Because of the expression of inhibitory receptors (KIR) for major histocompatibility complex (MHC) class I allotypes, a person’s natural killer (NK) cells will not recognize and will, therefore, kill cells from individuals lacking his/her KIR epitopes. This study investigated the role of NK cell alloreactivity in human HLA haplotype-mismatched hematopoietic stem cell transplantation and, specifically, the role of the three major NK specificities, ie, those for HLA-C group 1, HLA-C group 2, and HLA-Bw4 alleles. In 20 of 60 donor-recipient pairs, KIR epitope incompatibility and functional analyses of donor NK cell clones predicted donor NK cells could cause graft-versus-host (GVH)/graft-versus-leukemia (GVL) reactions. NK cell clones of donor origin were obtained from transplanted recipients and tested for lysis of recipient’s cryopreserved pretransplant lymphocytes. Despite the absence of GVH disease, we detected high frequencies of NK clones which killed recipient’s target cells. Lysis followed the rules of NK cell alloreactivity, being blocked only by the MHC class I KIR epitope which was missing in the recipient. The alloreactive NK clones also killed the allogeneic leukemia. Transplants from these KIR epitope incompatible donors had higher engraftment rates. Therefore, a GVL effector and engraftment facilitating mechanism, which is independent of T-cell–mediated GVH reactions, may be operational in HLA mismatched hematopoietic cell transplants.
INHIBITION OF natural killer (NK) cell lysis is signaled through specific receptors which bind to polymorphic determinants of major histocompatibility complex (MHC) class I molecules.1,2 In humans, one receptor is the lectin-like heterodimer CD94-NKG2A, which recognizes human leukocyte antigen (HLA)-E, a nonclassical MHC class Ib molecule whose expression is, in turn, upregulated by the binding of signal sequence peptides of other MHC class I molecules.3,4 Other receptors are a family of Ig-like molecules known as killer cell inhibitory receptors (KIR).5-7 The KIRs with two Ig domains (KIR2D) identify HLA-C allotypes: KIR2DL2 (formerly designated p58.1) recognizes an epitope shared by group 1 HLA-C allotypes (Cw2, 4, 5, and 6), whereas KIR2DL1 (p58.2) recognizes an epitope shared by the reciprocal group 2 HLA-C allotypes (Cw1, 3, 7, and 8). One KIR with three Ig domains KIR3DL1 (p70) recognizes an epitope shared by HLA-Bw4 alleles. Finally, a homodimer of molecules with three Ig domains KIR3DL2 (p140) recognizes HLA-A3 and -A11.8
KIR genes, each expressed by some of the individual’s NK cells, vary considerably among individuals.9 It is believed that during development each NK cell precursor makes a random choice of which KIR genes it will express, and the different combinations of HLA class I molecules select NK cells that express receptors for self HLA class I. Consequently, the NK cells from any given individual will be alloreactive toward cells from others which lack their KIR ligands and, conversely, will be tolerant of cells from another individual who has the same or additional KIR ligands.
Although KIR epitope mismatches are well-known causes of NK cell alloreactivity,10-17 their role in human transplantation have not been evaluated. Full haplotype-mismatched hematopoietic stem cell transplants have recently been used for treatment of bad-risk leukemia patients lacking a matched donor.18 19 In this context, host and/or donor NK cells may be responsible for three situations: (1) a potential for graft-versus-host (GVH) reactions, ie, when the recipient fails to express the donor’s KIR epitopes; (2) a potential for NK cell-mediated graft rejection, ie, when the donor fails to express the recipient’s KIR epitopes; (3) no NK cell alloreactions, ie, when the donor and recipient mismatched alleles express the same KIR epitopes. One aim of the present study was, therefore, to evaluate whether mismatches for the three major KIR epitopes, ie, those of the HLA-C group 1, HLA-C group 2, and HLA-Bw4 alleles, have any impact on transplantation outcome.
Moreover, the very fast donor-type NK cell recovery20 (and this report) strongly suggests that postgrafting NK cells derive, not from expansion of mature NK cells contaminating the stem cell graft, but from large-scale maturation of the engrafted stem cells. This maturation occurs under the influence of KIR epitopes expressed in the donor’s hematopoietic cells and the host’s marrow stromal cells.21 Because little is known about the cell types which drive selection of the NK cells, KIR epitope mismatched transplants appear to be a convenient model for analyzing the selection of human NK cell repertoires.
PATIENTS AND METHODS
Patients
A total of 60 consecutive high-risk leukemia patients, 22 with acute lymphoblastic leukemia (ALL), 25 with acute myeloid leukemia (AML), and 13 with chronic myeloid leukemia (CML), all without a matched donor, received hematopoietic cell transplants from mismatched family donors (Table 1).
Patient Characteristics According to Status of Leukemia at Transplantation
| Status of Disease . | No. of Patients (N = 60) . | Median Age (range) yr . | Median Time Since Diagnosis (range) mo . |
|---|---|---|---|
| ALL | 22 | 16 (4-27) | 17 (4-66) |
| First complete remission | 4* | ||
| Second or third complete remission | 10 | ||
| Advanced relapse | 8 | ||
| AML | 25 | 33 (7-53) | 10 (4-55) |
| First complete remission | 4† | ||
| Second or third complete remission | 14 | ||
| Advanced relapse | 7 | ||
| CML | 13 | 28 (18-45) | 31 (12-177) |
| Accelerated phase | 8 | ||
| Blastic crisis | 5 |
| Status of Disease . | No. of Patients (N = 60) . | Median Age (range) yr . | Median Time Since Diagnosis (range) mo . |
|---|---|---|---|
| ALL | 22 | 16 (4-27) | 17 (4-66) |
| First complete remission | 4* | ||
| Second or third complete remission | 10 | ||
| Advanced relapse | 8 | ||
| AML | 25 | 33 (7-53) | 10 (4-55) |
| First complete remission | 4† | ||
| Second or third complete remission | 14 | ||
| Advanced relapse | 7 | ||
| CML | 13 | 28 (18-45) | 31 (12-177) |
| Accelerated phase | 8 | ||
| Blastic crisis | 5 |
For patients with ALL in first remission the prognosis was poor; in 1 first-line induction therapy had failed, 2 had the t(9;22) translocation, and 1 had the T phenotype with bulky disease.
For 3 patients with AML in first remission, the prognosis was poor; 2 had myelodysplasia and in 1 first-line induction therapy had failed.
HLA Typing
Donors were either parents, siblings, or children of the recipients. They were assessed for HLA compatibility by serologic typing. All pairs of donors and recipients were identical only for one HLA haplotype (haploidentical) and incompatible at the HLA-A, B, C, and DR loci of the unshared haplotype. Consequently, 100% of the pairs were at risk of T-cell–mediated host-versus-graft (HVG) and GVH reactions. An additional level of alloreactivity was provided by mismatches at the KIR epitopes expressed by HLA-C group 1, HLA-C group 2, and HLA-Bw4 alleles.5-7 Twenty pairs were KIR epitope mismatched in the GVH direction, 17 pairs in the HVG direction.
Transplantation
Conditioning regimen, peripheral blood progenitor cell granulocyte colony-stimulating factor (G-CSF) mobilization, selection of donor peripheral blood CD34+ cells, and assessment of engraftment have been described elsewhere.19 Briefly, patients underwent conditioning with total-body irradiation (TBI) followed by thiotepa, ATG, and fludarabine. Donor G-CSF–mobilized peripheral blood cells, collected by leukapheresis, were T-cell depleted by sheep red blood cell rosetting and positive selection of CD34+ cells (with immuno-adsorption biotin-avidin Ceprate SC columns; Cell Pro Inc, Bothell WA). The inoculum contained 9 × 106 ± 5 × 106 CD34+ and 3 × 104 ± 2 × 104 CD3+ cells/kg body weight. No postgrafting immuno-suppressive treatment was given for GVH disease (GVHD) prophylaxis.
Assessment of Chimerism
Starting on day +12 after transplantation, chimerism of peripheral blood and bone marrow cells was determined by bimonthly assessment of polymerase chain reaction (PCR) amplification of a panel of variable number tandem repeat regions with different DNA polymorphism patterns in donor and recipient cells. All postengraftment blood samples used for the NK cell studies shown here exhibited 100% donor chimerism.
Immunofluorescence and Flow Cytometry
Indirect immunofluorescence with primary monoclonal antibodies (MoAbs) plus secondary fluorochrome-conjugated goat anti-mouse Ig antibodies (Southern Biotechnology Associates, Birmingham, AL) and flow cytometry (FACSCalibur; Becton Dickinson, San Jose, CA) determined NK cell phenotypes. NK cell clones were identified using an anti-CD16 MoAb (Immunotech, Marseille, France). Expression of KIRs recognizing group 1 HLA-C alleles (KIR2DL2), group 2 HLA-C alleles (KIR2DL1), and HLA-Bw4 alleles (KIR3DL1) was determined with MoAbs EB6, GL183 and Z27 respectively22 (kindly provided by L. Moretta, University of Genova, Genova, Italy). LFA-1 expression by allogeneic NK targets was measured by indirect immunofluorescence and flow cytometry with an anti-CD11a MoAb from Immunotech.
Preparation of NK Cell Clones
Peripheral blood mononuclear cells depleted of T cells by negative immuno-magnetic selection with anti-CD3 MoAb (OKT3 obtained from American Tissue Culture Collection [ATCC], Manassas, VA) were plated at the concentration of 10 cells per well in 96-well microtiter plates, activated with phytohemagglutinin (PHA), and cultured with interleukin-2 (IL-2) and irradiated feeder cells as described elsewhere.20 Cloned NK cells were used as effectors in standard 51Cr release cytotoxicity assays using as targets allogeneic PHA lymphoblasts or Epstein-Barr virus–transformed B-lymphoblastoid cell lines (BLCL), leukemia cells, and cell lines expressing single class I alleles. Effector to target (E:T) ratio was 10:1.
Analysis of NK Cell Allospecificity
Standard 51Cr release cytotoxicity assays against cell lines transfected with class I genes (kindly provided by L. Moretta) determined the three main NK allospecificities. Specificities for group 1 (Cw4-related) and group 2 (Cw3-related) HLA-C allotypes were analyzed using the HLA class I–negative cell line 721.221 and the same cell line transfected with Cw*0401 or Cw*0302 genes, respectively.23 Specificity for HLA-Bw4 allotypes (such as HLA-B27) was analyzed using untransfected C1R cells and C1R cells transfected with the B*2705 gene.22 Specificity for the nonclassical MHC class Ib molecule HLA-E was analyzed using C1R cells expressing HLA-B7 (Bw6)21 (cell-surface expression of HLA-E is regulated by the binding of peptides derived from the signal sequence of some other MHC class I molecules, such as HLA-B7).3 4 E:T ratio was 10:1. Results are presented as percentage inhibition compared with lysis of the untransfected 721.221 or C1R cells. NK clones lysed the untransfected cells at levels exceeding 60%, and were considered specific for a given allotype when this was reduced by at least 50%.
RESULTS
Transplantation Into Recipients Failing to Express the Donor’s KIR Epitopes, ie, With a Potential for NK Cell-Mediated GVH Reactions
Eight of the 20 patients in this group had AML, 5 CML, and 7 ALL. All patients engrafted successfully and reached 1,000 neutrophils/μL in 8 to 19 days (median, 11 days) and 50,000 platelets in 13 to 124 days (median, 29 days). No GVHD was observed. One hundred percent donor-type chimerism was detected in peripheral blood and bone marrow on all assessments (except at relapse). At a median follow-up of 6 months (range, 1 to 39 months), 0 of 8 AML, 0 of 5 CML, and 5 of 7 ALL patients relapsed.
Postgrafting emergence of donor-versus-recipient alloreactive NK cells.
Table 2 illustrates the HLA typing of these 20 donor-recipient pairs. Specific donor KIR ligands were missing in the recipient. Some of the donor’s NK cells could, therefore, lyse the recipient’s cells and cause GVHD. GVHD potential was assessed directly in 14 pairs by pretransplant analysis of donor NK cell clones. Table3 (left column) shows some donor NK cell clones killed recipient target cells (PHA lymphoblasts or BLCLs), indicating that donors possessed antirecipient NK cells in their repertoires before transplant. Lysis by these clones could only be blocked by transfected cell lines expressing the donor’s MHC class I allotypic group which was missing in the recipient (data not shown).
Donor-Recipient Pairs With Potential for Donor-Versus-Recipient NK Cell Allorecognition
| Transplant No. . | Donor . | Recipient . | Donor KIR Ligand Missing in Recipient . |
|---|---|---|---|
| 1 | Group 2 HLA-C | ||
| A24, B62, Cw3* | A24, B35, Cw4 | ||
| 2 | HLA-Bw4 | ||
| A24,B51, Cw− | A1, B35, Cw− | ||
| 3 | HLA-Bw4 | ||
| A3,B44, Cw4 | A2, B60, Cw3 | ||
| 4 | Group 1 HLA-C | ||
| A3, B18, Cw6 | A3, B52, Cw8 | ||
| 5 | Group 1 HLA-C | ||
| A30, B18, Cw6 | A3, B7, Cw7 | ||
| 6 | Group 2 HLA-C | ||
| A3, B45, Cw7 | A30, B14, Cw4 | ||
| 7 | Group 1 HLA-C | ||
| A2, B44, Cw5 | A25, B51, Cw3 | ||
| 8 | Group 1 HLA-C | ||
| A3, B39, Cw4 | A2, B35, Cw3 | ||
| 9 | HLA-Bw4 | ||
| A2,B51, Cw7 | A2, B41, Cw7 | ||
| 10 | Group 2 HLA-C | ||
| A2, B35, Cw3 | A10, B51, Cw5 | ||
| 11 | Group 1 HLA-C | ||
| A74, B18, Cw6 | A33, B14, Cw7 | ||
| 12 | A32, B44, Cw5 | A3, B62, Cw7 | Group 1 HLA-C + HLA-Bw4 |
| 13 | Group 2 HLA-C | ||
| A29, B8, Cw7 | A24, B35, Cw4 | ||
| 14 | Group 1 HLA-C | ||
| A3, B52, Cw2 | A2, B27, Cw1 | ||
| 15 | Group 1 HLA-C | ||
| A−, B35, Cw4 | A24, B18, Cw8 | ||
| 16 | HLA-Bw4 | ||
| A−,B51, Cw7 | A1, B8, Cw7 | ||
| 17 | Group 1 HLA-C | ||
| A31, B13, Cw6 | A11, B52, Cw7 | ||
| 18 | Group 2 HLA-C | ||
| A23, B17, Cw7 | A24, B38, Cw4 | ||
| 19 | Group 1 HLA-C | ||
| A3, B37, Cw6 | A3, B52, Cw3 | ||
| 20 | A11, B51, Cw7 | A3, B35, Cw4 | Group 2 HLA-C + HLA Bw4 |
| Transplant No. . | Donor . | Recipient . | Donor KIR Ligand Missing in Recipient . |
|---|---|---|---|
| 1 | Group 2 HLA-C | ||
| A24, B62, Cw3* | A24, B35, Cw4 | ||
| 2 | HLA-Bw4 | ||
| A24,B51, Cw− | A1, B35, Cw− | ||
| 3 | HLA-Bw4 | ||
| A3,B44, Cw4 | A2, B60, Cw3 | ||
| 4 | Group 1 HLA-C | ||
| A3, B18, Cw6 | A3, B52, Cw8 | ||
| 5 | Group 1 HLA-C | ||
| A30, B18, Cw6 | A3, B7, Cw7 | ||
| 6 | Group 2 HLA-C | ||
| A3, B45, Cw7 | A30, B14, Cw4 | ||
| 7 | Group 1 HLA-C | ||
| A2, B44, Cw5 | A25, B51, Cw3 | ||
| 8 | Group 1 HLA-C | ||
| A3, B39, Cw4 | A2, B35, Cw3 | ||
| 9 | HLA-Bw4 | ||
| A2,B51, Cw7 | A2, B41, Cw7 | ||
| 10 | Group 2 HLA-C | ||
| A2, B35, Cw3 | A10, B51, Cw5 | ||
| 11 | Group 1 HLA-C | ||
| A74, B18, Cw6 | A33, B14, Cw7 | ||
| 12 | A32, B44, Cw5 | A3, B62, Cw7 | Group 1 HLA-C + HLA-Bw4 |
| 13 | Group 2 HLA-C | ||
| A29, B8, Cw7 | A24, B35, Cw4 | ||
| 14 | Group 1 HLA-C | ||
| A3, B52, Cw2 | A2, B27, Cw1 | ||
| 15 | Group 1 HLA-C | ||
| A−, B35, Cw4 | A24, B18, Cw8 | ||
| 16 | HLA-Bw4 | ||
| A−,B51, Cw7 | A1, B8, Cw7 | ||
| 17 | Group 1 HLA-C | ||
| A31, B13, Cw6 | A11, B52, Cw7 | ||
| 18 | Group 2 HLA-C | ||
| A23, B17, Cw7 | A24, B38, Cw4 | ||
| 19 | Group 1 HLA-C | ||
| A3, B37, Cw6 | A3, B52, Cw3 | ||
| 20 | A11, B51, Cw7 | A3, B35, Cw4 | Group 2 HLA-C + HLA Bw4 |
Switched alleles involving possibility of NK allorecognition are indicated in bold.
Frequencies of Donor-Versus-Recipient Alloreactive NK Cell Clones
| Transplant No. . | Before Transplant (isolated from donor) . | 1-3 mo After Transplant (isolated from recipient) . | >4 mo After Transplant (isolated from recipient) . |
|---|---|---|---|
| 1 | 4/24 | 0/120 | ND |
| 2 | 6/11 | 0/25 | ND |
| 3 | 6/19 | 0/37 | ND |
| 4 | 2/14 | 0/25 | ND |
| 5 | 2/12 | 0/22 | ND |
| 6 | 1/19 | 0/44 | ND |
| 7 | 6/10 | 2/11 | 0/48 |
| 8 | 2/32 | 2/14 | 0/35 |
| 9 | 2/50 | 2/50 | 0/52 |
| 10 | 1/40 | 1/42 | 0/55 |
| 11 | ND | 2/44 | 0/60 |
| 12 | 3/26 | 2/16 | ND |
| 13 | 2/10 | 2/38 | ND |
| 14 | 1/40 | 2/40 | ND |
| Transplant No. . | Before Transplant (isolated from donor) . | 1-3 mo After Transplant (isolated from recipient) . | >4 mo After Transplant (isolated from recipient) . |
|---|---|---|---|
| 1 | 4/24 | 0/120 | ND |
| 2 | 6/11 | 0/25 | ND |
| 3 | 6/19 | 0/37 | ND |
| 4 | 2/14 | 0/25 | ND |
| 5 | 2/12 | 0/22 | ND |
| 6 | 1/19 | 0/44 | ND |
| 7 | 6/10 | 2/11 | 0/48 |
| 8 | 2/32 | 2/14 | 0/35 |
| 9 | 2/50 | 2/50 | 0/52 |
| 10 | 1/40 | 1/42 | 0/55 |
| 11 | ND | 2/44 | 0/60 |
| 12 | 3/26 | 2/16 | ND |
| 13 | 2/10 | 2/38 | ND |
| 14 | 1/40 | 2/40 | ND |
Number of alloreactive NK clones over total number of NK clones tested.
Abbreviation: ND, not determined.
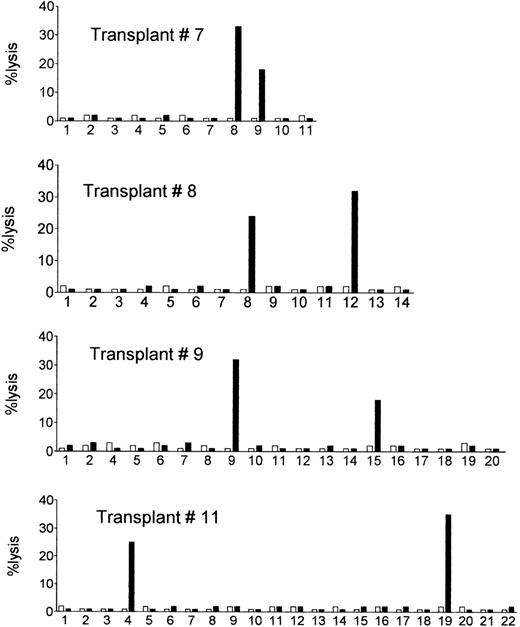
The postgrafting NK cell recovery was very efficient, as normal counts of 200 NK cells per microliter were reached within the first 3 weeks posttransplant. NK cell clones of donor origin (ie, obtained from 100% donor-type blood samples) were obtained from the engrafted recipients at different time points, and were tested for their ability to lyse recipient’s target cells (PHA lymphoblasts or BLCLs obtained from cryopreserved pretransplant lymphocytes). The results are shown in Table 3 (middle and right columns) and in Fig1. Despite the absence of GVHD, we detected high frequencies (ie, in the range detected in the donors) of donor NK clones which killed recipient’s target cells in 8 of the 14 recipients (see box in Table 3) during the first three postengraftment months. Donor-versus-recipient alloreactive clones were detected also in transplant no. 11, which had not been analyzed before transplant. No lysis whatsoever was detected when autologous (donor) cells were used as targets (Fig 1). In patients 1 through 6, donor alloreactive clones were not detected, ie, their frequencies were below 1/22 to 1/120.
Posttransplant NK cell clones of donor origin lyse the allogeneic recipient’s target cells. Donor NK cell clones obtained from the circulation of the engrafted recipients were tested for lysis of the allogeneic recipient’s target cells (PHA lymphoblasts or BLCLs obtained from cryopreserved pretransplant recipient’s lymphocytes) (▪), and of the autologous donor’s cells (□). E:T ratio = 10:1. Shown are cytotoxicity assays with NK cell clones from four representative recipients (see Table 3 for a summary of data).
Posttransplant NK cell clones of donor origin lyse the allogeneic recipient’s target cells. Donor NK cell clones obtained from the circulation of the engrafted recipients were tested for lysis of the allogeneic recipient’s target cells (PHA lymphoblasts or BLCLs obtained from cryopreserved pretransplant recipient’s lymphocytes) (▪), and of the autologous donor’s cells (□). E:T ratio = 10:1. Shown are cytotoxicity assays with NK cell clones from four representative recipients (see Table 3 for a summary of data).
MHC specificity of donor-versus-recipient alloreactive NK cells.
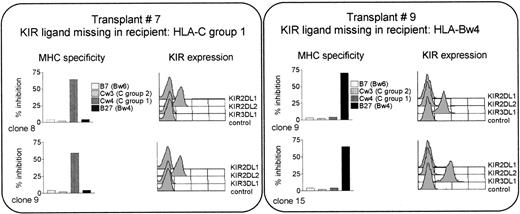
Posttransplant killing of recipient cells might be triggered by nonrecognition of recipient MHC. To test this hypothesis, clones were analyzed for MHC specificity using target cells expressing the HLA-Bw4 allele B27, the group 2 HLA-C allele Cw3, and the group 1 HLA-C allele Cw4. Expression of the corresponding KIR was determined by immunofluorescence. Control targets, not recognized by KIRs, were cells expressing the HLA-Bw6 allele B7 (see refs 3 and 4). As illustrated in Fig 2 for the alloreactive NK clones from transplants 7 and 9 (shown as examples), lysis could only be blocked by target cells expressing the donor allele group that was missing in the recipient. If the missing allele belonged to HLA-C group 1 (as in transplant 7), the postgrafting donor-versus-recipient alloreactive NK clones were only equipped with KIR2DL2, the receptor for HLA-C group 1. Their lysis could be blocked only by cells expressing an HLA-C group 1 allele (such as Cw4), and not by cells expressing allele groups recognized by other KIRs, such as HLA-C group 2, or HLA-Bw4 (and obviously not by cells bearing alleles, such as B7, which are not recognized by KIRs). This phenomenon was observed in all cases analyzed, ie, in transplants no. 7 and 9 (shown in the figure), as well as in transplants 12, 13, and 14 (not shown).
The postgrafting killing of recipient’s cells is mediated by clones expressing a single specificity, for the MHC class I KIR ligand which is missing in the recipient. Shown are the alloreactive NK clones from transplants 7 and 9. KIR expression was analyzed by immunofluorescence and MHC specificity by cytotoxicity assays against cell lines transfected with the indicated class I allotypes. Results are presented as percentage inhibition compared with lysis of the untransfected cells (E:T ratio = 10:1).
The postgrafting killing of recipient’s cells is mediated by clones expressing a single specificity, for the MHC class I KIR ligand which is missing in the recipient. Shown are the alloreactive NK clones from transplants 7 and 9. KIR expression was analyzed by immunofluorescence and MHC specificity by cytotoxicity assays against cell lines transfected with the indicated class I allotypes. Results are presented as percentage inhibition compared with lysis of the untransfected cells (E:T ratio = 10:1).
This analysis was repeated more than 4 months postgrafting (Table 3, right column). Alloreactive NK clones could no longer be detected, indicating that, by this time, all NK cells were able to recognize the recipient MHC.
Antileukemic effect of alloreactive NK cells.
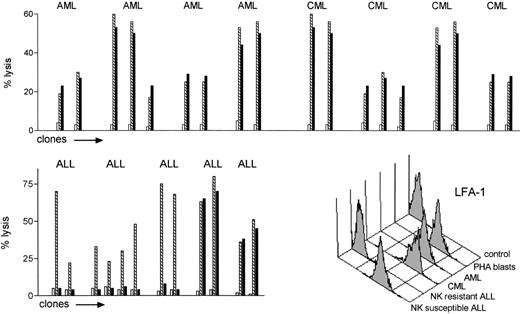
To test the hypothesis that NK allorecognition may serve as a graft-versus-leukemia (GVL) effector, we evaluated the susceptibility to allogeneic NK lysis of a set of leukemic cells and compared it with reference targets (PHA lymphoblasts and BLCLs from the same leukemia patients). NK clones that lysed allogeneic PHA lymphoblasts or BLCLs from leukemia patients were tested for their ability to lyse leukemic cells from the same individuals. Leukemia cells from 4 CML, 4 AML, and 5 ALL (calla+) patients were used as targets. The alloreactive NK clones efficiently killed all the acute and chronic myeloid leukemias (top panel in Fig 3). Only two of five ALLs were killed (bottom left panel Fig 3). Killing of susceptible leukemias followed the rules of NK cell allorecognition, as leukemias were killed by 100% of the alloreactive NK clones (24 of 24, E:T ratio = 10:1), and were not killed by 50 of 50 nonalloreactive NK clones (even at E:T ratio of 50:1). Immunofluorescence analysis of several adhesion molecules showed all NK-resistant ALLs exhibited lower surface expression of lymphocyte function antigen-1 (LFA-1) compared with the NK-susceptible ALLs and with all the CMLs and AMLs (bottom right panel in Fig 3).
Leukemia cells are targets for NK cell alloreactivity. NK cell clones which lysed allogeneic PHA lymphoblasts or BLCLs from leukemia patients () were tested for their ability to lyse pretransplant crypreserved leukemic cells from the same individuals (▪), (□ shows autologous targets). CML, AML, and ALL (calla+) cells were used as targets (E:T ratio = 10:1). The alloreactive NK clones killed all the CML and AML targets (top panel). Only a minority of the ALLs were killed (bottom left panel). None of 50 nonalloreactive NK clones killed any allogeneic leukemia (not shown). Bottom right panel: immunofluorescence analysis LFA-1 expression of allogeneic NK cell susceptible and resistant leukemic targets.
Leukemia cells are targets for NK cell alloreactivity. NK cell clones which lysed allogeneic PHA lymphoblasts or BLCLs from leukemia patients () were tested for their ability to lyse pretransplant crypreserved leukemic cells from the same individuals (▪), (□ shows autologous targets). CML, AML, and ALL (calla+) cells were used as targets (E:T ratio = 10:1). The alloreactive NK clones killed all the CML and AML targets (top panel). Only a minority of the ALLs were killed (bottom left panel). None of 50 nonalloreactive NK clones killed any allogeneic leukemia (not shown). Bottom right panel: immunofluorescence analysis LFA-1 expression of allogeneic NK cell susceptible and resistant leukemic targets.
Transplantation From Donors Failing to Express the Recipient’s KIR Epitopes, ie, With a Potential for NK Cell-Mediated Graft Rejection
In 17 pairs HLA typing indicated the donors failed to express a major KIR ligand (HLA-C group 1, HLA-C group 2, HLA-Bw4) that could engage the inhibitory receptors of all recipient’s NK cells. Analysis of 142 host NK cell clones performed before conditioning in four recipients who afterward accepted the graft showed high frequency (ranging from 1 cell in 2 to 1 cell in 10) antidonor alloreactive NK cell clones, as expected from the HLA typing.
One rejection occurred; immunofluorescence and cell-culture analyses failed to detect NK cells at the end of conditioning or at time of rejection. Hematologic recovery in the 16 engrafted patients was similar to the patients in the previous section. No GVHD cases were observed. Chimerism was 100% donor on all assessments (except at rejection or relapse). Relapses were 1 of 6 AML, 0 of 4 CML, and 4 of 7 ALL.
KIR Epitope-Matched Transplants, ie, With No Potential for NK Cell Alloreactions
Twenty-three donor-recipient pairs were matched for all known KIR ligands, that is, HLA-A3/A11 in addition to HLA-C group 1, HLA-C group 2, HLA-Bw4. Six rejections (2 reversed by a second transplant) occurred. However, (1) none of 208 NK cell clones obtained before conditioning from three recipients who afterward rejected the graft lysed the donor’s target cells; (2) at the end of conditioning, no host NK cells were detected by immunofluorescence, nor could they be cultured even by processing large (100 mL) samples of the recipient’s blood; (3) at the time of rejection, no NK cells could be identified by immunofluorescence nor could they be cultured from the bone marrow or peripheral blood (not shown). Consequently, these tests failed to detect any role for NK cells in rejection in KIR epitope-matched transplants.
Hematologic recovery of the 17 primary engrafted patients was similar to the other patients. GVHD occurred in 3 patients. Chimerism was 100% donor on all assessments (except at rejection or relapse). Relapses were 3 of 11 AML, 0 of 4 CML, and 8 of 8 ALL.
DISCUSSION
This study investigated the role of NK cell alloreactivity in clinical hematopoietic cell transplantation and, specifically, the role of the three major NK allospecificities, ie, those for the KIR epitopes of the HLA-C group 1, HLA-C group 2, and HLA-Bw4 alleles. Rejection and GVHD mediated by these NK specificities could have occurred in 37 of 60 high-risk leukemia patients who underwent mismatched hematopoietic stem cell transplants.
Although KIR epitope incompatibility and direct pretransplant functional analysis indicated the risk of NK cell-mediated graft rejection in 17 donor-recipient pairs, rejection occurred in only one case in this group, indicating the strong conditioning regimen had ablated NK cell ability to reject the graft.
Another group of 20 recipients failed to express one of the donor’s KIR ligands and pretransplant donor NK cell clones readily killed recipient target cells. Consequently, mature NK cells in the graft could have caused GVHD. As we had no case of GVHD, donor-versus-recipient alloreactive NK cells were presumably absent from the postgrafting repertoire. It should be noted that, on repeated bimonthly chimerism assessments, all blood samples from these recipients were 100% donor. Remarkably, in approximately 60% of patients we observed a postgrafting persistence of the NK cell repertoire which killed recipient target cells. Lysis followed the rules of NK cell allorecognition, ie, it was mediated by clones only bearing the KIR for which the recipient MHC was not a ligand.
The speed of the NK cell recovery suggested these donor-versus-recipient alloreactive NK clones had differentiated from the engrafted stem cells. In theory, differentiating NK cells could have been selected by KIR epitopes on donor hematopoietic cells, and on host stromal cells.21 However, the present data indicate the host MHC failed to select a compatible NK cell repertoire. Therefore, as shown in mice,24 hematopoietic cells, rather than stromal cells, appear to play a major role in the selection of the NK cell repertoire. The mechanisms involved in the disappearance of the alloreactive clones require further studies to establish whether they are deleted from the repertoire or whether they survive in a state of anergy as in MHC class I–deficient mice.25
The absence of GVHD despite the donor-versus-recipient NK cell alloreactivity is unexpected. The same biological phenomenon, although in the opposite sense, is found in the hybrid resistance mouse model,26 where the host F1 mouse NK cells reject parental bone marrow cells but tolerate parental skin and organ grafts. Both the transient failure of host tissues to educate the donor NK cell repertoire and the lack of GVHD may be two facets of the same phenomenon, ie, NK cell sensitivity to class I polymorphism is restricted to hematopoietic cells. Tissues may lack the ligands which bind to and/or activate NK cells.
In addressing the GVL potential of donor-versus-recipient NK cell alloreactivity we found that 100% myeloid leukemias, but only a minority of lymphoblastic leukemias, were killed by alloreactive NK clones. Only the NK-resistant ALL cells failed to express LFA-1, an adhesion molecule which is essential for effector-target conjugate formation and/or effector cell activation.27 Myeloid leukemia killing followed the rules of class I recognition by KIR, because it (1) occurred when the corresponding normal blood cells were also killed, (2) was only exerted by alloreactive NK clones, and (3) was related to the missing expression of donor MHC class I KIR ligands on the recipient’s target cells. Instances of leukemia killing or growth inhibition (notably CML) by autologous or allogeneic matched NK cells have been reported.28,29 The mechanisms of these interesting phenomena are unknown and consequently their occurrence is unpredictable. Conversely, killing of KIR epitope-mismatched myeloid leukemias can be predicted by the lysis of corresponding normal blood cells and by specific HLA disparities. Interestingly, strong antitumor effects and lack of,30-32 or even suppression of,33 GVHD have been observed in mice after infusion of allogeneic NK cells that do not recognize the recipient MHC.
Does donor-versus-recipient NK cell alloreactivity have any impact on the postgrafting emergence of susceptible host targets? Obviously, postgrafting detection of alloreactive NK clones may be an underestimate because of the limited number of clones screened, but for the sake of argument we will presume a donor is able to transfer antirecipient NK cell alloreactivity every time the HLA typing predicts it. Potential in vivo targets are myeloid leukemias, as they were susceptible to allogeneic NK killing in vitro, and host lymphocytes mediating rejection, because assessment of allogeneic NK lysis was routinely performed against activated lymphocytes. On the other hand, the ALL in vitro resistance to allogeneic NK killing predicts this leukemia will not be targeted in vivo. No myeloid relapses or graft rejections were observed in the 20 patients transplanted from donors with HLA-based potential for transfer of antirecipient NK cell alloreactivity. To date, the 4 myeloid relapses and the 7 rejections have occurred in the other 40 patients. Relapses of the NK cell–resistant target ALL were distributed equally in the two groups (5 of 20 v 12 of 40). Therefore, the potential for donor-versus-recipient NK cell alloreactivity, as predicted by the HLA disparity, might in itself predict a favorable outcome.
In conclusion, the present study uncovers one biological aspect of mismatched hematopoietic transplantation, ie, the unexpected postgrafting emergence of donor NK cells which do not recognize host alloantigens and which, in the absence of GVHD, kill recipient target cells in accordance with the laws of NK cell alloreactivity. KIR epitope-mismatching in the GVH direction may confer unique potential for GVL effect and for engraftment.
ACKNOWLEDGMENT
We thank Lorenzo Moretta (University of Genova, Genova, Italy) for antibodies and cell lines, Antonella Santucci for statistical analyses, and Geraldine Ann Boyd for assistance in the writing of the manuscript.
Supported by a grant from Associazione Italiana per la Ricerca sul Cancro (AIRC). L.R. and I.V. are supported by fellowships from Fondazione Italiana per la Ricerca sul Cancro (FIRC).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Andrea Velardi, MD, Sezione di Ematologia e Immunologia Clinica, Università di Perugia, Policlinico Monteluce, 06122-Perugia, Italy; e-mail: velardi@unipg.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal