During mouse embryogenesis, macrophage-like cells arise first in the yolk sac and are produced subsequently in the liver. The onset of liver hematopoiesis is associated with the transition from primitive to definitive erythrocyte production. This report addresses the hypothesis that a similar transition in phenotype occurs in myelopoiesis. We have used whole mount in situ hybridization to detect macrophage-specific genes expressed during mouse development. The mouse c-fms mRNA, encoding the receptor for macrophage colony-stimulating factor (CSF-1), was expressed on phagocytic cells in the yolk sac and throughout the embryo before the onset of liver hematopoiesis. Similar cells were detected using the mannose receptor, the complement receptor (CR3), or the Microphthalmia transcription factor (MITF) as mRNA markers. By contrast, other markers including the F4/80 antigen, the macrophage scavenger receptor, the S-100 proteins, S100A8 and S100A9, and the secretory product lysozyme appeared later in development and appeared restricted to only a subset of c-fms–positive cells. Two-color immunolabeling on disaggregated cells confirmed that CR3 and c-fmsproteins are expressed on the same cells. Among the genes appearing later in development was the macrophage-restricted transcription factor, PU.1, which has been shown to be required for normal adult myelopoiesis. Mice with null mutations in PU.1 had normal numbers of c-fms–positive phagocytes at 11.5dpc. PU.1(−/−) embryonic stem cells were able to give rise to macrophage-like cells after cultivation in vitro. The results support previous evidence that yolk sac–derived fetal phagocytes are functionally distinct from those arising in the liver and develop via a different pathway.
THE MONONUCLEAR phagocyte system is defined as a family of cells that arise from hematopoietic progenitors in bone marrow and progress through monoblasts and promonocytes to monocytes.1 At this stage, they enter the circulation and migrate into the tissues to become mature macrophages. The production of monocytes and macrophages in adult mice is controlled by macrophage colony-stimulating factor (M-CSF or CSF-1),2 which acts through a specific plasma membrane receptor encoded by c-fmsproto-oncogene.3,4 Granulocyte-macrophage colony-stimulating factor, interleukin-3 (IL-3), and many other cytokines/lymphokines can regulate monocytopoiesis in vitro or in vivo.5 6
Electron microscopic studies of early embryonic development have identified the first cells with the ultrastructural appearance of tissue macrophages in the yolk sac before they appear in the embryo. Putative phagocytes infiltrate the head and much of the rest of the body at the same time as the first pluripotent hematopoietic progenitor cells can be detected in the aorta-gonad-mesonephros (AGM) and subsequently in the liver7,8; that is, around 10 days postcoitum (dpc). The origin of early embryonic phagocytes in the yolk sac has been demonstrated most convincingly via the use of chick-quail chimeras.9 Yolk sac “macrophages” appear to develop without passing through an obvious monocyte stage, as evidenced by the lack of markers such as peroxidase and the monocyte surface marker, ER-MP20, and continue to proliferate actively once they have infiltrated the embryo.10,11 Within the embryo, macrophage-like cells are associated with the rapid removal of apoptotic cells,12-15 perhaps the most striking example being in the region between the developing digits.16Removal of dying cells by specialized, migratory phagocytes is a function conserved across evolution; mechanisms and receptors identified in Drosophila and Caenorhabditis eleganshave clear parallels in mice. Drosophila hemocytes that remove dying cells, like mammalian phagocytes, express a CD36 family member and scavenger receptors.17-19 The mouse homolog of C elegans cell death gene ced-7 is expressed in fetal phagocytes and is required for removal of dying cells.20Evidence from Drosophila21,22 to mouse12 indicates that cell death precedes macrophage infiltration, and that dying cells can influence migration and differentiation of the macrophage-like cells. Apart from the obvious role in clearance, fetal phagocytes are very closely associated with the developing vasculature23 and may control angiogenesis as they do in adults.
The first documented surface marker for the appearance of macrophage-like cells in the mouse was the F4/80 antigen, an unusual member of the serpentine receptor family of unknown function.24 Immunocytochemical localization of F4/80 showed positive cells in the yolk sac and head around 10.5 dpc and subsequently identified phagocytes in regions of tissue turnover and cell death.13 We subsequently applied the technique of whole mount in situ hybridization to localize c-fms mRNA. Cells with detectable levels of c-fms mRNA appeared simultaneously in the yolk sac and the embryo around 9.5 dpc. Later they were not only associated with presumptive hematopoietic cells in the developing liver,25 but were also observed throughout the body of the developing embryo, with particularly high frequency found in the sites of known tissue turnover including interdigital spaces and the brain. More recently, the β2 integrin, CD11b (Mac-1), was localized in the embryo in a pattern that appears roughly consistent withc-fms.26
The differentiation of myeloid cells is controlled by specific transcription factors. The first exon of the murine and humanc-fms gene that is transcribed in macrophages is flanked by a promoter that contains binding sites for the myeloid-restricted transcription factor PU.1.27-30 We have recently shown that these sites alone constitute a macrophage-specific minimal promoter.31 Similar binding sites are present in promoters of other macrophage-restricted genes, including tartrate resistant acid phosphatase (TRAP), lysozyme M, macrophage scavenger receptor, IL-1β, Fc receptors (FcγRI and FcγRIIIA) and CD11b (integrin adhesion molecule).32-38 Two groups have independently shown that a targeted disruption of the PU.1 gene causes gross reduction in myeloid differentiation.39,40 Although the formation of macrophage colonies from fetal liver progenitor cells was disrupted and there was no evidence of mature cells defined by markers such as the F4/80 antigen in tissues of knockout animals, expression of c-fmsmRNA was still detectable by reverse transcriptase-polymerase chain reaction (RT-PCR) in early embryos.41 In one of these knockouts at least, null mice are able to proceed to birth and morphogenesis is normal.39 The knockout phenotype was difficult to interpret in the absence of studies on PU.1 expression during development. In this report, we present evidence that the onset of hematopoiesis in the liver marks a clear transition in the phenotype and pathway of differentiation of phagocytes in the embryo, and we show that expression of PU.1 is associated with this transition.
MATERIALS AND METHODS
Animals
Naturally mated outbred mice (CD1 or Swiss Quackenbush) were killed by cervical dislocation on days 9.5 to 13.5 of development (day 0.5, morning of vaginal plug). The embryos were delivered by cesarean section, then dissected out of the decidua with the yolk sac and the amnion pulled away from the embryo, but retained attached. In embryos of 10.5 dpc and older, the head and heart were punctured with a 27-gauge needle to prevent the trapping of reagents in the lumen. Viable PU.1(+/−) mice were also naturally mated and killed by cervical dislocation on days 10.0 to 12.5 dpc. The embryos were genotyped by PCR on genomic DNA using one of the embryonic limbs as the source of DNA. The primers used were: PuKO 5′ primer: 5′ GCC CCG GAT GTG CTT CCC TTA TCA AAC C 3′, Pu920 3′ primer: 5′ TGC CTC GGC CCT GGG AAT GTC 3′, neo.1 3′ primer: 5′ CGC ACG GGT GTT GGG TCG TTT TGT TGG G 3′.42
Embryonal stem (ES) cells culture and differentiation.
D3 parent ES cells and PU.1(+/−) and (−/−) lines derived from them were described previously.42 They were cultured in flasks precoated with 0.1% (wt/vol) gelatin (Sigma, St Louis, MO) and seeded with neoR mitotically inactivated embryonic fibroblast feeders (a gift from Dr S. Delaney, CMCB, University of Queensland, Queensland, Australia). ES cells were plated at low density in these flasks in Dulbecco’s modified Eagle’s medium (DMEM) containing high glucose and Na pyruvate 15% fetal calf serum (FCS), 2 mmol/L L-glutamine, 30 μg/mL penicillin, 100 μg/mL streptomycin, 0.1 mmol/L β-mercaptoethanol, and 1 × 103 U leukemia inhibitory factor (LIF) (ESGRO, Amrad, Melbourne)/mL. ES cells were passaged by digestion with 0.25% trypsin (37°C; 3 to 5 minutes). The cells were washed in LIF-containing medium. Cells that were to be differentiated were resuspended in differentiation medium (DMEM supplemented with batch-selected 15% FCS (CSL, Melbourne, Australia), 0.1 mmol/L nonessential amino acids (GIBCO), 0.1 mmol/L β-mercaptoethanol, 5 × 103 U/mL of recombinant human CSF-1 (Chiron Corp, Emeryville, CA), 10 U/mL of human recombinant IL-3 (a gift from Dr A. Hapel, ANU, Canberra, Australia), at the final concentration of 1 × 105 cells/mL of differentiation medium and plated out into 10-cm bacteriological grade culture dishes. Medium was changed every 2 to 3 days as embryoid bodies developed and subsequently macrophage-like cells appeared attached to the surface.43
Latex beads phagocytosis assay.
Latex beads (1.16 μm in diameter) (Sigma) were purchased as 10% (wt/vol) aqueous suspension. They were washed and resuspended in sterile phosphate-buffered saline (PBS) at the same concentration. The beads were added to the cells at a final concentration of 0.01% (wt/vol), and the cells were incubated at 37°C, 5% CO2, and monitored for phagocytosis between 1 and 4 hours. The cells were then washed twice with PBS and fixed in 1% glutaraldehyde (Sigma) for 30 minutes at room temperature, washed twice with PBS, and stored in fresh PBS at 4°C.
In situ hybridization.
The probes were made and labeled using digoxygenin (DIG) RNA labeling mix according to the manufacturer’s instructions (Boehringer Mannheim, Mannheim, Germany), using T7 and T3 polymerases from Promega (Madison, WI), and Sp6 from Boehringer Mannheim. Probes were stored at −70°C until required. The probes used, their sizes, and the enzymes required for linearization are listed in Table 1. Mannose receptor cDNA was obtained from A. Ezekowitz (Harvard University, Boston, MA), scavenger receptor cDNA from M. Krieger (Massachusetts Institute of Technology, Boston, MA), lysozyme cDNA from R. Renkawitz (Max Planck Institute, Martinsried, Germany), and CP10 and MRP14 (S100A8 and S100A9) from C. Geczy (University of NSW, Sydney, Australia). Vectors cut withPstI were end-filled using Klenow polymerase at 12°C.
Plasmids and Probes Used for In Situ Hybridization
| Probe . | Plasmid . | Size (bp) . | Fragment . | |
|---|---|---|---|---|
| Antisense . | Sense . | |||
| c-fms | pGEM2 | ∼800 | NdeI or HindIII | EcoRI |
| MMR | pManR431 | 431 | AvaI | HincII |
| MSR | pJA18 | 1,444 | HindIII | XbaI |
| PU.1 | pOK PU.1S | 1,100 | — | XhoI |
| pOK PU.1A | XhoI | — | ||
| S100A8 | pCP 14 | 300 | EcoRI | — |
| Lysozyme | pBS | 300 | PstI | EcoRI |
| S100A9 | pGEM | 300 | XbaI | PstI |
| F4/80 | pBS F4/80 | 630 | KpnI | SpeI |
| Probe . | Plasmid . | Size (bp) . | Fragment . | |
|---|---|---|---|---|
| Antisense . | Sense . | |||
| c-fms | pGEM2 | ∼800 | NdeI or HindIII | EcoRI |
| MMR | pManR431 | 431 | AvaI | HincII |
| MSR | pJA18 | 1,444 | HindIII | XbaI |
| PU.1 | pOK PU.1S | 1,100 | — | XhoI |
| pOK PU.1A | XhoI | — | ||
| S100A8 | pCP 14 | 300 | EcoRI | — |
| Lysozyme | pBS | 300 | PstI | EcoRI |
| S100A9 | pGEM | 300 | XbaI | PstI |
| F4/80 | pBS F4/80 | 630 | KpnI | SpeI |
The whole-mount in situ hybridization was performed as described previously.25 Embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4°C and permeabilized with proteinase K (10 μg/mL) for 15 minutes at 9.5 dpc and 5 minutes more for each extra day of development. Embryos were prehybridized for at least 2 hours at 65°C and then hybridized overnight using 0.5 μg/mL of labeled probe. Posthybridization washes were also performed at 65°C. Washed embryos were blocked using 10% sheep serum, 2% bovine serum albumin (BSA) in TBTX (Tris-HCl pH 7.5, 150 mmol/L NaCl, and 0.1% Triton X-100) for at least 3 hours and then incubated overnight with anti-DIG F(ab′)2 fragments conjugated to alkaline phosphatase (Boehringer Mannheim). Labeled probes were detected using a colorimetric method using NBT (4-nitroblue tetrazolium chloride) and BCIP (5-bromo-4-chloro-3-indolyl-phosphate) (Boehringer Mannheim). As a negative control, the embryos were incubated with the corresponding sense probe. In none of the cases shown was there any specific signal obtained with the sense probe.
Histology
The stained embryos were refixed in 4% PFA, cleared in xylene, and embedded in paraffin wax. A total of 7- to 8-μm sections were floated in a waterbath onto slides, dried, dewaxed, and mounted after counterstaining with neutral red or hematoxylin as required. Unstained slides were viewed with Nomarski optics and the stained sections with bright field on an Olympus microscope (AX70).
Immunocytochemistry
ES cell-derived macrophages were fixed in 4% PFA/PBS (Sigma) for 30 minutes at room temperature. The cells were then washed in PBS and incubated with ice-cold methanol containing 0.5% (vol/vol) H2O2 to quench any endogenous peroxidase activity. Nonspecific protein binding was blocked by incubating the cells with 10% FCS in PBS for 10 minutes at room temperature. The primary rat antimouse F4/80 antibody (as rat hybridoma supernatant) was diluted 1:100 in PBS containing 10% FCS and then added to the cells for 60 minutes at room temperature. The primary antibody was detected with a secondary antirat horseradish peroxidase-linked antibody for 60 minutes at room temperature. A solution of 0.5 mg diaminobenzidine (DAB) substrate/mL (Sigma) supplemented with 1 μL/mL of 30% H2O2 was added to detect the antibody for a maximum of 10 minutes. Cells were monitored during this time for the emergence of an orange-brown stain, which indicated the presence of the surface antigen F4/80.
Photography
Photographs of whole mounts were taken on a Leica dissecting microscope with cold lamp side illumination using Kodak T64 film. Photographs of sections were taken on an Olympus AX70 microscope using Kodak daylight film under Nomarski optics (unstained) or bright field (stained sections), and finally the photographs of cells in culture were taken on an inverted Olympus microscope using Kodak T160 film and Nomarski optics.
Flow Cytometry
Embryos were disaggregated using a modified method of Yoder et al.44 Briefly, the yolk sacs (10 to 13.5 dpc), embryos (10.0 and 11.5 dpc), or fetal livers (10.5 to 12.5 dpc) were dissected free and washed in PBS. They were then drawn through an 18G (yolk sacs 11.5 to 13.5 dpc) or 23-gauge needle and transferred to a petri dish and incubated with 0.1% collagenase D; 0.2% dispase (Boehringer) in PBS/20%FCS for 60 minutes at 37°C. Dispersed cells were drawn through a 23-gauge needle into a syringe, pelleted by centrifugation, washed in PBS, and counted. If required, the cells were cultured in medium containing 5 × 103 Cetus U/mL of recombinant human CSF-1 (Chiron Corp) in bacteriological petri dishes.45
The cells were stained as described previously46 except that 5% rat serum was used for blocking rather than purified IgG. Anti–c-fms monoclonal antibody was used as 1/100 dilution of hybridoma supernatant,47 commercial reagents were goat antirat FITC F(ab′)2 fragments (Serotec, Oxford, UK), anti-F4/80 PE/Cy5 antibody (Serotec), and anti-Mac-I PE antibody (Caltag, Burlingame, CA). Note that the three rat antibodies are each IgG2B and effectively act as isotype controls for each other. Analyses were performed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA), and samples were analyzed using the associated CellQuest software.
RESULTS
Location of c-fms–Positive Cells by Whole-Mount In Situ Hybridization Shows a Large Population of Phagocytes in the Embryo
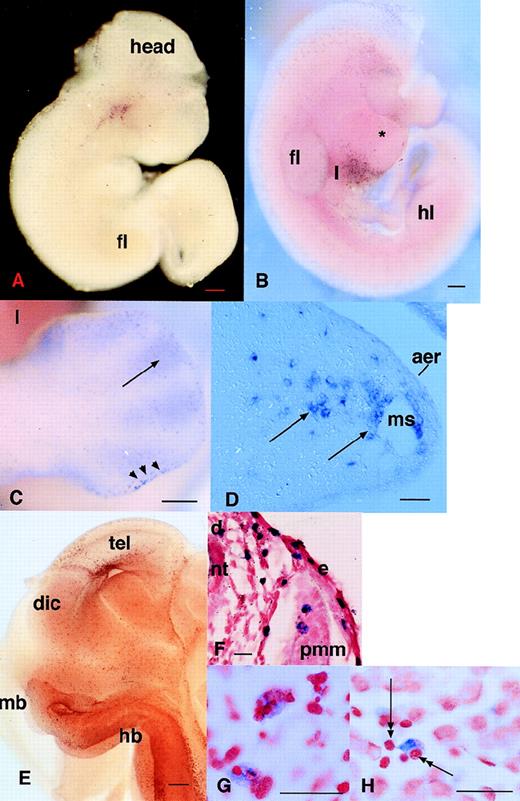
The detection of c-fms–positive cells in the developing mouse embryo by in situ hybridization was reported previously.25Although it was inferred, no independent marker or phenotypic characteristic confirmed the identity of c-fms–positive cells as phagocytes. As a baseline for the examination of other putative macrophage markers, we performed a more extensive analysis of the appearance of this marker. Cells expressing c-fms mRNA were detected first in the yolk sac at 9.5 dpc as isolated individual cells rather than clusters associated with blood islands, which develop in the yolk sac at 8.0 to 8.5 dpc. Among numerous embryos examined, we have never observed any in which the first appearance ofc-fms–positive cells in the head of the embryo is clearly delayed relative to the yolk sac. Assuming the cells are first formed in the yolk sac, the infiltration into the embryo must occur almost immediately. Figure 1A shows an embryo at 10 dpc, in which positive cells are evident in the head. In some embryos at this time, c-fms–positive cells are particularly concentrated in a band overlying the developing heart (not shown; but see a similar pattern with the MITF gene in Fig 4F). By 10.5 dpc, c-fms–positive cells were first detected in the liver (Fig 1B), consistent with the appearance of macrophage-like cells defined by other criteria.13 Macrophages have been shown previously to be actively involved in removing apoptotic cells to form a foot with separate digits.16 20 To assess whetherc-fms mRNA was associated with fetal phagocytes, we concentrated particularly on the distribution in this site. The infiltration of the limb buds by c-fms–positive cells was actually evident well before the onset of cell death between the digits, at 10.5 dpc, when they accumulate in the apical ectodermal ridge (AER). At 12.5 dpc, c-fms–positive cells still delineated the AER and they also accumulated in the anterior “necrotic” zone (Fig 1C). When the cell death commences between the digits, c-fms provides a clear marker for phagocytic cells that infiltrate the interdigital region from the marginal sinus (Fig1D). At higher magnification, many c-fms–positive cells in this site contain inclusions of ingested pyknotic nuclei (not shown).
Localization of cells expressing c-fmsmRNA in the embryo. In all panels, c-fms mRNA was localized by whole mount in situ hybridization as described in Materials and Methods. The blue-purple reaction product indicates sites of expression of the c-fms mRNA. In panels F, G, and H, the stained embryos were embedded, sectioned, and counterstained with neutral red. Bar in panels A through C and E represents 250 μm; in panels D, F through H, 20 μm; heart (*), forelimb (fl), hindlimb (hl), neural tube (nt). (A) Embryo 10.5 dpc. Localization of c-fms mRNA shows a very extensive speckled pattern throughout the embryo. Each spot is a single-labeled cell. Labeled cells are particularly concentrated in the head, around the branchial arches, and along the dorsal midline, although this is not evident in a single focal plane. (B) Embryo 11 dpc. The head of the embryo was removed before staining. At a slightly more advanced stage than (A), the rapid appearance ofc-fms–labeled cells is evident in the liver (l). The fine speckled pattern of labeled cells is more prominent throughout the embryo than in (A), but is less readily demonstrated in a single focal plane. (C) Hindlimb of 12.5 dpc embryo showing accumulation ofc-fms–positive cells at the apical ectodermal ridge (aer) (arrowheads) and infiltration between the digits (arrows). (D) A section through the limb bud of 12.5 dpc embryo showing the positive cells leaving the marginal sinus (ms) and infiltrating the mesenchyme. (E) Whole mount of the head of a hemisected 12.5-dpc embryo. Numerousc-fms–positive cells are lining the ventricular surfaces of the brain, including telencephalon (tel), diencephalon (dic), midbrain (mb), and hindbrain (hb). (F) Section through the body wall of an 11.5-dpc embryo. Numerous c-fms–positive cells accumulate particularly along the dorsal midline (d). In this field, they can be seen to be closely associated with the vascular spaces adjacent to the neural tube, but are also infiltrated within the premuscle mass (pmm) and the epidermis (e). (G) High power view of c-fms–positive cells from a section of brain at 11.5 dpc shows clear evidence that many labeled cells have engulfed dying cells and contain pyknotic nuclei stained intensely with neutral red. (H) High-power view ofc-fms–positive cell present in the liver shows stellate morphology and intimate association with smaller hematopoietic cells (double arrows).
Localization of cells expressing c-fmsmRNA in the embryo. In all panels, c-fms mRNA was localized by whole mount in situ hybridization as described in Materials and Methods. The blue-purple reaction product indicates sites of expression of the c-fms mRNA. In panels F, G, and H, the stained embryos were embedded, sectioned, and counterstained with neutral red. Bar in panels A through C and E represents 250 μm; in panels D, F through H, 20 μm; heart (*), forelimb (fl), hindlimb (hl), neural tube (nt). (A) Embryo 10.5 dpc. Localization of c-fms mRNA shows a very extensive speckled pattern throughout the embryo. Each spot is a single-labeled cell. Labeled cells are particularly concentrated in the head, around the branchial arches, and along the dorsal midline, although this is not evident in a single focal plane. (B) Embryo 11 dpc. The head of the embryo was removed before staining. At a slightly more advanced stage than (A), the rapid appearance ofc-fms–labeled cells is evident in the liver (l). The fine speckled pattern of labeled cells is more prominent throughout the embryo than in (A), but is less readily demonstrated in a single focal plane. (C) Hindlimb of 12.5 dpc embryo showing accumulation ofc-fms–positive cells at the apical ectodermal ridge (aer) (arrowheads) and infiltration between the digits (arrows). (D) A section through the limb bud of 12.5 dpc embryo showing the positive cells leaving the marginal sinus (ms) and infiltrating the mesenchyme. (E) Whole mount of the head of a hemisected 12.5-dpc embryo. Numerousc-fms–positive cells are lining the ventricular surfaces of the brain, including telencephalon (tel), diencephalon (dic), midbrain (mb), and hindbrain (hb). (F) Section through the body wall of an 11.5-dpc embryo. Numerous c-fms–positive cells accumulate particularly along the dorsal midline (d). In this field, they can be seen to be closely associated with the vascular spaces adjacent to the neural tube, but are also infiltrated within the premuscle mass (pmm) and the epidermis (e). (G) High power view of c-fms–positive cells from a section of brain at 11.5 dpc shows clear evidence that many labeled cells have engulfed dying cells and contain pyknotic nuclei stained intensely with neutral red. (H) High-power view ofc-fms–positive cell present in the liver shows stellate morphology and intimate association with smaller hematopoietic cells (double arrows).
By 11.5 to 12.5 dpc, the degree of penetration of the probe in whole mount in situ hybridization starts to become limiting. To improve the penetration, some embryos were cut midsagitally before hybridization. This approach allowed the visualization of c-fms–positive cells lining all of the ventricular surfaces in the brain (Fig 1E). Sectioning of the uncut whole mounts showed the association ofc-fms–positive cells with developing neuronal cells throughout the body, including developing ganglia along the body wall (Fig 1F), the developing eye, and along the dorsal midline in close apposition with the neural tube (not shown). The pattern of expression ofc-fms in the vicinity of the dorsal midline is reminiscent of migrating neural crest cells identified with markers such asc-kit and trp-2,48 raising the possibility that at least some of the c-fms–positive cells could be melanocytes. However, the distribution of these cells was not affected by the dominant mi/mi mutation in the microphthalmic mouse, which prevents migration of c-kit and trp-2–positive melanocytes (K.M. and D.A.H., unpublished results).
To confirm further the likely identity of c-fms–positive cells and phagocytes, we analyzed serial sections of the whole mounts. As in the footpad, throughout the body c-fms–labeled cells were stellate in appearance and either within vessels or very closely associated with them, indicating their likely origin in the vasculature (Fig 1D and F). In most sections examined, many labeled cells were clearly involved in active phagocytosis and contained ingested pyknotic nuclei that stained intensely with neutral red (Fig 1G). In the liver, macrophages are known to be associated with hematopoietic islands.13 Consistent with restriction of c-fms to the macrophage lineage, most stellate c-fms–positive cells in this organ were associated with clusters of hematopoietic cells that are clearly distinct from parenchymal cells (Fig 1H).
The c-fms–positive cells that were detected in this study were more numerous and widespread than the cells that were previously reported to express F4/80 antigen.13 Previous immunocytochemical localization of F4/8011 13 (D.A.H., unpublished results) detected small numbers of positive cells in the yolk sac around 9.5 dpc and the first positive cells in the liver associated with the onset of erythropoiesis in that site at 10.5 dpc. We were first able to detect F4/80 mRNA in the liver at 11.5 dpc, but the level of expression remained low and did not contrast well with background staining due to the long exposure times required to detect the signal (not shown). On this basis, the expression of detectable F4/80 mRNA appears to be a relatively late marker for monocytopoiesis associated with the onset of hematopoiesis in the liver. An internal positive control confirming the relatively low expression of F4/80 mRNA was the detection of significant numbers of strongly positive maternal macrophage in the placenta (not shown).
c-FMS Protein Is Coexpressed on Isolated Embryonic Cells With Other Myeloid Markers, Mac-1 (CR3) and F4/80
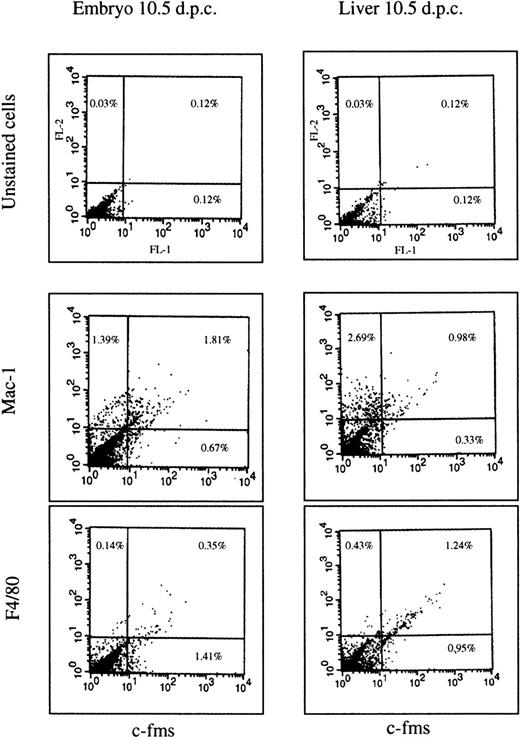
Whole-mount in situ hybridization is difficult to adapt to simultaneous localization of more than one marker on a dispersed population of cells. As an independent way to confirm that the protein product ofc-fms mRNA is expressed and that it is coexpressed with other myeloid markers, we digested embryos with collagenase and dispase and examined the expression of surface markers by flow cytometry. Cells were isolated from the embryonic liver and the remainder of the embryo at 10.5 dpc and analyzed by double-labeling with an anti–c-fmsmonoclonal antibody in combination with either anti-F4/80 or anti-Mac-1 (which recognizes the type III complement receptor, CR3). At 10.5 dpc, in the liver (Fig 2), just over 50% of thec-fms–positive cells also expressed detectable F4/80, and more than 60% coexpressed Mac-1. In nonhepatic tissues, the proportion ofc-fms–labeled cells (1.5% to 2.0% of the total) was greater than in the liver as expected from the mRNA localization. Consistent with the localization of the F4/80 mRNA, less than 20% coexpressed detectable F4/80 antigen, and even in these, the level of expression was very low. By contrast, more than 70% coexpressed high levels of Mac-1. Compared with the remainder of the embryo, the liver was enriched for a population of cells that expressed Mac-1, but notc-fms, which probably reflects early stages of granulocyte differentiation. In cells isolated from both the liver and embryo, the percentage of cells coexpressing c-fms with F4/80 or Mac-1 was increased by overnight culture in CSF-1 containing medium (data not shown). This observation could be due to cell growth and/or reversal of the partial loss of these surface markers during enzymatic digestion and isolation. For this reason, the analysis cannot be viewed as quantitative and serves primarily to prove that the majority ofc-fms–positive cells also express other myeloid cell markers.
FACS profiles of embryonic cells. The cells were isolated from the liver of 10.5 dpc embryo or from the rest of the body and stained by immunofluorescence for each of the antibody markers shown as described in Materials and Methods. Three antibodies are the same rat IgG2b isotype. No staining above the autofluorescence background was observed with an irrelevant antibody of the same isotype. The unstained cells in the upper two panels were stained with second antibody alone. Numbers on each panel represent the percentage of the total cell population falling within the quadrant indicated. A total of 104 cells was counted in each experiment. The overall pattern is representative of two independent cell isolations from a litter of pooled embryos at 10.5 dpc.
FACS profiles of embryonic cells. The cells were isolated from the liver of 10.5 dpc embryo or from the rest of the body and stained by immunofluorescence for each of the antibody markers shown as described in Materials and Methods. Three antibodies are the same rat IgG2b isotype. No staining above the autofluorescence background was observed with an irrelevant antibody of the same isotype. The unstained cells in the upper two panels were stained with second antibody alone. Numbers on each panel represent the percentage of the total cell population falling within the quadrant indicated. A total of 104 cells was counted in each experiment. The overall pattern is representative of two independent cell isolations from a litter of pooled embryos at 10.5 dpc.
Mannose Receptor mRNA Colocalizes With c-fms, Whereas the Scavenger Receptor mRNA Is Restricted to a Subset of Phagocytes
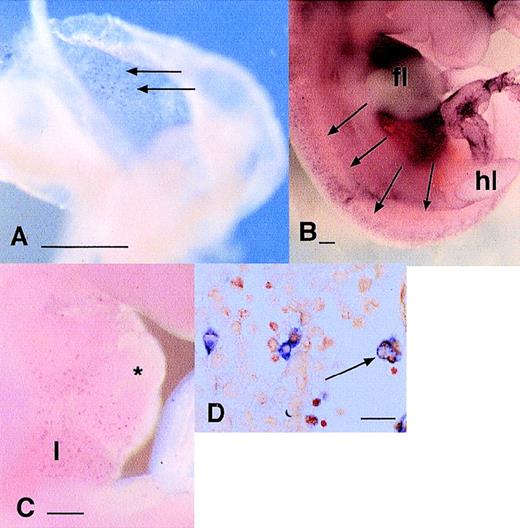
The coexpression of Mac-1 antigen (CR3) with c-FMS antigen shows that at least some of the embryonic phagocytes have endocytic receptors in common with adult macrophages. To extend knowledge of the endocytic capacity of embryonic phagocytes, we localized the macrophage mannose receptor (MMR)49 and macrophage scavenger receptor (MSR)50 in embryos from 9.5 to 13.5 dpc. At all stages examined, MMR-positive cells were as abundant in the yolk sac and embryo as c-fms–positive cells and the perivascular locations and cellular morphologies of labeled cells were indistinguishable either at the gross level (Fig 3A and B) or on examination of sections (not shown). By contrast, MSR mRNA expression was much more restricted. Positive cells were detected in the heart, liver (Fig 3C), limb buds, and all over the body, but the numbers were clearly less than observed using c-fms or MMR probes and the expression appeared restricted to larger cells, more closely resembling the abundance of F4/80. Examination of sections of these embryos confirmed a distribution and morphology of MSR-positive cells consistent with restriction to larger c-fms–positive cells that are actively involved in phagocytosis of pyknotic cells (Fig3D; compare with Fig 1G and H).
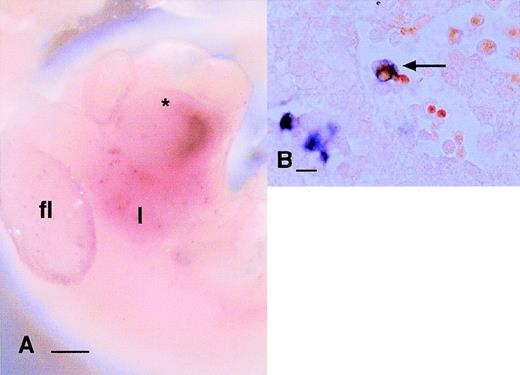
Localization of cells expressing mRNAs encoding endocytic receptors in the embryo. (A) Localization of MMR mRNA at 9.5 dpc. Individual labeled cells appear in a diffuse speckled pattern (arrows) throughout the yolk sac. (B) Localization of MMR mRNA in 11.5 dpc embryo. MMR-positive cells are distributed very similarly toc-fms–positive cells (Fig 1A) at the same time point as diffuse speckled pattern throughout the embryo. In this focal plane, the particular concentration of MMR-positive cells along the dorsal midline flanking the neural tube is evident. The darkened appearance of the liver, immediately below the forelimb bud (fl) is due to numerous MMR-positive cells, which are out of focus in this plane (hl, hindlimb bud). (C) Localization of the MSR mRNA at 10.5 dpc. The first labeled cells can be detected in the liver (l) at this time, immediately below the heart (*). At this time point, few MSR-positive cells could be detected elsewhere in the embryo. (D) The section through the liver of an 11.5-dpc embryo stained for MSR mRNA. Some positive cells are associated with aggregates of hematopoietic cells (to the left of field), but many are clearly filled with engulfed cells (arrow). Most MSR-positive cells elsewhere in this embryo are large and contain neutral red-positive inclusions (not shown). Bar in panels A through C represents 250 μm and in panel D, 20 μm.
Localization of cells expressing mRNAs encoding endocytic receptors in the embryo. (A) Localization of MMR mRNA at 9.5 dpc. Individual labeled cells appear in a diffuse speckled pattern (arrows) throughout the yolk sac. (B) Localization of MMR mRNA in 11.5 dpc embryo. MMR-positive cells are distributed very similarly toc-fms–positive cells (Fig 1A) at the same time point as diffuse speckled pattern throughout the embryo. In this focal plane, the particular concentration of MMR-positive cells along the dorsal midline flanking the neural tube is evident. The darkened appearance of the liver, immediately below the forelimb bud (fl) is due to numerous MMR-positive cells, which are out of focus in this plane (hl, hindlimb bud). (C) Localization of the MSR mRNA at 10.5 dpc. The first labeled cells can be detected in the liver (l) at this time, immediately below the heart (*). At this time point, few MSR-positive cells could be detected elsewhere in the embryo. (D) The section through the liver of an 11.5-dpc embryo stained for MSR mRNA. Some positive cells are associated with aggregates of hematopoietic cells (to the left of field), but many are clearly filled with engulfed cells (arrow). Most MSR-positive cells elsewhere in this embryo are large and contain neutral red-positive inclusions (not shown). Bar in panels A through C represents 250 μm and in panel D, 20 μm.
PU.1 mRNA Is Not Expressed at Detectable Levels in All Embryonic Phagocytes
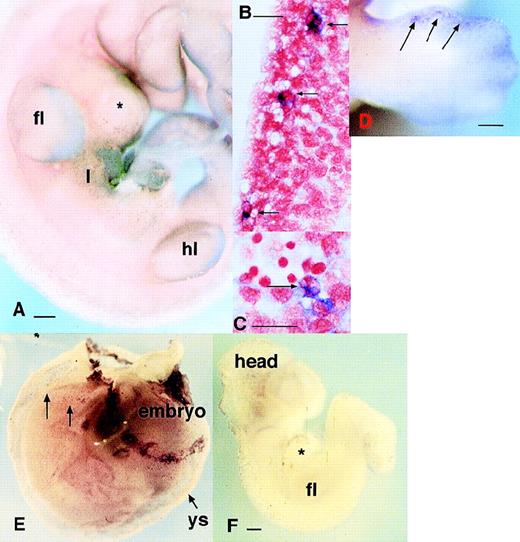
As noted earlier in this report, the Ets family of transcription factors is implicated in myeloid differentiation. Clearly, if PU.1 controls the differentiation of fetal phagocytes, it should be expressed before c-fms and other myeloid markers. Whole-mount in situ hybridization failed to detect any PU.1-positive cells at 9.5 dpc in the yolk sac or embryo (not shown) where c-fms is readily detected (Fig 1A). In fact, cells expressing PU.1 mRNA were not detectable until the onset of liver hematopoiesis (Fig 4A and B). It must be emphasized that the absence of detectable PU.1 mRNA in the earlier embryo does not indicate total absence and is clearly constrained by the sensitivity of the method. In all experiments, embryos are incubated in the staining reagents until any signal detected becomes maximal, or the background “nonspecific” staining becomes evident. A number of cells detected in the liver at this time were clearly phagocytic, suggesting that the populations of PU.1 and c-fms–positive cells do overlap, but c-fms is more widespread (Fig 4C). Apart from the absence of detectable PU.1 in the yolk sac, PU.1 mRNA was detectable in considerably fewer cells in the brain thanc-fms, although the signal intensity was comparable where it was expressed (not shown). Another site in which it was clear that PU.1 was not detectable in all c-fms–expressing cells was the limb. The first detectable PU.1-positive cells in the distal part of the limbs appeared at 12.5 dpc (c-fms cells were detectable there from 10.5 dpc) and remained at the margins (Fig 4D) at the time whenc-fms–positive phagocytes had already invaded the interdigital spaces (see Fig 1C). This pattern of PU.1 expression in the limb buds was very similar to that of MSR, which also accumulates at the limb bud margins comparatively late at 12.5 dpc. In summary, PU.1 mRNA is not expressed at detectable levels in all c-fms–positive cells, and the pattern of expression is consistent with restriction to a subset of cells arising later in development.
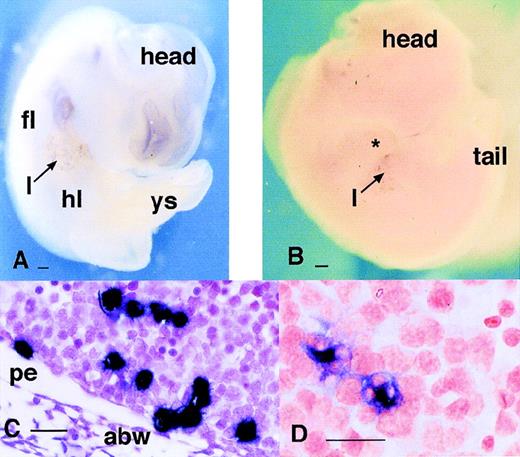
Localization of cells expressing MITF and PU.1 mRNAs in the embryo. (A) Localization of cells expressing PU.1 mRNA in the 11.5-dpc embryo. PU.1-positive cells are particularly concentrated in the liver (l). They can be detected as a fine speckled pattern throughout the embryo, especially flanking the neural tube at the bottom of the field. (B) Localization of PU.1-positive cells in a sagittal section of 12.5 dpc embryo confirming that each focus of stain represents a single labeled cell, in this case located at intervals along the dorsal midline in mesenchyme surrounding the spinal cord. (C) A section through the same embryo as in (B) at higher magnification. Each of the cells expressing PU.1 mRNA in this region contains multiple foci of neutral red staining indicating phagocytosis of pyknotic cells. (D) Localization of cells expressing PU.1 mRNA in an embryo at 12.5 dpc showing accumulation of the positive cells at the limb margins. Note the distinction from the pattern obtained with c-fms in Fig 1D. (E) Localization of MITF mRNA in the embryo. This panel shows an embryo still surrounded by the yolk sac at 9.5 dpc. The yolk sac contains numerous scattered positive cells. The band of positive staining flanking the yolk sac represents expression of MITF in trophoblasts. At higher magnification, the pattern is indistinguishable from MMR (3A) orc-fms.25 (F) At 10 dpc, mi-positive cells spread from the brain throughout the body. Bar in panels A, D, and F represents 250 μm, and in panels B and C, 20 μm.
Localization of cells expressing MITF and PU.1 mRNAs in the embryo. (A) Localization of cells expressing PU.1 mRNA in the 11.5-dpc embryo. PU.1-positive cells are particularly concentrated in the liver (l). They can be detected as a fine speckled pattern throughout the embryo, especially flanking the neural tube at the bottom of the field. (B) Localization of PU.1-positive cells in a sagittal section of 12.5 dpc embryo confirming that each focus of stain represents a single labeled cell, in this case located at intervals along the dorsal midline in mesenchyme surrounding the spinal cord. (C) A section through the same embryo as in (B) at higher magnification. Each of the cells expressing PU.1 mRNA in this region contains multiple foci of neutral red staining indicating phagocytosis of pyknotic cells. (D) Localization of cells expressing PU.1 mRNA in an embryo at 12.5 dpc showing accumulation of the positive cells at the limb margins. Note the distinction from the pattern obtained with c-fms in Fig 1D. (E) Localization of MITF mRNA in the embryo. This panel shows an embryo still surrounded by the yolk sac at 9.5 dpc. The yolk sac contains numerous scattered positive cells. The band of positive staining flanking the yolk sac represents expression of MITF in trophoblasts. At higher magnification, the pattern is indistinguishable from MMR (3A) orc-fms.25 (F) At 10 dpc, mi-positive cells spread from the brain throughout the body. Bar in panels A, D, and F represents 250 μm, and in panels B and C, 20 μm.
MITF Is an Early Phagocyte Marker
Of the markers examined thus far, only c-fms, MMR, and CR3 appear to be expressed on early phagocytes. Presumably, the expression of these genes is controlled by transcription factors expressed specifically in these cells. Other than PU.1, there are few transcription factors known to be restricted to the macrophage lineage. Other studies in our laboratory have implicated factors in the basic helix-loop-helix-ZIP family in transcriptional regulation of macrophage-specific genes, including c-fms. Among this family, mutations in the MITF have been shown to cause osteopetrosis, a phenotype also associated with deficiency in CSF-1. Like the PU.1 transcription factor, MITF has been shown to be expressed in adult macrophages and osteoclasts.51 We, therefore, investigated whether MITF mRNA is also expressed in the embryo. Figure 4E and F shows that MITF mRNA, like c-fms mRNA, was detectable on numerous cells in yolk sac and at the same time in the head and in the characteristic band of cells above the early developing heart. The location, morphology, and apparent phagocytic activity of the labeled cells was consistent with identity or substantial overlap of the MITF and c-fms–expressing populations.
Identification of Additional Markers Associated With the Onset of Hematopoiesis in the Embryonic Liver
The secretory product, lysozyme, is widely expressed in macrophages in adult mice. In the embryo, lysozyme mRNA was not detectable in the yolk sac or embryo until 10.5 dpc, when its expression was restricted to sparse large cells in the liver (Fig 5A). From 11.5 dpc, lysozyme-positive cells were present outside the liver, but were not as abundant as even MSR-positive cells. Sections of the stained embryos indicated heterogeneous levels of expression of the gene, with strongly and weakly positive cells. The former were generally larger cells, examples were observed in greatest concentration in the pericardial wall, the peritoneal cavity, and also in the sinusoids of the liver, and many were clearly actively involved in phagocytosis of dying cells (Fig 5B).
Localization of cells expressing lysozyme mRNA in the embryo. (A) Panel shows a 10.5-dpc embryo in which lysozyme mRNA has been detected by whole mount in situ hybridization. The majority of labeled cells at this stage of development can be seen in the liver (l); the band observed in the heart is due to nonspecific trapping. A small number of cells can be seen in this focal plane in the forelimb bud, but few cells are detected elsewhere in the embryo in any focal plane. (B) A section through the liver of one of the embryos stained in (A) demonstrates that the majority of labeled cells have inclusions that stain with neutral red (arrow) and are evidently involved in phagocytosis of dying cells. Bar in (A) represents 250 μm and in (B), 20 μm.
Localization of cells expressing lysozyme mRNA in the embryo. (A) Panel shows a 10.5-dpc embryo in which lysozyme mRNA has been detected by whole mount in situ hybridization. The majority of labeled cells at this stage of development can be seen in the liver (l); the band observed in the heart is due to nonspecific trapping. A small number of cells can be seen in this focal plane in the forelimb bud, but few cells are detected elsewhere in the embryo in any focal plane. (B) A section through the liver of one of the embryos stained in (A) demonstrates that the majority of labeled cells have inclusions that stain with neutral red (arrow) and are evidently involved in phagocytosis of dying cells. Bar in (A) represents 250 μm and in (B), 20 μm.
Two members of the S-100 gene family, S100A8 and S100A9, have been identified as markers of an early stage of differentiation in bone marrow macrophage differentiation in vitro. The initial report of cloning of these genes in mice (then referred to as MRP8 and MRP14) reported the expression of both mRNAs in fetal liver by RT-PCR52 so we considered them also as possible markers for later stages of myeloid development. As with lysozyme, neither of these S-100 genes was detectable anywhere in the embryo or yolk sac until 11.5 dpc (Fig 6) when the first positive cells appeared; at 11.5 dpc, the level of expression in individual cells in the liver was so high that the staining was evident to the naked eye. Sections of the 11.5-dpc liver showed that the dense foci of stain observed in the whole mounts with S100A8 reflected aggregates of small intensely staining cells (Fig 6C). Conversely, staining for S100A9 mRNA, which was clearly less intense in whole mounts, appeared in sections to be restricted to phagocytic cells resembling those expressing lysozyme or MSR mRNA. Without double-labeling, it is not possible to make a definitive conclusion, but it would appear that the expression of the two genes is not coincident. The lack of S100A8 or S100A9-positive cells outside the liver, at all embryonic stages analyzed, suggests that monocytopoiesis in the liver resembles that in bone marrow.53 54 These two S-100 markers therefore provide the clearest distinction between monocytopoiesis in the liver and production of phagocytes from the yolk sac.
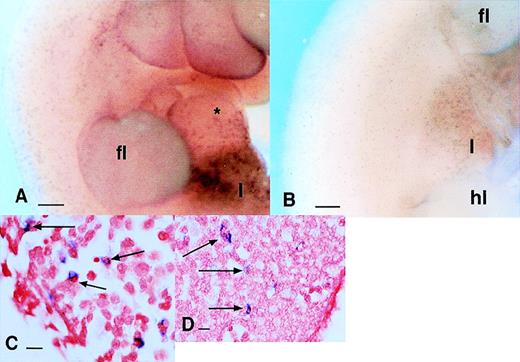
Expression of S-100 proteins in the embryo. (A and B) Localization of mRNA encoding the S100 proteins MRP8 (A) and MRP14 (B) showing that both are expressed only in the liver of 11.5 dpc embryo. (C and D) Sections through the liver of embryos probed for MRP8 (C) and MRP14 (D) expression. Panel (C) is counterstained with hematoxylin. Note the difference between the two markers. In (C), MRP8-positive cells are small and clustered, whereas in (D), cells expressing MRP14 mRNA appear fewer in number, larger, and often contain inclusions suggesting active involvement in phagocytosis. Bar in (A) and (B) represents 250 μm and in (C) and (D), 20 μm; yolk sac (ys), abdominal wall (abw), and peritoneum (pe).
Expression of S-100 proteins in the embryo. (A and B) Localization of mRNA encoding the S100 proteins MRP8 (A) and MRP14 (B) showing that both are expressed only in the liver of 11.5 dpc embryo. (C and D) Sections through the liver of embryos probed for MRP8 (C) and MRP14 (D) expression. Panel (C) is counterstained with hematoxylin. Note the difference between the two markers. In (C), MRP8-positive cells are small and clustered, whereas in (D), cells expressing MRP14 mRNA appear fewer in number, larger, and often contain inclusions suggesting active involvement in phagocytosis. Bar in (A) and (B) represents 250 μm and in (C) and (D), 20 μm; yolk sac (ys), abdominal wall (abw), and peritoneum (pe).
The Presence of Phagocytes in PU.1(−/−) Embryos
The expression of c-fms mRNA has been detected previously in PU.1(−/−)39,41 mice despite the observation that fetal liver cells from these animals are severely compromised in their ability to form colonies in CSF-1 and lack expression of more mature macrophage markers such as F4/80.55 We analyzed the knockout embryos by whole mount in situ hybridization usingc-fms as a probe at 10.5 dpc. In keeping with the absence of apparent overlap between PU.1 and c-fms expression at this time (see above), there was no effect of the null mutation on the number or location of c-fms–positive cells anywhere in the embryo at this time (Fig 7A and B). Thec-fms–positive cells were present in the same areas as in the wild-type mice: liver, heart, along the dorsal midline, limbs, brain, branchial arches, and under the skin. In sections of the embryos, the morphology and location of the labeled cells was also unaltered by the null mutation (Fig 7C and D).
Localization of c-fms mRNA in PU.1 knockout animals. (A and B) Comparison of c-fms mRNA localization in wild-type (A) or PU.1(−/−) (B) 11.5 dpc littermates. Note the speckled pattern throughout the embryo representing individualc-fms–positive cells, particularly concentrated in the liver (l). The apparent difference in intensity of staining of individual cells is not significant and is due to the focal plane and lighting. (C and D) Saggital sections through a PU.1(−/−) embryo stained as in (B), showing that cells expressing c-fms in the liver and limb buds have similar morphology and location to those in wild-type mice (see Fig 1). Bar in (A) and (B) represents 250 μm and in (C) and (D), 20 μm.
Localization of c-fms mRNA in PU.1 knockout animals. (A and B) Comparison of c-fms mRNA localization in wild-type (A) or PU.1(−/−) (B) 11.5 dpc littermates. Note the speckled pattern throughout the embryo representing individualc-fms–positive cells, particularly concentrated in the liver (l). The apparent difference in intensity of staining of individual cells is not significant and is due to the focal plane and lighting. (C and D) Saggital sections through a PU.1(−/−) embryo stained as in (B), showing that cells expressing c-fms in the liver and limb buds have similar morphology and location to those in wild-type mice (see Fig 1). Bar in (A) and (B) represents 250 μm and in (C) and (D), 20 μm.
Differentiation of PU.1(−/−) ES Cells
Totipotent embryonic stem cells can form cystic embryoid bodies and partly recapitulate normal fetal hematopoiesis in appropriate culture conditions. Previous studies demonstrated that production of macrophages from PU.1 null ES cells in the presence of pokeweed mitogen spleen-conditioned medium (a source of multiple different colony-stimulating factors) was grossly deficient compared with heterozygous or wild-type parent ES cells.42 We described an optimized method for differentiating wild-type ES cells into macrophages in liquid culture using recombinant CSF-1 and IL-3.43 We therefore examined whether this approach would permit PU.1(−/−) ES cells to give rise to macrophage-like cells. The analysis was performed on two independent clones: D3.9 and D3.15.42 Differentiation of the two clones was performed for 31 days and the adherent cells deposited by embryoid bodies were assayed by immunohistochemistry with F4/80 antibody and phagocytosis of latex beads. In both knockout clones, the production of cystic embryoid bodies was delayed by up to a week relative to the parent ES cells (Fig 8A), but the numbers of clones examined is clearly not sufficient to allow determination of whether this was due to the cloning/transfections of the ES lines or is a consequence of the null mutation. In both of the ES cell lines, the embryoid bodies eventually produced significant numbers of cells that adhered to the bacteriological plastic dish and spread with a polar morphology that was indistinguishable from the cells produced in much larger numbers by the heterozygous or wild-type cell (not shown). The cells produced from PU.1 null clones resembled macrophages morphologically and were able to ingest latex particles (Fig 8B). Immunoperoxidase staining detected F4/80 antigen on the adherent cells from null ES cells (Fig 8C), but the expression was much weaker than in wild-type or heterozygous cells, and only some cells expressed the marker.
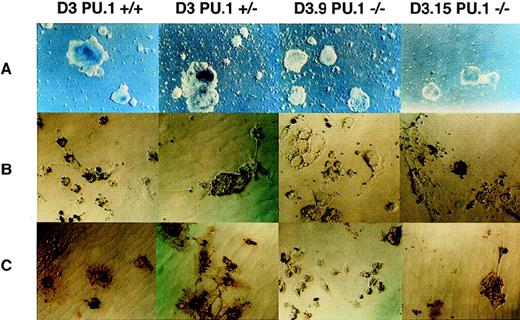
Differentiation of PU.1(−/−) ES cells. ES cells were cultivated under conditions optimized for production of macrophages as described in Materials and Methods. (A) Comparison of the development of embryoid bodies at day 8 of differentiation. Note that the two PU.1(−/−) ES cells have generated smaller embryoid bodies than either the (+/+) or (+/−) lines at this stage of differentiation. (B) After 31 days of differentiation, each of the lines of ES cells produced adherent cells with morphology resembling macrophages. The yield of macrophage-like cells was considerably lower at this time point in the (−/−) cells. The cells were harvested, replated, and incubated with latex beads as described in Materials and Methods. Panel shows adherent cells from each culture as indicated; black staining represents the uptake of latex beads. (C) An independent experiment to (B). In this case, adherent cells generated from differentiated ES cells were incubated first with latex beads, then fixed, and stained using an indirect immunoperoxidase method for expression of F4/80 antigen. In the (+/+) and (+/−) cultures, the brown reaction product demonstrating positive staining for F4/80 antigen is clearly evident on all phagocytic cells. In the two (−/−) lines, staining can be seen particularly in larger cells (eg, in the panel showing D3.15 cells), but was generally not sufficiently strong to permit photographic reproduction. Original magnification for (A) was 4× and for (B) and (C), 20×.
Differentiation of PU.1(−/−) ES cells. ES cells were cultivated under conditions optimized for production of macrophages as described in Materials and Methods. (A) Comparison of the development of embryoid bodies at day 8 of differentiation. Note that the two PU.1(−/−) ES cells have generated smaller embryoid bodies than either the (+/+) or (+/−) lines at this stage of differentiation. (B) After 31 days of differentiation, each of the lines of ES cells produced adherent cells with morphology resembling macrophages. The yield of macrophage-like cells was considerably lower at this time point in the (−/−) cells. The cells were harvested, replated, and incubated with latex beads as described in Materials and Methods. Panel shows adherent cells from each culture as indicated; black staining represents the uptake of latex beads. (C) An independent experiment to (B). In this case, adherent cells generated from differentiated ES cells were incubated first with latex beads, then fixed, and stained using an indirect immunoperoxidase method for expression of F4/80 antigen. In the (+/+) and (+/−) cultures, the brown reaction product demonstrating positive staining for F4/80 antigen is clearly evident on all phagocytic cells. In the two (−/−) lines, staining can be seen particularly in larger cells (eg, in the panel showing D3.15 cells), but was generally not sufficiently strong to permit photographic reproduction. Original magnification for (A) was 4× and for (B) and (C), 20×.
DISCUSSION
The Expression of c-fms Is a Marker for Early Fetal Phagocytes
The first part of this study extends an earlier examination of the distribution of the mature macrophage marker, c-fms, in mouse development. We have presented evidence that c-fms–positive cells probably coexpress other myeloid markers, the MMR, and the MITF transcription factor. Each of the mRNAs was detected in cells that are concentrated in the same areas of the embryo both at a gross anatomical level and in sections, resemble each other morphologically, and are clearly involved in the phagocytosis of dying cells. The FACS analysis of cells isolated from 10.5-dpc embryos (Fig 2) showed that the majority of c-fms–positive cells also expressed Mac-1 antigen (CD11b/CR3), further indicating their likely identity and suggesting that the type III complement receptor is another early marker expressed by the majority of these cells. Recent immunocytochemical study on Mac-1 expression in the embryo confirms the presence of Mac-1–positive cells in the yolk sac and liver from as early as 9.5 dpc.26Similarly, MMR-positive cells have also been identified in 9.0 dpc yolk sacs.56 Some of the MMR expression we have observed could be attributed to selected endothelial cells,56 but we have seen no evidence of expression by these cells in sections. The classical endothelial cell marker, flk-1, gives the clear reticular pattern of expression expected from its association with a capillary network. The pattern is quite distinct from the “speckled” pattern of MMR mRNA localization in whole mount in situ hybridization of embryos at the same age and in sections the capillary endothelial cells labeled with flk-1 are morphologically distinct and located differently from the perivascular phagocytes labeled with MMR orc-fms (P. Koopman, personal communication, November 1998). Hence, we feel that few, if any, of the cells expressing detectable MMR mRNA at 9.5 to 11.5 dpc are endothelial cells. This conclusion does not exclude the possibility of expression of MMR by vascular cells at later stages of development.
Figure 1E and F, combined with the immunofluorescence data on isolated cells, indicates the very large numbers of these phagocytes in the embryo. The proportion of cells expressing c-FMS–antigen in disaggregated 10.5 dpc embryos, approximately 2% on FACS analysis (Fig2), is certainly an underestimate, and the numbers increase substantially in the ensuing 2 days. Based on counting cells in sections, we estimate that 5% to 10% of total cells in the embryo at 11.5 to 12.5 dpc are c-fms–expressing phagocytes; the proportion is likely to be higher in some areas such as the brain (Fig1E). The PU.1-independent early mouse phagocytes are so numerous that it is difficult to believe that their function in development is redundant or dispensible.
The Onset of Myelopoiesis in Liver Correlates With Altered Expression of Macrophage-Specific Genes
In contrast to c-fms, MMR, MITF, and CR3, several markers characteristic of adult macrophages, were only detected once hematopoiesis was established in the liver. The mRNA encoding the S-100 proteins, S100A8 and S100A9, was shown to be transiently expressed in presumptive myeloid progenitor cells only in the liver, providing a striking marker of this transition. F4/80 antigen was apparently another late marker. Previous descriptions of embryonic mononuclear phagocytes used F4/80 antibody.13 The gene was cloned only recently and encodes an integral membrane protein of unknown function.24 Both FACS analysis (Fig 2) and whole mount in situ hybridization indicated that c-fms, MMR, and Mac-1 expression precedes the appearance of F4/80, and that the F4/80 antigen is probably absent, or at very low levels, on yolk sac–derived phagocytes. The patterns of expression of two other late markers, the secretory product lysozyme and MSR (Figs 3 and 5) were broadly similar to that of F4/80, although both mRNAs appeared particularly evident on large cells involved in active phagocytosis. The function of MSR in embryonic phagocytosis appears to be conserved during evolution, as the phagocytes in Drosophila also express a form of scavenger receptor.18,19 This function is not indispensable because scavenger receptor null mice do not have developmental abnormalities.57,58 Association of lysozyme expression with phagocytosis is compatible with its apparent function in adult mice59 and the low expression of lysozyme in the fetal macrophages has also been reported by others.60
PU.1 Transcription Factor Is a Late Marker and Is Not Required for Embryonic Phagocyte Production
The PU.1 null mutation in mice had no discernible effect on the number or distribution of c-fms–positive phagocytes at 11.5 dpc (Fig7). Olson et al41 used semiquantitative RT-PCR to examine gene expression in an independent line of PU.1 null animals and found not only c-fms, but also CD11b and CD18 (which make up Mac-1 antigen, shown to be coexpressed with c-fms in our study) and GM-CSF receptor were unaltered at this early stage of development. Hence, although there are some differences in the hematopoietic phenotypes and viability of the two published lines of PU.1(−/−) mice,39-41 both probably retain the phagocytes we have defined. We have not performed marker studies later in development, but the morphogenic processes that appear to involve fetal phagocytes occur normally in the PU.1(−/−) null embryos. For example, the digits are formed normally, and in histological sections at 12.5 to 13.0 dpc, phagocytes can be seen between the digits internalizing pyknotic nuclei (not shown). The lack of effect of the PU.1 null mutation on early embryonic phagocyte production is consistent with the distribution of PU.1 mRNA, which was not detected until the onset of liver hematopoiesis and even thereafter, in locations such as the footpad, was detected in only a subset of cells expressing c-fms and other early markers. Our conclusion that PU.1 is not required for embryonic phagocyte production is not incompatible with published evidence that it is absolutely required for “definitive” myelopoiesis. In fact, there is general agreement that the ability of the fetal liver to produce monocyte-macrophage progenitor cells and to respond to hematopoietic growth factors such as G-CSF, GM-CSF, and CSF-1 is almost completely compromised by the PU.1 null mutation.61-64 These studies involve in vitro colony assays, and the analysis of embryos later in development (>14.5 dpc) when PU.1 is normally expressed at readily detectable levels in the liver.
Consistent with the view that embryonic phagocytes can develop without PU.1, we produced adherent phagocytic cells from PU.1(−/−) ES cells. Olson et al41 also produced some adherent cells in cultures containing a mix of colony-stimulating factors, but dismissed their identity as macrophages. Our system for differentiation of ES cells, using recombinant IL-3 and CSF-1 rather than a mixed conditioned medium, is biased towards macrophage differentiation, but there was still a clear reduction in the number of adherent cells produced in null cells compared with heterozygous or wild-type cells. It is important to recognize that the ES cell differentiation system proceeds via the formation of IL3/CSF-1–responsive colony-forming units (CFU).39,41 The two independent PU.1 null mutations in mice have been shown to disrupt the appearance of such (CFU) from the yolk sac and liver,39,41 60-64 so the reduced yield of cells seen when PU.1 null ES cells are differentiated in vitro probably reflects a defect in production of the committed macrophage progenitors. The in situ hybridization studies, particularly of S100A8 and S100A9, imply that early phagocytes differentiate via a separate pathway (see above), and our ES cell differentiation system has not been developed specifically for their production. If the conditions could be optimized for embryonic as opposed to “definitive” phagocyte production, we would predict that production of phagocytes would be unaffected by the PU.1 null mutation.
Transcription of Macrophage-Specific Genes Is Not Absolutely PU.1-Dependent
The presence of c-fms–expressing phagocytes in the PU.1 (−/−) mice indicates that PU.1 cannot be absolutely required for transcription of the c-fms gene itself. By contrast, deKoter et al64 reported thatc-fms expression was greatly reduced in myeloid progenitor cells isolated from 14.5 dpc PU.1(−/−) fetal livers, and that retroviral transduction of c-fms partly rescued the ability of the cells to proliferate, but not to differentiate, in response to CSF-1. The apparent reduction in c-fms expression in these studies was measured by PCR in purified lin−progenitor cells. It remains possible that the investigators could have detected normal expression of c-fms in the lin+population of fetal liver cells from PU.1(−/−) animals, as demonstrated from our observations. Furthermore, the possibility that the PU.1 null mutation alters the distribution of c-fmsexpressing cells between lin+ and lin−populations was not excluded. The low c-fms expression and CSF-1 unresponsiveness of the lin− cells in these studies might be an indirect consequence of lack of expression of the GM-CSFR. We and others have shown that optimal proliferation of murine macrophage progenitors requires cooperation between GM-CSF and CSF-1.65 Hence, the published data do not demonstrate unequivocally that PU.1 directly controls c-fms transcription even in adult hematopoiesis. Both the mouse and human c-fmspromoters have multiple functional binding sites for this factor in the proximal promoter,28,30,31,66 but neither promoter is absolutely PU.1-dependent, and both can be activated by other members of the Ets transcription factor family such as Ets-2.31Additionally, both human67 and mouse (J. Pollard, personal communication, July 1997) c-fms genes are expressed in trophoblasts from separate promoters that might also be used by fetal phagocytes.
MITF emerges from the current study as an alternative transcription factor that could contribute to regulation of the c-fmspromoter. Our demonstration that MITF is coexpressed with c-fmsis consistent with reports of others. We find (Rehli M., D.A.H., unpublished results) that mouse macrophages, like those of the rat,51 use upstream promoter(s) of the MITF gene to produce a longer form of the protein than is expressed in melanocytes, and we have obtained evidence that an MITF expression plasmid can transactivate the murine and human c-fms promoters. One argument against a function for MITF in macrophages is that the microphthalmia mutation (mi/mi) has no effect on c-fmsexpression68 or on its distribution in the embryo (not shown), but clearly does effect melanocytes (the animals are albino). The difference may lie in the fact that macrophages also express all three of the other members of the MITF family, TFE3, TFEC, and TFEB; in fact, TFEC is macrophage-restricted in its expression and shares many proximal promoter elements with c-fms.69Additionally, there are still other candidate regulators. For example, the murine (manuscript submitted) and human70c-fms promoters contain essential conserved motifs that bind members of the C/EBP transcription factor family, which might also contribute to expression of c-fms before induction of PU.1.
What Happens to PU.1-Independent Phagocytes?
Both lines of PU.1(−/−) mice lack mature macrophages as evidenced by localization of mature macrophage markers such as F4/80,39-41,61-64 but embryonic development and organogenesis occurs relatively normally. Based on the data presented here, we hypothesize that PU.1-independent phagocytes are retained throughout development and carry out their function normally in the null mice. If that view is correct, PU.1-independent macrophages could be retained in the adult. In fact, there is no published evidence that PU.1 is expressed in all adult tissue macrophages. The PU.1-independent macrophages could be the same as the so-called CSF-1–independent macrophages that develop in the op/op mouse.71 If they retain their ability to proliferate locally within tissues,72-75 they could represent a pool of precursors of resident tissue macrophages in the adult and therefore be quite distinct from the inflammatory macrophages derived from circulating blood monocytes in the normal steady state. Recent data using marrow transplanted from mice with an integrated lacZ reporter gene supports the view that some macrophage pools are very slowly infiltrated by cells derived from the blood.76 Such a proposal clearly undermines the basic concept of the mononuclear phagocyte system, namely that tissue macrophages derive from blood monocytes, which in turn come from marrow progenitors,1 but it might provide some additional insight into the origins of macrophage heterogeneity.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Professor David A. Hume, PhD, Department of Microbiology, University of Queensland, Queensland 4072, Australia; e-mail: D.Hume@cmcb.uq.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal