To the Editor:
Recently, Garcı́a-Sanz et al1 reported a series of 26 patients with plasma cell leukemia (PCL) in whom a number of biological parameters were studied, with particular emphasis about measure of DNA cell content by flow cytometry and detection of numeric chromosomal aberrations by interphase fluorescent in situ hybridization (FISH). For the purposes of comparison, the same parameters were tested in a series of 56 multiple myeloma (MM) patients. Their data, as presented, showed clear differences between the PCL and MM patients studied.
Firstly, all except 1 PCL patient studied showed a normal DNA content, whereas most MM patients (57%) showed a hyperdiploid DNA content.
Secondly, FISH studies using 15 chromosome-specific probes performed in 13 PCL and 56 MM cases showed statistically significant differences between PCL and MM for the following chromosomes: −13 (84% in PCL and 26% in MM), +9 (0% in PCL and 52% in MM), and +6 (0% in PCL and 32% in MM). Although chromosome 3, 7, 11, and 15 were found trisomic in MM cases only, the results of the statistical analysis regarding these defects are not mentioned. On the other hand, no difference was found for chromosome 1 involvement (43% in PCL and 37% in MM). The used chromosome 1 probe hybridized to the 1q12 heterochromatic region, making trisomy of chromosome 1 indistinguishable from the presence of a 1q chromosome derivative, a frequent finding in these pathologies.2
The investigators concluded that, in PCL and MM, the DNA cell content and cytogenetic characteristics are different, leading to a different disease evolution.
We do not agree with these results, because we think that the techniques used, ie, flow cytometry and FISH with centromeric probes, are not appropriate to compare genetic aberrations in PCL and MM patients because they missed a number of aberrations detected by other techniques.
Whereas flow cytometry is a good technique to detect hyperdiploidy, it is not able to detect a minor error in DNA content such as a small hypodiploid clone or, moreover, pseudodiploid aberrations. Zandecki et al,3 using a different technique to appreciate the plasma cell DNA content (histochemical staining with computed analysis), found that 58.7% of patients were hyperdiploid, as reported by Garcı́a-Sanz et al,1 but they also detected 24% patients with hypodiploidy and 4.3% with biclonality among a series of 46 MM patients. Elsewhere, conventional cytogenetics (CC) in stage III MM and in PCL cases lead to informations in more than 70% of cases studied.4-7 Using CC, we studied a series of 81 MM patients with an abnormal karyotype and found 2 different cytogenetic patterns based on chromosome number: a hyperdiploid pattern (54%) with recurrent trisomies 3, 5, 7, 9, 11, 15, and 19 and a second pattern (46%) showing either pseudoploid, hypoploid, or near tetraploid karyotypes. In this study, a prognostic correlation was found between the cytogenetic pattern and overall survival: hyperdiploid patients had a longer survival than patients belonging to the pseudo/hypo/near-tetraploid group.8
In the same way, a recent conventional cytogenetic study of 13 primary PCL patients showed that 12 of 13 had an abnormal clone. A hypodiploid karyotype was found in 9 cases and a pseudodiploid was found in 2 cases. It was concluded that PCL patients share a poor prognosis with MM patients with a hypodoploid karyotype.7
From a cytogeneticist point of view, we do not find any difference between plasma cell leukemia and hypo/pseudo/near-tetraploid multiple myeloma associated with poor prognosis. We think that PCL and these MM are, in fact, the same disease. PCL could be is a fulminant form of MM and could be compared with those rare cases of chronic myeloid leukemia showing an acute phase at presentation.
Response
We are pleased for the interest that Smadja et al have demonstrated on our report,1-1 in which we report on the clinical and biological characteristics of a series of primary PCL. In their letter, they add cytogenetic information on 81 multiple myelomas and give a reference submitted for publication concerning 13 primary PCL, concluding that MM with pseudoploid, hypoploid, or near tetraploid karyotypes show similar genetic characteristics as compared with PCL. According to this information, they conclude that both diseases are the same, which would be in contrast to the conclusion that presumably we support in our report. Actually, in our previous publication, we just concluded that, because PCL and MM show overall distinct clinical, immunophenotypic, DNA ploidy, and cytogenetic characteristics, the link between both diseases remains unclear. In this sense, the apparent discrepancies between these investigators and us would be merely based on data interpretation.
Actually, a careful analysis of the information reported by Smadja et al would support our own data, even considering that different technical approaches were used for the analysis of chromosomal aberrations. They report on 81 MM, in which 54% are hyperdiploid (this prevalence is almost exactly the same as the 56% of MM with a DNA index >1 identified in our series). In contrast, no hyperdiploid PCL were found by Brigadeau et al1-2 among 13 cases tested, which is also coincidental with our experience (series 0/22 PCL had a DNA index >1). It is obvious that these differences favor the notion that, overall, PCL and MM are different diseases.
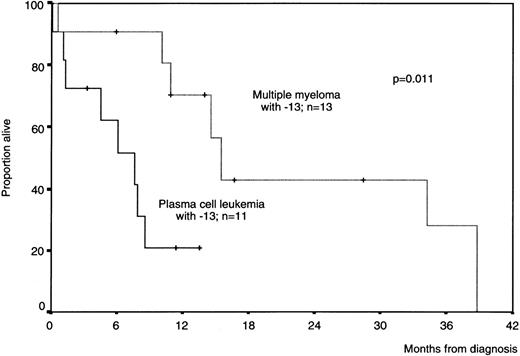
The most relevant cytogenetic molecular marker of PCL is the deletion of the Rb gene, located at chromosome 13, because it was present in the great majority of our PCL cases (86%). Nevertheless, this is not a specific marker of PCL, because a subset of myeloma patients also share this chromosomal change. This subset of patients display an unfavorable prognosis.1-3-1-5 Therefore, it could be argued that these MM patients are tightly related to PCL. However, upon analyzing other chromosomal changes, we observed that a high proportion of MM cases with monosomy 13 concomitantly show trisomy 9 (67% of cases) and trisomy 6 (33%), whereas none of these changes was observed in our PCL patients. Moreover, as shown in Fig1-1, the survival of MM patients with monosomy 13 was significantly better than in those PCL patients with the same chromosome abnormality (median survival, 16 and 7 months, respectively; P = .011). Based on these results, it could be speculated that the occurrence of a deletion in the closer area of the Rb gene would be a critical alteration that changes the clinical behavior of the disease. However, this alteration would appear in MM in a different moment of the clonal evolution as compared with de novo PCL. In addition, from the biological point of view, we described in our report several other parameters, including the proliferative activity, extramedulary involvement, LDH serum level, immunophenotype, response to therapy, etc, that were also different between MM and PCL, suggesting that MM and PCL are different.
Overall survival curves of patients with monosomy 13 according to the presence of plasma cell leukemia or multiple myeloma criteria.
Overall survival curves of patients with monosomy 13 according to the presence of plasma cell leukemia or multiple myeloma criteria.
Despite these data, we think that the answer to the question on whether PCL and MM are the same disease remains open. In this sense, we still can recognize overlapping cases that accomplish MM or PCL criteria but display clinical or biological characteristics of the other form. This was already reported in our publication. This fact could lead to the hypothesis that MM and PCL belong to the same group of diseases (here we should use the unspecific term monoclonal gammopathies), but there is a different pattern in the accumulation of genetic alterations leading to a different clinical behavior in the 2 forms of the disease. Because the clonal evolution and the genetic way to acquire aberrations depend on many circumstances,1-6 this could explain why there exist some overlapping cases.