 72 (81.8) | 23.8 ± 4.8 | .0013 |
| Monosomy 7 | 9 (1.9) | 10.4 | 7/2 | 20.1 | 6 (66.7) | ||
| Trisomy 8* | 10 (2.1) | 14.9 | 8/2 | 53.7 | 8 (80.0) | ||
| Trisomy 21† | 7 (1.5) | 1.9 | 2/5 | 12.1 | 6 (85.7) | ||
| Rare recurrent translocations | 27 (5.6) | 6.9 | 14/13 | 18.1 | 22 (81.5) | 19.7 ± 8.8 | .086 |
| t(10;11)(p13;q21) | 9 (1.9) | 8.7 | 6/3 | 23.8 | 8 (88.9) | ||
| t(6;9)(p23;q34) | 6 (1.3) | 11.2 | 2/4 | 15.4 | 5 (83.3) | ||
| t(3;5)(q25;q34) | 5 (1.1) | 4.5 | 4/1 | 22.5 | 4 (80.0) | ||
| t(1;22)(p13;q13) | 4 (0.8) | 0.4 | 1/3 | 19.5 | 3 (75.0) | ||
| t(8;16)(p11;p13) | 3 (0.6) | 7.2 | 1/2 | 10.8 | 2 (66.7) | ||
| Miscellaneous | 89 (18.6) | 3.6 | 47/42 | 17.4 | 77 (86.5) | 34.8 ± 5.6 | .85 |
| Single clonal abnormality | 36 (7.5) | 10.5 | 20/16 | 22.4 | 31 (86.1) | 38.9 ± 9.2 | |
| Multiple clonal abnormalities | 53 (11.1) | 2.2 | 27/26 | 13.8 | 46 (86.8) | 32.1 ± 7.1 | |
| Cytogenetic Subgroup . | No. of Cases (% of population) . | Median Age (yr) . | Sex (M/F) . | Median WBC Count (×109/L) . | Remission Rate (%) . | 4-yr EFS (% ± SE, P Value) . | |
|---|---|---|---|---|---|---|---|
| All patients | 478 (100.0) | 7.8 | 259/219 | 20.1 | 406 (84.9) | 33.8 ± 2.4 | |
| Normal karyotype | 109 (22.8) | 11.3 | 57/52 | 24.5 | 93 (85.3) | 43.1 ± 5.0 | .012 |
| t(8;21)(q22;q22) | 56 (11.7) | 10.7 | 29/27 | 15.9 | 54 (96.4) | 45.1 ± 7.7 | .014 |
| t(15;17)(q21;q12-21) | 55 (11.5) | 12.8 | 32/23 | 8.4 | 41 (74.5) | 19.6 ± 8.0 | .033 |
| inv(16)(p13q22)/t(16;16) | 28 (5.9) | 7.8 | 19/9 | 60.5 | 27 (96.4) | 58.2 ± 10.9 | .0071 |
| 11q23 Abnormalities | 88 (18.4) | 3.0 | 44/44 | 26.7 | 72 (81.8) | 23.8 ± 4.8 | .0013 |
| Monosomy 7 | 9 (1.9) | 10.4 | 7/2 | 20.1 | 6 (66.7) | ||
| Trisomy 8* | 10 (2.1) | 14.9 | 8/2 | 53.7 | 8 (80.0) | ||
| Trisomy 21† | 7 (1.5) | 1.9 | 2/5 | 12.1 | 6 (85.7) | ||
| Rare recurrent translocations | 27 (5.6) | 6.9 | 14/13 | 18.1 | 22 (81.5) | 19.7 ± 8.8 | .086 |
| t(10;11)(p13;q21) | 9 (1.9) | 8.7 | 6/3 | 23.8 | 8 (88.9) | ||
| t(6;9)(p23;q34) | 6 (1.3) | 11.2 | 2/4 | 15.4 | 5 (83.3) | ||
| t(3;5)(q25;q34) | 5 (1.1) | 4.5 | 4/1 | 22.5 | 4 (80.0) | ||
| t(1;22)(p13;q13) | 4 (0.8) | 0.4 | 1/3 | 19.5 | 3 (75.0) | ||
| t(8;16)(p11;p13) | 3 (0.6) | 7.2 | 1/2 | 10.8 | 2 (66.7) | ||
| Miscellaneous | 89 (18.6) | 3.6 | 47/42 | 17.4 | 77 (86.5) | 34.8 ± 5.6 | .85 |
| Single clonal abnormality | 36 (7.5) | 10.5 | 20/16 | 22.4 | 31 (86.1) | 38.9 ± 9.2 | |
| Multiple clonal abnormalities | 53 (11.1) | 2.2 | 27/26 | 13.8 | 46 (86.8) | 32.1 ± 7.1 | |
Sole chromosomal abnormality.
Sole chromosomal abnormality, non–Down-associated.
Miscellaneous Clonal Chromosomal Abnormalities Identified in 89 Patients With AML
| Case No. . | Karyotype . |
|---|---|
| A. One miscellaneous clonal chromosome abnormality (n = 36) | |
| 1 | 45,X,−X |
| 2 | 45,XY,−19 |
| 3-4 | 47,XY,+X |
| 5-6 | 47,XY,+4 |
| 7 | 46,XY,+6,t(14q15q)c |
| 8 | 47,XX,+11 |
| 9 | 47,XX,+19 |
| 10-11 | 47,XY,+22 |
| 12 | 46,XY,del(3)(p13p14) |
| 13 | 46,XX,del(5)(q13) |
| 14 | 46,XX,del(5)(q13q22) |
| 15 | 46,XX,del(9)(q13q22) |
| 16 | 47,XX,+del(5)(q13q22.3) |
| 17 | 46,XX,del(16)(q22) |
| 18 | 46,XX,?inv(19)(p13q12) |
| 19-20 | 46,XX,i(7)(p10) |
| 21 | 46,XX,i(17)(q10) |
| 22 | 45,XX,dic(16;21)(q11;p11) |
| 23 | 47,XX,+add(8)(p23) |
| 24 | 46,XY,add(14)(q24) |
| 25 | 45,XY,−7,der(19)t(7;19)(p13;p11) |
| 26 | 46,XX,der(20)t(1;20)(q21;q13) |
| 27 | 46,X,t(X;5)(p22;p14) |
| 28 | 46,Y,t(X;7)(q13;p15) |
| 29 | 46,X,ins(X;8)(q22;q22q24.1)/45,idem,−X |
| 30 | 46,XY,t(1;8)(8;11;16;8;1)(p32;q24q13;q13;p11;p36) |
| 31 | 46,XY,t(2;10)(p22;p13) |
| 32 | 46,XY,t(3;7)(p23;p15) |
| 33 | 46,XX,t(3;21)(q26.3;q22) |
| 34 | 46,XY,t(6;11)(p21;p15) |
| 35 | 46,XX,t(16;17)(q24;q11) |
| 36 | 46,XY,t(11;20)(p15;q11) |
| B. More than one miscellaneous clonal chromosomal abnormality (n = 53) | |
| 37 | 41,X,−Y,add(5)(q11),add(8)(q24),−10,−11,−14,−17, −20,−21,+2mar |
| 38 | 45,X,−Y/46,idem,+21/47,XY,+21 |
| 39 | 45,XX,add(9)(p13),−11 |
| 40 | 45,X,−X,t(1;8)(p22;q21),t(7;21)(p14;q22),del(9)(q13q22) |
| 41 | 45,XX,t(2;12)(12;?)(q11;q24p12;?),−12,−13,+mar/45,idem,−X,+der(12)t(X;12)(p11;p11) |
| 42 | 46,XY,t(5;19)(q15;p13.1)/45,idem,del(1)(p34),der(11)t(11;15)(p11;q11),−15 |
| 43 | 46,XX,der(2)t(1;2)(q21;q37),del(3)(q25q28) |
| 44 | 46,XX,−8,der(16)t(8;16)(q11;p13)×2,+mar |
| 45 | 46,XX,?inv(8)(p12p22),t(8;17)(q22;q25) |
| 46 | 46,XX,del(1)(p21)/46,idem,del(15)(q23) |
| 47 | 46,XX,dup(1)(q21q44)/46,idem,t(18;21)(q21;q22) |
| 48 | 46,XX,t(1;10)(10;?)(q12;p12q25;?)/47,idem,+i(21)(q10) |
| 49 | 46,XX,t(1;8)(p32;p11),t(9;10)(q34.3;p12.2) |
| 50 | 46,XY,t(11;18)(p15;q23),der(17)t(1;17)(q12;p13) |
| 51 | 46,XY,?inv(10)(p11p13)/47,idem,+del(1)(p22)/47,idem,+del(9)(q31) |
| 52 | 46,XY,t(2;12;12;2)(q23;q13;p13;q21),t(7;11)(p15;q21),del(17)(p12) |
| 53 | 46,XY,del(6)(q13q21),t(16;21)(p11.2;q22) |
| 54 | 46,XY,inv(8)(p23q22)/46,idem,der(20)t(1;20)(q25;q13) |
| 55 | 46,XY,t(2;12)(q33;p13),r(16)(p13q24) |
| 56 | 46,XY,t(9;11)(p22;q13)/48,idem,+16,+18 |
| 57 | 46,Y,t(X;11)(11;6)(q28;q14.2p11;p23),del(20)(q11) |
| 58 | 46,XX,−5,r(6)(p25q21),del(12)(q24),?inv(17)(p11q21), +der(?)t(?;5)(?;p13)/ 45,idem,add(1)(p13),del(16)(q13),−17,add(21)(q22) |
| 59 | 46,X,idic(Yq)c,der(18)t(11;18)(q13;p11)/46,idem, del(3)(p13),add(4)(p14),add(6)(p23), add(7)(p36),del(13)(q12q14),t(18;19)(q11.2;q13.3) |
| 60 | 46,XX,t(12;21)(q12;q21)/46,XX,dup(1)(q21q42), r(7)(p22q31),add(13)(q34),der (21)t(7;21)(q32;q22) |
| 61 | 46,XY,del(7)(q11)/46,idem,del(1)(q21),der(4)t(1;4)(q21;q21),der(6)t(6;6)(p23;q15)/45,XY, ins(2)(p23q33q37),t(3;8)(p23;p23), r(7)(p15q35),−13 |
| 62 | 46,XY,t(6;12)(p15;q13),del(7)(q32q35)/46,idem,t(3;14)(q12;q23)/46,idem,t(3;14),t(15;19)(p11;p11) |
| 63 | 47,XX,del(12)(p13),+19/49,idem,+X,+8 |
| 64 | 47,XX,t(7;12)(p36;p13.1),+19 |
| 65 | 47,XX,+21/47,idem,?inv(11)(p13q11) |
| 66 | 47,XX,+8,dup(10)(q24q26) |
| 67 | 47,XX,+8/46,XX,t(6;8)(q21;q24)/50,idem, +11,+19,+21 |
| 68 | 47,XX,add(1)(q24),add(5)(q11),+8 |
| 69 | 47,XX,−8,+2mar |
| 70 | 47,XX,del(8)(q22),+mar |
| 71 | 47,XY,+21/48,idem,+8/47,idem,−6,+mar |
| 72 | 47,XY,+22/48,idem,+6 |
| 73 | 47,XY,+der(2)t(2;15)(q31;q15),add(5)(q33), +i(10)(q10),+11,−13,del(15)(q15),−17,−20,+mar |
| 74 | 47,Y,der(X)t(X;6)(p11.2;q15),add(1)(q32),der(2)t(X;2)(p11.2;p11.2),der(6)t(1;6)(q31;q13),+13,der(16)t(16;?;2)(q23;?;p15) |
| 75 | 47,XX,der(17)t(11;17)(q13;p13),+21/46,XX,−7, +der(21)t(7;21)(q11;p11)/47,X X,add(7)(p15),+21 |
| 76 | 47,XX,der(6)inv(6)(p21p25)del(6)(q23q25),del(7)(p15), +10,del(11)(q21),der(14)t(7;14)(q11;p11),der(16)add(16)(p11)add(16)(q22),add(18)(p11)/55,idem,+6,+8 ,+11,+15,+19,+20,+21,+22 |
| 77 | 48,XX,der(9)t(1;9)(q23;q34),+11,+21 |
| 78 | 48,XY,i(7)(q10),dup(8)(p11p23)×2,+8,+21 |
| 79 | 48,XY,+4,+8 |
| 80 | 48,XY,t(6;8)(q21;p23),+6,inv(17)(p11q25),+19/48,idem,−t(6;8) |
| 81 | 48,XY,add(2)(q33),+6,+der(11)t(2;11)(q21;p13) |
| 82 | 48,Y,−X,add(6)(q23),+9,add(10)(q22),add(19)(p13), +20,+r |
| 83 | 49,XX,+Y,+8,+21 |
| 84 | 50,XX,t(6;16)(q21;q24),+14,+17,+19,+21 |
| 85 | 51,XX,+8,+10,+19,+2mar |
| 86 | 52,XY,+2,+6,+7,+8,inv(11)(p13q14),+19,+19 |
| 87 | 55,XX,+2,+6,+7,+8,+10,+13,+19,+19,+21 |
| 88 | 58,XX,+X,+2,+4,+6,+8,+10,+13,+17,+20,+21,+21, +mar |
| 89 | 92,XXYY,del(5)(q22q31)×2/91,idem,−17 |
| Case No. . | Karyotype . |
|---|---|
| A. One miscellaneous clonal chromosome abnormality (n = 36) | |
| 1 | 45,X,−X |
| 2 | 45,XY,−19 |
| 3-4 | 47,XY,+X |
| 5-6 | 47,XY,+4 |
| 7 | 46,XY,+6,t(14q15q)c |
| 8 | 47,XX,+11 |
| 9 | 47,XX,+19 |
| 10-11 | 47,XY,+22 |
| 12 | 46,XY,del(3)(p13p14) |
| 13 | 46,XX,del(5)(q13) |
| 14 | 46,XX,del(5)(q13q22) |
| 15 | 46,XX,del(9)(q13q22) |
| 16 | 47,XX,+del(5)(q13q22.3) |
| 17 | 46,XX,del(16)(q22) |
| 18 | 46,XX,?inv(19)(p13q12) |
| 19-20 | 46,XX,i(7)(p10) |
| 21 | 46,XX,i(17)(q10) |
| 22 | 45,XX,dic(16;21)(q11;p11) |
| 23 | 47,XX,+add(8)(p23) |
| 24 | 46,XY,add(14)(q24) |
| 25 | 45,XY,−7,der(19)t(7;19)(p13;p11) |
| 26 | 46,XX,der(20)t(1;20)(q21;q13) |
| 27 | 46,X,t(X;5)(p22;p14) |
| 28 | 46,Y,t(X;7)(q13;p15) |
| 29 | 46,X,ins(X;8)(q22;q22q24.1)/45,idem,−X |
| 30 | 46,XY,t(1;8)(8;11;16;8;1)(p32;q24q13;q13;p11;p36) |
| 31 | 46,XY,t(2;10)(p22;p13) |
| 32 | 46,XY,t(3;7)(p23;p15) |
| 33 | 46,XX,t(3;21)(q26.3;q22) |
| 34 | 46,XY,t(6;11)(p21;p15) |
| 35 | 46,XX,t(16;17)(q24;q11) |
| 36 | 46,XY,t(11;20)(p15;q11) |
| B. More than one miscellaneous clonal chromosomal abnormality (n = 53) | |
| 37 | 41,X,−Y,add(5)(q11),add(8)(q24),−10,−11,−14,−17, −20,−21,+2mar |
| 38 | 45,X,−Y/46,idem,+21/47,XY,+21 |
| 39 | 45,XX,add(9)(p13),−11 |
| 40 | 45,X,−X,t(1;8)(p22;q21),t(7;21)(p14;q22),del(9)(q13q22) |
| 41 | 45,XX,t(2;12)(12;?)(q11;q24p12;?),−12,−13,+mar/45,idem,−X,+der(12)t(X;12)(p11;p11) |
| 42 | 46,XY,t(5;19)(q15;p13.1)/45,idem,del(1)(p34),der(11)t(11;15)(p11;q11),−15 |
| 43 | 46,XX,der(2)t(1;2)(q21;q37),del(3)(q25q28) |
| 44 | 46,XX,−8,der(16)t(8;16)(q11;p13)×2,+mar |
| 45 | 46,XX,?inv(8)(p12p22),t(8;17)(q22;q25) |
| 46 | 46,XX,del(1)(p21)/46,idem,del(15)(q23) |
| 47 | 46,XX,dup(1)(q21q44)/46,idem,t(18;21)(q21;q22) |
| 48 | 46,XX,t(1;10)(10;?)(q12;p12q25;?)/47,idem,+i(21)(q10) |
| 49 | 46,XX,t(1;8)(p32;p11),t(9;10)(q34.3;p12.2) |
| 50 | 46,XY,t(11;18)(p15;q23),der(17)t(1;17)(q12;p13) |
| 51 | 46,XY,?inv(10)(p11p13)/47,idem,+del(1)(p22)/47,idem,+del(9)(q31) |
| 52 | 46,XY,t(2;12;12;2)(q23;q13;p13;q21),t(7;11)(p15;q21),del(17)(p12) |
| 53 | 46,XY,del(6)(q13q21),t(16;21)(p11.2;q22) |
| 54 | 46,XY,inv(8)(p23q22)/46,idem,der(20)t(1;20)(q25;q13) |
| 55 | 46,XY,t(2;12)(q33;p13),r(16)(p13q24) |
| 56 | 46,XY,t(9;11)(p22;q13)/48,idem,+16,+18 |
| 57 | 46,Y,t(X;11)(11;6)(q28;q14.2p11;p23),del(20)(q11) |
| 58 | 46,XX,−5,r(6)(p25q21),del(12)(q24),?inv(17)(p11q21), +der(?)t(?;5)(?;p13)/ 45,idem,add(1)(p13),del(16)(q13),−17,add(21)(q22) |
| 59 | 46,X,idic(Yq)c,der(18)t(11;18)(q13;p11)/46,idem, del(3)(p13),add(4)(p14),add(6)(p23), add(7)(p36),del(13)(q12q14),t(18;19)(q11.2;q13.3) |
| 60 | 46,XX,t(12;21)(q12;q21)/46,XX,dup(1)(q21q42), r(7)(p22q31),add(13)(q34),der (21)t(7;21)(q32;q22) |
| 61 | 46,XY,del(7)(q11)/46,idem,del(1)(q21),der(4)t(1;4)(q21;q21),der(6)t(6;6)(p23;q15)/45,XY, ins(2)(p23q33q37),t(3;8)(p23;p23), r(7)(p15q35),−13 |
| 62 | 46,XY,t(6;12)(p15;q13),del(7)(q32q35)/46,idem,t(3;14)(q12;q23)/46,idem,t(3;14),t(15;19)(p11;p11) |
| 63 | 47,XX,del(12)(p13),+19/49,idem,+X,+8 |
| 64 | 47,XX,t(7;12)(p36;p13.1),+19 |
| 65 | 47,XX,+21/47,idem,?inv(11)(p13q11) |
| 66 | 47,XX,+8,dup(10)(q24q26) |
| 67 | 47,XX,+8/46,XX,t(6;8)(q21;q24)/50,idem, +11,+19,+21 |
| 68 | 47,XX,add(1)(q24),add(5)(q11),+8 |
| 69 | 47,XX,−8,+2mar |
| 70 | 47,XX,del(8)(q22),+mar |
| 71 | 47,XY,+21/48,idem,+8/47,idem,−6,+mar |
| 72 | 47,XY,+22/48,idem,+6 |
| 73 | 47,XY,+der(2)t(2;15)(q31;q15),add(5)(q33), +i(10)(q10),+11,−13,del(15)(q15),−17,−20,+mar |
| 74 | 47,Y,der(X)t(X;6)(p11.2;q15),add(1)(q32),der(2)t(X;2)(p11.2;p11.2),der(6)t(1;6)(q31;q13),+13,der(16)t(16;?;2)(q23;?;p15) |
| 75 | 47,XX,der(17)t(11;17)(q13;p13),+21/46,XX,−7, +der(21)t(7;21)(q11;p11)/47,X X,add(7)(p15),+21 |
| 76 | 47,XX,der(6)inv(6)(p21p25)del(6)(q23q25),del(7)(p15), +10,del(11)(q21),der(14)t(7;14)(q11;p11),der(16)add(16)(p11)add(16)(q22),add(18)(p11)/55,idem,+6,+8 ,+11,+15,+19,+20,+21,+22 |
| 77 | 48,XX,der(9)t(1;9)(q23;q34),+11,+21 |
| 78 | 48,XY,i(7)(q10),dup(8)(p11p23)×2,+8,+21 |
| 79 | 48,XY,+4,+8 |
| 80 | 48,XY,t(6;8)(q21;p23),+6,inv(17)(p11q25),+19/48,idem,−t(6;8) |
| 81 | 48,XY,add(2)(q33),+6,+der(11)t(2;11)(q21;p13) |
| 82 | 48,Y,−X,add(6)(q23),+9,add(10)(q22),add(19)(p13), +20,+r |
| 83 | 49,XX,+Y,+8,+21 |
| 84 | 50,XX,t(6;16)(q21;q24),+14,+17,+19,+21 |
| 85 | 51,XX,+8,+10,+19,+2mar |
| 86 | 52,XY,+2,+6,+7,+8,inv(11)(p13q14),+19,+19 |
| 87 | 55,XX,+2,+6,+7,+8,+10,+13,+19,+19,+21 |
| 88 | 58,XX,+X,+2,+4,+6,+8,+10,+13,+17,+20,+21,+21, +mar |
| 89 | 92,XXYY,del(5)(q22q31)×2/91,idem,−17 |
Modal Number
Pseudodiploidy was found in 245 cases, representing 51.3% of all cases. All but 1 of the 47 hypodiploid cases had 45 chromosomes; the remaining case had 41 chromosomes. In most of the cases with 45 chromosomes, hypodiploidy was the result of the loss of a sex chromosome (n = 26) or monosomy 7 (n = 9). Most of the hyperdiploid cases had 47 (n = 53) or 48 chromosomes (n = 12); an additional 2 cases each had 49 and 50 chromosomes. In addition, there were 5 patients with 51 to 58 chromosomes in the leukemic lines, and 3 with 91 to 93 chromosomes and structural abnormalities. Therefore, hyperdiploidy 47-50 accounted for 14.4% of the study population, hyperdiploidy 51-58 for 1.1%, and hyperdiploidy 91-93 for 0.6%.
Common Recurrent Chromosomal Abnormalities
t(8;21)(q22;q22).
This translocation, found in 56 patients (11.7%), was the most frequently observed translocation. Among the cases characterized by this aberration, 33 (58.9%) had 46 chromosomes, 21 (37.5%) had 45, and 2 (3.6%) had 47. This translocation was the sole structural abnormality in 22 (39.3%) cases, and 4 had a complex t(8;21). The 21 cases of t(8;21) with a modal number of 45 all had a loss of a sex chromosome. An additional 2 cases had a loss of a sex chromosome as well as other numerical abnormalities, and in another 2 cases, loss of the Y chromosome was found only in the sideline. Loss of the Y chromosome occurred more frequently (16 of 29 males) than loss of the X chromosome (9 of 27 females). An additional chromosome was identified in 4 cases (+4, n = 2; +8, n = 1; +15, n = 1). Secondary structural changes (n = 16) included deletion of 9q in 6 of the 56 cases (10.7%). Abnormalities of 7q were seen in 5 cases (8.9%); 2 cases each had a del(7q) or der(7q), and 1 had a dup(7)(q22q32). The remaining 5 cases had random secondary structural aberrations.
t(15;17)(q22;q12-21).
The t(15;17) was observed in 55 of the 478 cases (11.5%); the modal number was 45 (n = 3), 46 (n = 50), or 47 (n = 2). This rearrangement was the sole structural abnormality in 48 (87.3%) of the cases in this subgroup. The only additional recurrent structural aberration observed with the t(15;17) was del(9q), which was seen in a single case. The t(15;17) subgroup included 2 cases with i(der 17q), 1 of which had the isochromosome in the sideline (indicating clonal evolution), and 3 cases with complex t(15;17) rearrangements. Additional numerical abnormalities included +8 (n = 2), −Y (n = 1), and −20 (n = 1).
inv(16)(p13q22) or t(16;16)(p13;q22).
Twenty-eight cases (5.9% of the study population) were assigned to this subgroup: 26 with inv(16) and 2 with t(16;16). The inv(16)/t(16;16) was the sole abnormality in 22 of these 28 cases (78.6%). Associated numerical abnormalities were +8 (n = 1), +21 (n = 1), and +22 (n = 3); an additional case had a del(9q) as an associated aberration. Because of the strong association between +22 and inv(16), 2 additional cases, in which +22 was apparently the sole chromosomal abnormality, were reevaluated for inv(16). However, we were unable to document the presence of inv(16) in either of these cases.
11q23 abnormalities.
The most frequent chromosomal breakpoint in the present series occurred in chromosome 11 at q23; this abnormality was identified in 88 patients (18.4% of the study population). These cases were further classified into the following subgroups: t(9;11)(p22;q23) (n = 35), t(11;19)(q23;p13.1) (n = 9), t(11;19)(q23;p13.3) (n = 10), and other chromosomal aberrations involving the 11q23 region (n = 28).
t(9;11)(p22;q23).
Breakpoints at 9p22 and 11q23 were identified in 35 cases (7.3% of the study population). In 24 of these 35 cases, the t(9;11) was the sole chromosomal aberration, and most (32 cases) had a modal chromosome number of 46. Complex rearrangements of 9p22 and 11q23 with 14q24, 20p13, or 22q13 were seen in one case each. Only 1 of the 35 patients had an insertion [ins(9;11)(p21;q13q23)]; this case also showed another breakpoint at 11q23 [t(11;12)(q23;p13)]. An additional 2 cases each were associated with i(1)(q10) or +8; 2 other cases had +6, which was seen in the sidelines.
t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3).
The t(11;19) rearrangement was observed in 19 cases (4.0% of the study population), with breakpoints at 19p13.1 (n = 9) and at 19p13.3 (n = 10). The distribution of modal numbers was 46 (16 cases), 48 (1 case), 49 (1 case), and 50 (1 case). In 12 patients (63.2% of this subgroup), the t(11;19) was the only abnormality; in another 2 patients, the rearrangement was complex. None of the cases with a breakpoint at 19p13.1 had additional chromosomal aberrations. Three t(11;19)(q23;p13.3) cases had an extra copy of the der(19)t(11;19) as well as trisomy 8.
Other chromosomal aberrations involving the 11q23 region were identified in 34 cases and comprised t(11q23;V) (n = 26), inv(11) [n = 2; inv(11)(p13q23) and inv(11)(q21q23)], and del(11)(q23) (n = 6). The chromosomes that exchanged with 11q23 were variable, and 4 recurrent aberrations were observed: t(10;11)(p11.2;q23) (n = 5), ins(10;11)(p13;q13q23) (n = 3), and t(1;11)(q21;q23) or t(11;17)(q23;q21) (n = 2). The 2 cases with t(11;17)(q23;q21) had FAB-M1/M2 or M4 morphology and were not associated with acute promyelocytic leukemia. No additional aberrations were noted in 47% (16 of 34) of the cases with a variable change at 11q23.
del(11)(q23).
Of the 6 cases with deletion at 11q23 (the region of the MLLgene), it was the sole chromosomal aberration in 3, and in none of these 6 cases could a translocation be identified by conventional G-banding techniques. Currently, it is recommended that newer molecular cytogenetic techniques be used, like chromosome 11 painting, and fluorescence in situ hybridization (FISH) or Southern assay with theMLL probe, to rule out a subtle translocation and to assess whether the MLL is rearranged.
Rare Recurrent Chromosomal Abnormalities
t(10;11)(p13;q21).
Assigning the specific breakpoint occasionally was problematic for the 9 cases (1.9% of the study population) with a t(10;11)(p13;q21); some karyotypes were read as t(10;11)(p13 or p14;q14.2 or q21). The t(10;11)(p13;q21) was the sole chromosomal aberration in 2 cases, and +8 and −Y were present in 2 cases each. The remaining patients showed rather simple additional chromosomal changes.
t(6;9)(p23;q34).
Seen in 6 patients (1.3% of the study population), the t(6;9) was the sole abnormality in 5 cases. The remaining case had a sideline with +4.
t(3;5)(q25;q34).
In all 5 cases, the t(3;5) was the sole chromosomal aberration. In addition, 1 patient had a sideline that contained extra chromosomes.
t(1;22)(p13;q13).
In 2 of the 4 cases in this subgroup, the t(1;22) was the only abnormality; the other 2 had hyperdiploid karyotypes (54 and 58 chromosomes), both with an extra der(1)t(1;22). Consistent with this cytogenetic finding, 3 of the cases had FAB-M7 morphology, and 1 patient presented with granulocytic sarcoma.
t(8;16)(p11;p13).
The t(8;16) was the sole abnormality in 1 of the 3 cases with this aberration.
Abnormalities of chromosomes 7 and 5.
Monosomy 7 was the sole abnormality in 6 of the 9 cases in this subgroup (1.9% of the study population); 2 additional patients also had +12 or +21. The remaining patient had a very complex karyotype that included monosomy 5. Other abnormalities resulting in partial loss of 7q were identified in 12 cases. Deletion accounted for 4 of these cases. The del(7)(q31) or del(7)(q21), seen in 1 case each, were secondary changes associated with t(8;21). An additional case had a del(7)(q32q35) as a component of a complex karyotype. The remaining case characterized by a deletion in 7q had del(7)(q11) in the main line and r(7q) in the sideline. Ring formations of chromosome 7 were seen in 2 additional cases, 1 of which had r(7)(p15q35) as a secondary change. In the other case, the ring was one of the many aberrations observed in a patient with Fanconi anemia (Table 2, case no. 60). An isochromosome of 7p resulted in the loss of 7q in 2 patients. In 3 cases, DNA of unknown origin was attached to the q arm of chromosome 7, resulting in add(7q). The breakpoints differed in all 3 patients, of which 2 also had t(8;21), and 1 also had t(15;17). The 7q loss in the remaining case was due to der(19)t(7;19)(p13;p11). Therefore, these 12 cases were not included in this series as examples of −7/7q− syndrome because of the apparent secondary nature of the 7q aberrations.
Abnormalities resulting in total or partial loss of 5q were identified in 6 cases. Only 1 case had monosomy 5 as part of a complex karyotype, which included monosomy 7. Two patients in this subgroup had a deletion of 5q as the sole chromosomal abnormality, and 2 others had a deletion of 5q as well as additional chromosomal aberrations. The remaining case was characterized by add(5)(q11). Contrary to its frequency in adult patients, the −5/5q− syndrome is very rare in childhood AML.8,9 23
Trisomy 8.
An extra chromosome 8 was observed in 40 cases (8.4% of the study population); in 10 (2.1%) patients, it was the sole chromosomal abnormality. Therefore, trisomy 8 was the most frequent numerical chromosomal abnormality seen in the present series of patients with childhood AML. The +8 was seen with other recurrent (18 cases) and nonrecurrent (12 cases) changes, but these 30 cases were not incorporated in the +8 subgroup for further analysis because they were interpreted as secondary changes.
Trisomy 21.
Acquisition of an extra chromosome 21 occurred in 26 cases and was the sole chromosomal aberration in 7. The karyotype of 1 of these 7 cases was 93,XXXX,+21. Trisomy 21 was seen with other recurrent (7 cases) and nonrecurrent (12 cases) changes; like that for the +8 subgroup, the analysis was restricted to cases with a single extra 21 chromosome.
Other types of trisomy and monosomy.
Trisomy of chromosome X, 4, or 22 was the sole abnormality in 2 cases each, and 1 case each showed gains of chromosomes 6, 11, or 19. Cases in which the loss of a chromosome was the sole aberration were rare and included 1 case each of monosomy X or 19 (Table 2).
Miscellaneous Clonal Abnormalities
This group includes 89 patients with various clonal abnormalities not represented by other subgroups (Table 2). A single abnormal clone was seen in 36 cases; 2 or more abnormal clones occurred in the remaining 53. Most of these cases are discussed in the following section. Noteworthy are rare abnormalities that have been previously reported, which include t(3;21)(q26.3;q22) (Table 2, case no. 33), and t(16;21)(p11.2;q22) (case no. 53).24 25
Deletion.
A total of 67 deletions were observed, the majority of which were secondary changes. Deletion was the sole acquired abnormality in only 8 cases: 3 cases with del(11)(q23), del(5q) and del(9q) in 2 cases each, and 1 case each with a deletion of 3p or 16q. No corresponding molecular or FISH analysis was done to determine whether subtle translocations were present in any of these cases. Patients who had deletion of 5q (n = 4) or 7q (n = 4) were discussed previously.
The most frequently identified recurrent deletion (n = 10) in this series of childhood AML was del(9q), which was the sole abnormality in only 1 case. This aberration was associated with t(8;21) in 5 patients and with t(15;17) or inv(16) in 1 case each. Therefore, most 9q deletions among our study population were associated with recurrent structural chromosomal aberrations. In contrast to their frequency in childhood ALL, del(9p) occurred in only 1 of our patients with childhood AML, and del(12p) was seen in only 3 cases. Deletion of the 11q arm was identified in 9 cases, 6 of which had a breakpoint in the 11q23 region. Of the 2 cases in which the breakpoint occurred at 11q13, 1 had a t(15;17) and the other a t(6;11)(q15;q23). An additional patient had a del(11)(q21) as well as multiple other chromosomal aberrations.
Duplication.
In 4 of the 9 cases with duplication, the abnormality occurred in the q arm of chromosome 1. The remaining cases showed duplications of 6q, 7q, 8p, 10q, and 11q (1 each). All of these 9 cases had additional chromosomal abnormalities.
Isochromosomes.
Of the 9 cases with an isochromosome, 2 had an i(1q), which was associated with t(9;11) and FAB M5 morphology. An additional 2 cases had i(7)(p10) as the sole abnormality; because the consequence of this isochromosome is loss of the 7q arm, these cases were considered to be 7q−. Another case had an i(7)(q10), leading to loss of the 7p region. The aberration i(17)(q10) was the sole abnormality in a single patient, and in 2 patients with APL, an isochromosome of der(17)t(15;17) was acquired as a secondary event during the process of clonal evolution. The remaining case had i(21)(q10).
Inversion.
Inversions were observed in 41 cases: inv(16) (n = 26; discussed previously), inv(11) [n = 4; inv(11)(p13q11), inv(11)(p13q14), inv(11)(p13q23), or inv(11)(q21q23)], inv(8) (n = 3), inv(17) (n = 2). Other chromosomes were involved in the remaining 6 cases. Identical breakpoints were detected only in the inv(16). The inv(3)(p25q21) seen in our patients differs from the recurrent inv(3)(q21q26) that is typically seen in adults with AML.8Among the 15 non-inv(16) cases, the inversion was the sole abnormality only in the patient with inv(19)(p13q12). Therefore, other than those involving chromosome 16, most of the inversions appear to be secondary changes.
Insertion.
Of the 8 cases with this type of abnormality, 4 were included in the 11q23 subgroup. The remaining 4 insertions affected various chromosomes and were part of complex karyotypes.
Dicentric chromosomes.
Only 2 cases had dicentric chromosomes. A dic(16;21)(q11;p11) was the sole chromosomal abnormality in 1 case.
Ring chromosomes.
A total of 5 cases had a ring chromosome: r(6), r(16)(p13q24), r(7)(p22q31), r(7)(p15q35), and r(?).
Presenting Features and Outcome
The 478 patients in our study population were assigned to 1 of 7 groups [ie, normal karyotype, t(8;21), t(15;17), inv(16), 11q23 abnormalities, rare recurrent abnormalities, and miscellaneous aberrations] according to the primary chromosomal characteristic. Except when noted, reported differences in clinico-biological features reflect the comparison between patients with or without 1 of these 7 cytogenetic characteristics (Table 1).
The proportion of male or female patients in our series included more male patients than female (259 v 219), and a slightly greater proportion of the patients with t(15;17), inv(16), −7, or +8 alone were male. The median age at diagnosis for all patients was 7.8 years. Patients with +8 alone, t(15;17), normal karyotype, or t(6;9) tended to be older (median age, 14.9 years, 12.8 years, 11.3 years, and 11.2 years, respectively). The median age of those with 11q23 abnormalities was 3.0 years; for those with miscellaneous abnormalities the median age was 3.6 years. The median white blood cell (WBC) count for all patients was 20.1 × 109/L; the median WBC count was lower (8.4 × 109/L) for patients with t(15;17) but was higher for those with inv(16)/t(16;16) (60.5 × 109/L) or +8 alone (53.7 × 109/L).
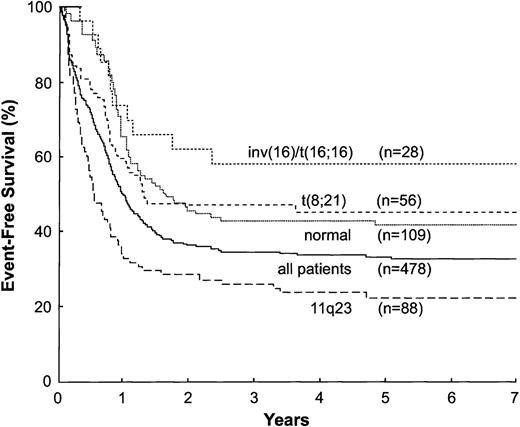
The remission rate for the entire study population was 84.9%. This rate was higher (96.4%, P = .011) for patients with t(8;21) and inv(16)/t(16;16) but lower (74.5%, P = .022) for those with t(15;17). The 4-year overall survival rate for all patients was 43.5% ± 2.4%; for those with an inv(16)/t(16;16), 75.0% ± 8.6%; a normal karyotype, 53.8% ± 4.9%; a t(8;21), 51.6% ± 7.3%; a t(15;17), 39.8% ± 6.9%; and 11q23 abnormalities, 32.9% ± 5.1%. The overall 4-year estimated EFS rate was 33.8% ± 2.4% (Fig2). The 4-year EFS was higher for patients with an inv(16)/t(16;16) (58.2% ± 10.9%, P = .0071), t(8;21) (45.1% ± 7.7%, P = .014), or normal karyotype (43.1% ± 5.0%, P = .012) but lower for those with t(15;17) (19.6% ± 8.0%, P = .033) or 11q23 abnormalities (23.8% ± 4.8%, P = .0013) (Fig 2). Remission rate and EFS did not differ significantly with the type of 11q23 abnormality (Fig 3). Furthermore, we found no difference in outcome between patients with the t(9;11) and other patients with other 11q23 abnormality.
Remission rates for patients with 1 versus 2 or more miscellaneous abnormalities were similar, but those with a single abnormality had a significantly longer 4-year EFS (38.9% ± 9.2% v32.1% ± 7.1%, P = .0003). Further, there was no significant difference between the outcome of patients with 5 or more miscellaneous clonal abnormalities and the outcome of those with 2 to 4 miscellaneous clonal abnormalities (data not shown). The estimates of EFS of patients with and without additional chromosomal abnormalities were not significantly different within each of the following cytogenetic subgroups: t(8;21), t(15;17), inv(16)/t(16;16), t(9;11), +8, and +21 (data not shown).
The distribution of various cytogenetic subgroups among the randomized treatment arms is shown in Table 3. Because of the small numbers within each subgroup, a meaningful evaluation of the impact of each of the three treatment options within cytogenetic subgroups was not possible.
Patients Randomized According to the Intended Treatment
| Randomized Treatment . | Normal Karyotype . | t(8;21) . | t(15;17) . | inv(16)/t(16;16) . | 11q23 . | Rare Translocations . | Miscellaneous . | Total . |
|---|---|---|---|---|---|---|---|---|
| Chemotherapy | 20 | 15 | 10 | 5 | 13 | 5 | 19 | 87 |
| Auto-BMT | 20 | 11 | 8 | 7 | 20 | 2 | 17 | 85 |
| Allo-BMT | 18 | 7 | 3 | 7 | 16 | 3 | 14 | 65 |
| Randomized Treatment . | Normal Karyotype . | t(8;21) . | t(15;17) . | inv(16)/t(16;16) . | 11q23 . | Rare Translocations . | Miscellaneous . | Total . |
|---|---|---|---|---|---|---|---|---|
| Chemotherapy | 20 | 15 | 10 | 5 | 13 | 5 | 19 | 87 |
| Auto-BMT | 20 | 11 | 8 | 7 | 20 | 2 | 17 | 85 |
| Allo-BMT | 18 | 7 | 3 | 7 | 16 | 3 | 14 | 65 |
Abbreviations: Auto-BMT, autologous bone marrow transplantation; Allo-BMT, allogeneic bone marrow transplantation.
DISCUSSION
We compared our findings with those of 5 previously reported series with cytogenetic data,1-5 representing a total of 926 cases of childhood AML (Table 4). Cytogenetic analysis showed clonal abnormalities in 75% of those earlier cases, in which t(8;21) is the most commonly observed single structural abnormality (13% of patients, similar to our finding of 12%). The t(15;17) was observed in 8% of the previously reported patients. However, patients with APL sometimes receive therapies that are different than those used to treat other types of AML.10 11Such patients may have been omitted from the earlier series, thereby skewing the reported incidence of t(15;17). In this study, patients with APL were not excluded from the analysis and accounted for 11% of our population.
Frequency of Chromosomal Abnormalities in Children (Compared With Adults) With AML
| Chromosome Abnormality . | Children . | Adults . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 19881 No. (%) . | 19892 No. (%) . | 19953 No. (%) . | 19964 No. (%) . | 199854-150 No. (%) . | Total1-54-151 No. (%) . | Present No. (%) . | Total4-151 No. (%) . | 19985‡ No. (%) . | 1998234-153 No. (%) . | Total5 23 No. (%) . | |
| Total | 130 | 121 | 115 | 220 | 340 | 926 | 478 | 1,404 | 1,272 | 609 | 1,881 |
| Normal | 41 (31) | 25 (21) | 17 (15) | 61 (28) | 91 (27) | 235 (25) | 109 (23) | 344 (24) | 589 (46) | 245 (40) | 834 (44) |
| t(8;21)(q22;q22) | 17 (13) | 14 (12) | 9 (8) | 36 (16) | 41 (12) | 117 (13) | 56 (12) | 173 (12) | 81 (6) | 40 (7) | 121 (6) |
| inv(16)(p13q22)/t(16;16) | 4 (3) | 15 (12) | 9 (8) | 19 (9) | 16 (6) | 63 (7) | 28 (6) | 91 (6) | 41 (3) | 49 (8) | 90 (5) |
| t(15;17)(q22;q12-21) | 10 (8) | 9 (7) | 12 (10) | 9 (4) | 31 (9) | 71 (8) | 55 (11) | 126 (9) | 167 (13) | 27 (4) | 194 (10) |
| t(9;11)(p22;q23) | 0 | 9 (7) | 10 (9) | 16 (7) | NA | 35 (6) | 35 (7) | 70 (7) | NA | NA | NA |
| t(11q23;V)/inv/del(11q23) | 12 (9) | 7 (6) | 11 (10) | 21 (9) | 26 (8) | 77 (8) | 53 (11) | 130 (9) | 34 (3)4-155 | 42 (7)4-155 | 76 (4) |
| t(6;9)(p23;q24) | 0 | 0 | 1 (<1) | NA | NA | 1 (<1) | 6 (1) | 7 (<1) | NA | 11 (2) | 11 (2) |
| t(1;22)(p13;q13) | 0 | 0 | 4 (3) | NA | NA | 4 (1) | 4 (<1) | 8 (<1) | NA | 0 | NA |
| −7/del(7q) | 6 (5) | 6 (5) | 6 (5) | NA | 19 (6) | 37 (5) | 21 (4) | 58 (5) | 74 (6) | 52 (8) | 126 (7) |
| −5/del(5q) | 2 (1) | 1 (<1) | 0 | NA | 6 (2) | 9 (1) | 6 (1) | 15 (1) | 48 (4) | 36 (6) | 84 (4) |
| Hyperdiploid | 14 (11) | 17 (14) | 24 (21) | NA | NA | 55 (15) | 77 (16) | 132 (16) | NA | NA | NA |
| 47-50 | 13 (10) | 14 (12) | 20 (17)4-154 | NA | NA | 47 (13) | 69 (14) | 116 (14) | NA | NA | NA |
| >50 | 1 (<1) | 3 (2) | 4 (3) | NA | NA | 8 (2) | 8 (2) | 16 (2) | NA | NA | NA |
| +8 Overall | 7 (5) | 7 (6) | 12 (10) | NA | 46 (14) | 72 (10) | 40 (8) | 112 (9) | 102 (8) | 28 (5) | 130 (7) |
| +21 Overall | 5 (4) | 5 (4) | 4 (3) | NA | 20 (5) | 34 (5) | 26 (5) | 60 (5) | 25 (2) | NA | 25 (2) |
| Chromosome Abnormality . | Children . | Adults . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 19881 No. (%) . | 19892 No. (%) . | 19953 No. (%) . | 19964 No. (%) . | 199854-150 No. (%) . | Total1-54-151 No. (%) . | Present No. (%) . | Total4-151 No. (%) . | 19985‡ No. (%) . | 1998234-153 No. (%) . | Total5 23 No. (%) . | |
| Total | 130 | 121 | 115 | 220 | 340 | 926 | 478 | 1,404 | 1,272 | 609 | 1,881 |
| Normal | 41 (31) | 25 (21) | 17 (15) | 61 (28) | 91 (27) | 235 (25) | 109 (23) | 344 (24) | 589 (46) | 245 (40) | 834 (44) |
| t(8;21)(q22;q22) | 17 (13) | 14 (12) | 9 (8) | 36 (16) | 41 (12) | 117 (13) | 56 (12) | 173 (12) | 81 (6) | 40 (7) | 121 (6) |
| inv(16)(p13q22)/t(16;16) | 4 (3) | 15 (12) | 9 (8) | 19 (9) | 16 (6) | 63 (7) | 28 (6) | 91 (6) | 41 (3) | 49 (8) | 90 (5) |
| t(15;17)(q22;q12-21) | 10 (8) | 9 (7) | 12 (10) | 9 (4) | 31 (9) | 71 (8) | 55 (11) | 126 (9) | 167 (13) | 27 (4) | 194 (10) |
| t(9;11)(p22;q23) | 0 | 9 (7) | 10 (9) | 16 (7) | NA | 35 (6) | 35 (7) | 70 (7) | NA | NA | NA |
| t(11q23;V)/inv/del(11q23) | 12 (9) | 7 (6) | 11 (10) | 21 (9) | 26 (8) | 77 (8) | 53 (11) | 130 (9) | 34 (3)4-155 | 42 (7)4-155 | 76 (4) |
| t(6;9)(p23;q24) | 0 | 0 | 1 (<1) | NA | NA | 1 (<1) | 6 (1) | 7 (<1) | NA | 11 (2) | 11 (2) |
| t(1;22)(p13;q13) | 0 | 0 | 4 (3) | NA | NA | 4 (1) | 4 (<1) | 8 (<1) | NA | 0 | NA |
| −7/del(7q) | 6 (5) | 6 (5) | 6 (5) | NA | 19 (6) | 37 (5) | 21 (4) | 58 (5) | 74 (6) | 52 (8) | 126 (7) |
| −5/del(5q) | 2 (1) | 1 (<1) | 0 | NA | 6 (2) | 9 (1) | 6 (1) | 15 (1) | 48 (4) | 36 (6) | 84 (4) |
| Hyperdiploid | 14 (11) | 17 (14) | 24 (21) | NA | NA | 55 (15) | 77 (16) | 132 (16) | NA | NA | NA |
| 47-50 | 13 (10) | 14 (12) | 20 (17)4-154 | NA | NA | 47 (13) | 69 (14) | 116 (14) | NA | NA | NA |
| >50 | 1 (<1) | 3 (2) | 4 (3) | NA | NA | 8 (2) | 8 (2) | 16 (2) | NA | NA | NA |
| +8 Overall | 7 (5) | 7 (6) | 12 (10) | NA | 46 (14) | 72 (10) | 40 (8) | 112 (9) | 102 (8) | 28 (5) | 130 (7) |
| +21 Overall | 5 (4) | 5 (4) | 4 (3) | NA | 20 (5) | 34 (5) | 26 (5) | 60 (5) | 25 (2) | NA | 25 (2) |
Abbreviations: NA, not available; V, variable chromosome.
Age group in years 0-14.
Percentages for specific abnormalities exclude unavailable cases.
Age group in years 15-55, included secondary leukemias due to chemotherapy or prior MDS.
SWOG/ECOG intergroup trial in AML, patients aged 16-55 years.
The t(9;11) cases were included in the 11q23 subgroup.
Hyperdiploid cases classified as 47-48 and 49.
Abnormalities of chromosome 16 were previously identified in 7% of patients,1-5 and we found abnormalities of chromosome 16 in 6% of our study population. Because these chromosomal rearrangements are subtle and difficult to detect with conventional cytogenetics, they likely are underrepresented. At present, reverse transcription-polymerase chain reaction (RT-PCR) is a sensitive technique used for detecting the CBFB-MYH11 fusion product generated by the inversion.26 27 Presumably, this method and FISH with the commercially available probe to detect inv(16) will detect larger numbers of aberrations of chromosome 16 in future prospective studies.
Translocations affecting the 11q23 are overall the most frequently involved chromosomal breakpoint observed in childhood AML (14%) in previous series1-5; 18.4% of the patients in the present study had 11q23 abnormalities. However, these rates are probably underestimated because some 11q23 rearrangements are subtle. FISH and Southern blotting are currently used to evaluate the MLL gene, which is rearranged in most 11q23 translocations in AML.28-30 The t(6;9) rearrangement is rare in children (0.1%) in the previous as well as present series (1%), as are cases with t(10;11)(p13;q21), t(1;22), t(8;16), or t(3;5), that together accounted for 5% of the cases in our series. The incidence of these abnormalities in earlier series is unknown because these abnormalities were not reported individually.
Most hyperdiploid cases of pediatric AML have 47 to 50 chromosomes; fewer than 3% of patients have more than 50 chromosomes. The most frequent gain is an extra chromosome 8, with an incidence of 10% overall in previously reported series and of 8% in our series. Approximately 4% to 5% of all cases in our population and in earlier series have an extra chromosome 21. The +8 or +21 are usually found together with other abnormalities1-5; in our series, we identified +8 alone in 2% of patients and +21 alone in 1.5%. Other trisomies in childhood AML are found in less than 1% of cases. Monosomy 7 has been observed in as many as 5% of all cases. However, in most of the previously reported series, cases of monosomy 7 were grouped with those with 7q−, as both abnormalities lead to the loss of a putative tumor suppressor gene on 7q. In our series, 7q abnormalities usually were accompanied by other recurrent changes [eg, t(8;21)]; therefore, these cases were not grouped with those with monosomy 7. As expected, −7 was associated with a poor response in our series. As shown previously1-5 and in the present study, cases of childhood AML with −5/5q− occur very infrequently. The rarity of other monosomies (ie, loss of chromosome 19 or either sex chromosome as the sole abnormality) precludes assessment of their effect on outcome in pediatric patients.
We compared the cytogenetic findings obtained from the 6 pediatric series with the series by Grimwade et al5 and Slovak et al23 with an estimated frequency based on reports for de novo adult AML patients (Table 4). The overall incidence of clonal chromosomal abnormalities was higher in children (76%) than in adults (56%). The differences regarding the type of chromosomal abnormalities identified between pediatric cases and adults were minor. However, the main difference noted between the distributions of chromosomal abnormalities was an increase in 11q23, t(8;21), and trisomies 8 and 21 in the children’s series, and an increase in the abnormalities of chromosomes 7 and 5 in the adult series.5,31 These findings suggest that these abnormalities in adults result from exposure to leukemogenic agents. Indirect support of this hypothesis includes the increased number of abnormalities of chromosomes 7 and 5 in patients older than 55 years with de novo AML31,32; the numbers are even greater in the leukemic cells of patients with treatment-related leukemia and in patients exposed to mutagenic agents.8 33Therefore, the reduced incidence of abnormalities of chromosomes 7 and 5 in children may be the result of their limited environmental exposures.
In our study population, various cytogenetic characteristics were associated with outcome. A significantly favorable prognostic impact was found for patients with inv(16)/t(16;16), t(8;21), and normal karyotypes, which were associated with 4-year EFS rates of 58.2% ± 10.9%, 45.1% ± 7.7%, and 43.1% ± 5.0%, respectively. Thus, our data confirm that despite its association with a high median leukocyte count at presentation, inv(16)/t(16;16) is one of the best indicators for prolonged survival after treatment for childhood AML.3,5 Furthermore, inv(16)/t(16;16) subgroup had the best overall survival (75.0%), suggesting that salvage postrelapse is frequently possible and transplant may not be necessary in this subgroup. The poor outcome observed for patients with the t(8;21) by Martinez-Climent et al3 and Leblanc et al4 may reflect differences in treatment regimens, such as addition of high-dose cytarabine in more recent protocols, including this study. In our analysis, patients with any type of 11q23 abnormality had a poor response to treatment, with an overall 4-year EFS rate of 23.8% ± 4.8%. Further, the favorable outcome of patients with t(9;11) rearrangements reported for other series was not supported by our data.5,12 34-37
In this study, patients with t(15;17) did not receive ATRA either during induction or in first remission because this treatment was not available until the latter period of this study. Thus, our patients received chemotherapy only. The initial response rate, the EFS rate, and the overall survival rate were all significantly lower than reports of patients who received both retinoid and chemotherapy.11Although the overall survival rate at 4 years (39%) was less than that of patients treated with retinoids, the rate was similar to that of patients in the control treatment arm who did not receive retinoid therapy.11
We observed a poor outcome among patients with rare recurrent translocations (4-year EFS, 19.7% ± 8.8%). However, despite the large size of our study population (478 children with de novo AML), the paucity of patients with rare recurrent chromosomal abnormalities prevents meaningful evaluation of the effect of chemotherapy in patients with individual translocations. As expected, patients with a single clonal abnormality fared slightly better than did those with multiple chromosomal changes (4-year EFS, 38.9% ± 9.2% v32.1% ± 7.1%, P = .0003). However, the finding of multiple chromosomal changes did not have a statistically significant adverse impact in our series. Our study confirms the identity of several rare recurrent structural chromosomal abnormalities, and it discloses an unusual heterogeneity of miscellaneous chromosomal abnormalities, which has not been reported by others (Table 2).
In summary, the present report focuses on the largest series of children for whom cytogenetics data are available and comprehensively delineates the incidence of chromosomal abnormalities in childhood AML. The types of abnormalities identified are similar to those seen in adults, although the frequency of the changes differs between the populations (Table 4). The majority of the children with AML can be assigned to a known cytogenetic subgroup. In the present study, 23% of our patients had apparently normal karyotypes, and approximately 60% had a known recurrent chromosomal abnormality and assigned independent prognostic impact to the most frequent recurrent changes. The most favorable prognoses were conferred by inv(16), t(8;21), and a normal karyotype; these patients do well with currently available chemotherapeutic regimens. The poorest outcomes were associated with t(15;17), 11q23 abnormalities, and rare recurrent translocations [eg, t(10;11), t(6;9), t(3;5), t(1;22), and t(8;16)]; patients with these chromosomal characteristics require more effective therapies. However, the effect of +8 on outcome is difficult to assess, because most cases (like those of +21) are associated with known recurrent chromosomal abnormalities. Survival rates among the different series vary, probably because of the lack of standardized treatment. For optimal results, future cooperative groups should compare prospectively the efficacy of individualized therapy strategies within defined cytogenetic subgroups of patients.
ACKNOWLEDGMENT
The authors thank V. Turner for her secretarial assistance in preparing this manuscript, and A. Frazier and J.C. Jones for editorial assistance. The participating Pediatric Oncology Group (POG) Institutions and their Grant support are listed in the.
Institutions and Grant Numbers of Participants
| Institution . | Grant No. . |
|---|---|
| Alberta Children’s Hospital | |
| All Children’s Hospital | |
| Baylor | CA-03161 |
| Boston Floating Hospital | |
| Cancer Center of Hawaii | |
| Carolinas Medical Center | CA-69177 |
| Children’s Hospital Greenville System | CA-69177 |
| Children’s Hospital Michigan | CA-29691 |
| Children’s Hospital New Orleans/LSU CCOP | |
| Children’s Memorial Hospital (Chicago) | CA-07431 |
| UC/San Diego | CA-28439 |
| Christ Hospital | CA-07431 |
| City of Hope | |
| Cook-Ft. Worth Children’s Medical Center | CA-33625 |
| Dana-Farber Cancer Institute | CA-41573 |
| Dartmouth Hitchcock | CA-29293 |
| Duke University | CA-15525 |
| East Carolina University | CA-69177 |
| Emory University | CA-20549 |
| Fairfax Hospital | CA-28476 |
| Hackensack Medical Center | |
| Hurley Medical Center | CA-29691 |
| Joe DiMaggio Children’s | |
| Johns Hopkins University | CA-28476 |
| Kaiser Permanente/San Diego | CA-28439 |
| Kaiser/Santa Clara | CA-33603 |
| Keesler AFB Hospital | CA-15898 |
| Medical University of South Carolina | CA-69177 |
| Maine Children’s | CA-41573 |
| Operations Office | CA-30969 |
| Statistical Office | CA-29139 |
| Massachusetts General Hospital | CA-29293 |
| McGill University | CA-33587 |
| Medical College Virginia | |
| Miami Children’s Hospital | |
| Midwest Children’s Cancer Center | CA-32053 |
| Mount Sinai Medical School (N.Y.) | CA-69428 |
| Naval Medical Center, SD | |
| Nemours Children’s Clinic | |
| Nemours/Orlando | |
| Oklahoma University | CA-11233 |
| Presbyterian Hospital | CA-69177 |
| Puerto Rico POG | |
| Rhode Island Hospital | CA-29293 |
| Roswell Park Cancer Institute | CA-28383 |
| Rush-Presbyterian | CA-07431 |
| SPOG Basel | |
| SPOG Bern | |
| SPOG Geneva | |
| SPOG Lausanne | |
| SPOG Zurich | |
| SUNY Syracuse | |
| Sacred Heart Hospital | |
| San Antonio MPC & BDC | |
| San Jorge Children’s Hospital | |
| St. Christopher’s Hospital | |
| St. Francis Regional | |
| St. Johns Hospital | CA-29691 |
| St. Mary’s Hospital | |
| St. Vincent Hospital | |
| Stanford University | CA-33603 |
| Tampa Children’s Hospital | |
| Tripler Army Medical Center | |
| University of Alabama | CA-25408 |
| University of Arizona | CA-33603 |
| University of Arkansas | |
| University of Florida | |
| University of Kansas | |
| University of Massachusetts | CA-69428 |
| University of Miami | |
| University of Mississippi Medical Center | CA-15989 |
| University of Missouri | CA-05587 |
| University of Rochester | |
| University of South Alabama | |
| University of South Florida | |
| University of Vermont | CA-29293 |
| University of Virginia | |
| UC/Davis | |
| UC/San Diego | CA-28439 |
| Southwestern Medical School | CA-33625 |
| UT/Galveston | CA-03161 |
| UT/San Antonio | |
| Wake Forest University School of Medicine | |
| West Virginia University, Charleston | CA-15525 |
| West Virginia University, Morgantown | CA-15525 |
| Walt Disney Memorial Cancer Institute | |
| Walter Reed Army Medical | |
| Warren Clinics | CA-11233 |
| Washington University | CA-05587 |
| Yale University | CA-69428 |
| Institution . | Grant No. . |
|---|---|
| Alberta Children’s Hospital | |
| All Children’s Hospital | |
| Baylor | CA-03161 |
| Boston Floating Hospital | |
| Cancer Center of Hawaii | |
| Carolinas Medical Center | CA-69177 |
| Children’s Hospital Greenville System | CA-69177 |
| Children’s Hospital Michigan | CA-29691 |
| Children’s Hospital New Orleans/LSU CCOP | |
| Children’s Memorial Hospital (Chicago) | CA-07431 |
| UC/San Diego | CA-28439 |
| Christ Hospital | CA-07431 |
| City of Hope | |
| Cook-Ft. Worth Children’s Medical Center | CA-33625 |
| Dana-Farber Cancer Institute | CA-41573 |
| Dartmouth Hitchcock | CA-29293 |
| Duke University | CA-15525 |
| East Carolina University | CA-69177 |
| Emory University | CA-20549 |
| Fairfax Hospital | CA-28476 |
| Hackensack Medical Center | |
| Hurley Medical Center | CA-29691 |
| Joe DiMaggio Children’s | |
| Johns Hopkins University | CA-28476 |
| Kaiser Permanente/San Diego | CA-28439 |
| Kaiser/Santa Clara | CA-33603 |
| Keesler AFB Hospital | CA-15898 |
| Medical University of South Carolina | CA-69177 |
| Maine Children’s | CA-41573 |
| Operations Office | CA-30969 |
| Statistical Office | CA-29139 |
| Massachusetts General Hospital | CA-29293 |
| McGill University | CA-33587 |
| Medical College Virginia | |
| Miami Children’s Hospital | |
| Midwest Children’s Cancer Center | CA-32053 |
| Mount Sinai Medical School (N.Y.) | CA-69428 |
| Naval Medical Center, SD | |
| Nemours Children’s Clinic | |
| Nemours/Orlando | |
| Oklahoma University | CA-11233 |
| Presbyterian Hospital | CA-69177 |
| Puerto Rico POG | |
| Rhode Island Hospital | CA-29293 |
| Roswell Park Cancer Institute | CA-28383 |
| Rush-Presbyterian | CA-07431 |
| SPOG Basel | |
| SPOG Bern | |
| SPOG Geneva | |
| SPOG Lausanne | |
| SPOG Zurich | |
| SUNY Syracuse | |
| Sacred Heart Hospital | |
| San Antonio MPC & BDC | |
| San Jorge Children’s Hospital | |
| St. Christopher’s Hospital | |
| St. Francis Regional | |
| St. Johns Hospital | CA-29691 |
| St. Mary’s Hospital | |
| St. Vincent Hospital | |
| Stanford University | CA-33603 |
| Tampa Children’s Hospital | |
| Tripler Army Medical Center | |
| University of Alabama | CA-25408 |
| University of Arizona | CA-33603 |
| University of Arkansas | |
| University of Florida | |
| University of Kansas | |
| University of Massachusetts | CA-69428 |
| University of Miami | |
| University of Mississippi Medical Center | CA-15989 |
| University of Missouri | CA-05587 |
| University of Rochester | |
| University of South Alabama | |
| University of South Florida | |
| University of Vermont | CA-29293 |
| University of Virginia | |
| UC/Davis | |
| UC/San Diego | CA-28439 |
| Southwestern Medical School | CA-33625 |
| UT/Galveston | CA-03161 |
| UT/San Antonio | |
| Wake Forest University School of Medicine | |
| West Virginia University, Charleston | CA-15525 |
| West Virginia University, Morgantown | CA-15525 |
| Walt Disney Memorial Cancer Institute | |
| Walter Reed Army Medical | |
| Warren Clinics | CA-11233 |
| Washington University | CA-05587 |
| Yale University | CA-69428 |
Supported in part by grants from the National Institutes of Health (CA-21765, CA-03161, CA-05587, CA-07431, CA-11233, CA-15525, CA-15898, CA-15989, CA-20549, CA-25408, CA-28383, CA-28439, CA-28476, CA-29139, CA-29293, CA-29691, CA-30969, CA-32053, CA-33587, CA-33603, CA-33625, CA-41573, CA-69177, and CA-69428), and by American Lebanese Syrian Associated Charities (ALSAC). Additional grant support is listed in the .
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Susana C. Raimondi, PhD, Department of Pathology, St Jude Children’s Research Hospital, 332 N Lauderdale, Memphis, TN 38105; e-mail: susana.raimondi@stjude.org.