Targeted mutagenesis was used to produce two mutations in the murine hemochromatosis gene (Hfe) locus. The first mutation deletes a large portion of the coding sequence, generating a null allele. The second mutation introduces a missense mutation (C282Y) into theHfe locus, but otherwise leaves the gene intact. This mutation is identical to the disease-causing mutation in patients with hereditary hemochromatosis. Mice carrying each of the two mutations were bred and analyzed. Homozygosity for either mutation results in postnatal iron loading. The effects of the null mutation are more severe than the effects of the C282Y mutation. Mice heterozygous for either mutation accumulate more iron than normal controls. Interestingly, although liver iron stores are greatly increased, splenic iron is decreased. We conclude that the C282Y mutation does not result in a null allele.
HEREDITARY HEMOCHROMATOSIS is a common, autosomal recessive disorder affecting approximately one million people in the United States. The gene responsible for most cases was identified by positional cloning in 1996.1 TermedHFE, it encodes an atypical member of the class I major histocompatibility protein family that heterodimerizes with β2-microglobulin. Most affected patients are homozygous for a unique missense mutation that results in a tyrosine for cysteine substitution at amino acid 282 (C282Y).1 It is not clear whether heterozygosity for the C282Y mutation, by itself, can predispose to iron overload. The C282Y mutation disrupts an intramolecular disulfide bond, and may interfere with β2-microglobulin binding.2Mice deficient in β2-microglobulin and mice deficient in Hfe both develop systemic iron overload.3 4 The function of HFE and its role in iron metabolism remain unknown. We created two mutant mouse strains to investigate whether the C282Y mutation results in total loss of protein function, and to develop a model system to study the pathogenesis of hemochromatosis.
MATERIALS AND METHODS
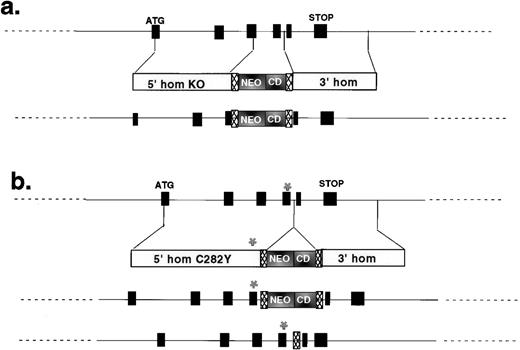
The murine Hfe gene resembles its human ortholog. Codon 282 is 49 nucleotides upstream of an exon-intron junction. We used site-directed mutagenesis (QuikChange kit; Stratagene, La Jolla, CA) to introduce the C282Y mutation into murine codon 282. Genomic HfeDNA was inserted into the pTKLNCL targeting vector (from R. Mortenson, Brigham and Women’s Hospital), such that the C282Y mutation was close to the neomycin resistance cassette (Fig1). The construct was electroporated into TC-1 embryonic stem (ES) cells,5 and recombinant clones were selected as previously described.6 We anticipated that most targeting events would involve recombination between homologous sequences located further from the selectable marker than the site of the mutation and would, therefore, result in introduction of the C282Y mutation into the mouse genome. This proved to be true. Targeting vector sequences were located within an intron, between loxP sites. A second targeting vector was similarly used to construct an Hfenull allele (Fig 1).
Targeting constructs used to create mutant Hfealleles. The structures of the two targeting constructs are shown, with reference to the murine Hfe locus. In each case, the intron/exon structure of the genomic clone is shown on the top line, the structure of the targeting construct is shown on the second line, and the structure of the correctly targeted mutant locus is shown on the third line. Black boxes are Hfe exons. Translational start (ATG) and stop (STOP) sites are indicated. 5′ homology (5′ hom) and 3′ homology (3′ hom) regions are indicated for each targeting vector. The locations of the neomycin resistance (NEO) and cytosine deaminase (CD) cassettes are shown. Hatched boxes represent loxP sites. The asterisk (*) shows the site of codon 282. (a) Summarizes the strategy used to make the null allele; (b) summarizes the strategy used to introduce the C282Y missense mutation. The final line in (b) shows the structure of the C282Y allele after vector sequences have been removed by Cre-mediated recombination between loxP sites.
Targeting constructs used to create mutant Hfealleles. The structures of the two targeting constructs are shown, with reference to the murine Hfe locus. In each case, the intron/exon structure of the genomic clone is shown on the top line, the structure of the targeting construct is shown on the second line, and the structure of the correctly targeted mutant locus is shown on the third line. Black boxes are Hfe exons. Translational start (ATG) and stop (STOP) sites are indicated. 5′ homology (5′ hom) and 3′ homology (3′ hom) regions are indicated for each targeting vector. The locations of the neomycin resistance (NEO) and cytosine deaminase (CD) cassettes are shown. Hatched boxes represent loxP sites. The asterisk (*) shows the site of codon 282. (a) Summarizes the strategy used to make the null allele; (b) summarizes the strategy used to introduce the C282Y missense mutation. The final line in (b) shows the structure of the C282Y allele after vector sequences have been removed by Cre-mediated recombination between loxP sites.
Correctly targeted ES cell clones were injected into mouse blastocysts to generate chimeric mice carrying each mutant allele. Inbred mouse strains vary markedly in parameters of iron metabolism7(and our unpublished data). For this reason, the original chimeric mice were bred to two different inbred strains. First, they were bred to 129/SvEvTac mice from which the TC-1 ES cell line was derived,5 to place the mutations on an inbred background. Because 129 mice load iron to a greater extent than many other strains (unpublished data), first-generation chimeric mice were also bred to C57BL/6J mice, to place each mutation on a mixed C57BL/6J × 129/SvEvTac background. Vector sequences were subsequently removed from the Hfe locus carrying the C282Y mutation by breeding to 129/SvEvTac mice expressing Cre recombinase.8This maneuver deleted vector sequences in the germline, leaving only a single loxP site within the intron. Reverse transcriptase-polymerase chain reaction analysis of mRNA confirmed that the C282Y allele was expressed and produced a properly spliced mRNA (data not shown).
Animals were maintained on standard mouse diet and iron status was evaluated at various ages as previously described.6
RESULTS AND DISCUSSION
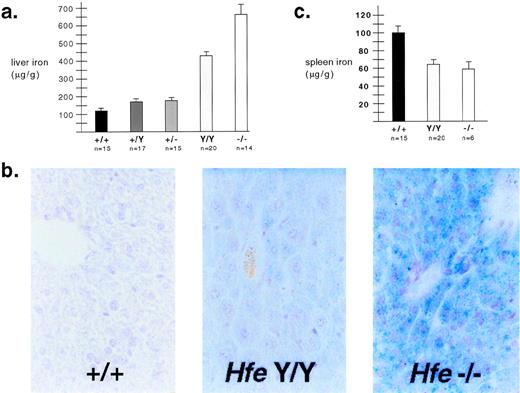
As shown in Fig 2, genotype correlated strongly with iron loading. Mice heterozygous for either mutant allele showed more iron loading than wild-type mice of the same genetic background (Fig 2a). Similar to previously reported results,4 mice homozygous for the null allele developed massive iron loading early in life. This occurred postnatally; wild-type and mutant iron levels were identical at 3 days of age, and iron loading occurred more rapidly on the 129/SvEvTac background (data not shown). C282Y homozygotes had a phenotype intermediate between null homozygotes and wild-type mice. Results were confirmed by Prussian blue staining of liver tissue (Fig 2b). These results indicate that the C282Y mutation does not completely disrupt the function of Hfe, and is not a null mutation. Interestingly, mice began to accumulate hepatic iron before circulating transferrin was fully saturated. At 4 weeks of age, on the 129/SvEvTac background, nonfasting transferrin saturations were 84.0 ± 1.1, 89.5 ± 1.3, and 88.3 ± 1.4 for wild type, C282Y homozygotes, and null homozygotes, respectively. These blood samples were obtained by retroorbital bleeding or cardiac puncture; it remains possible that portal transferrin saturations are higher.
Iron metabolism in Hfe mutant mice. (a) Liver iron content was determined for 4-week-old 129/SvEvTac mice as previously described10 and expressed as micrograms per gram wet weight ± standard error. Genotypes are abbreviated as follows: wild-type (+/+), Hfe C282Y homozygous (Y/Y ), andHfe null homozygous (−/−). All differences between genotypes were statistically significant when P values were determined by Welch correction of the unpaired t-test. (b) Hepatic iron was visualized by Prussian blue staining of tissue sections from wild-type and mutant F2 mice with a C57BL/6J × 129/SvEvTac background. Nonheme iron deposits appear blue. (c) Spleen iron was determined for 4-week-old 129/SvEvTac mice using the same method as was used for liver iron, and expressed as microgram per gram wet weight ± standard error. Differences between wild-type mice and each mutant strain were statistically significant according toP values determined by Welch correction of the unpairedt-test. The mutant strains were not significantly different from each other.
Iron metabolism in Hfe mutant mice. (a) Liver iron content was determined for 4-week-old 129/SvEvTac mice as previously described10 and expressed as micrograms per gram wet weight ± standard error. Genotypes are abbreviated as follows: wild-type (+/+), Hfe C282Y homozygous (Y/Y ), andHfe null homozygous (−/−). All differences between genotypes were statistically significant when P values were determined by Welch correction of the unpaired t-test. (b) Hepatic iron was visualized by Prussian blue staining of tissue sections from wild-type and mutant F2 mice with a C57BL/6J × 129/SvEvTac background. Nonheme iron deposits appear blue. (c) Spleen iron was determined for 4-week-old 129/SvEvTac mice using the same method as was used for liver iron, and expressed as microgram per gram wet weight ± standard error. Differences between wild-type mice and each mutant strain were statistically significant according toP values determined by Welch correction of the unpairedt-test. The mutant strains were not significantly different from each other.
Mice homozygous for each Hfe mutation had less splenic nonheme iron than wild-type mice, despite systemic iron overload (Fig 2c). This may be because mouse spleen is an active site of erythropoiesis, and most splenic storage iron is found in macrophages. This may be analogous to human hemochromatosis, where patients have been shown to have decreased iron in bone marrow macrophages.9
In conclusion, the C282Y mutation clearly predisposes to iron loading, but is not as severe as a null allele. Heterozygosity for either mutantHfe allele results in increased iron stores, consistent with the notion that the prevalence of the human mutation may result from heterozygote advantage. Analogous to human patients, mice carryingHfe mutations also have depleted splenic iron stores, further confirming that these mouse mutants offer a valid system for studying the human disease. Finally, liver iron loading begins before circulating transferrin is fully saturated, suggesting that it may occur in the absence of significant levels of nontransferrin-bound iron. This has implications for the mechanism of hepatic iron loading, and suggests that it may begin earlier than generally appreciated in hemochromatosis patients.
ACKNOWLEDGMENT
We thank P. Leder for providing TC1 ES cells, R. Mortenson for the targeting vector, Y. Fujiwara and S. Orkin for assistance with blastocyst injections, and H. Westphal and F. Alt for providing mice expressing Cre recombinase.
J.E.L. is supported by National Institutes of Health (NIH) Grant No. K08 HL03503; M.D.F. is supported by NIH Grant No. K08 HL03600 and a fellowship from the American Liver Foundation. N.C.A. is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
Author notes
Address reprint requests to Nancy C. Andrews, MD, PhD, Howard Hughes Medical Institute, Enders 720, Children’s Hospital, 300 Longwood Ave, Boston, MA; e-mail: nandrews@rascal.med.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal