Abstract
T-cell–rich B-cell lymphoma (TCRBCL) belongs to the group of diffuse large cell lymphomas (DLL). It is characterized by a small number of tumor B cells among a major population of nonmalignant polyclonal T cells. To identify the developmental stage of the tumor progenitor cells, we micromanipulated the putative neoplastic large CD20+ cells from TCRBCLs and amplified and sequenced immunoglobulin (Ig) V gene rearrangements from individual cells. In six cases, clonal Ig heavy, as well as light chain, gene rearrangements were amplified from the isolated B cells. All six cases harbored somatically mutated V gene rearrangements with an average mutation frequency of 15.5% for heavy (VH) and 5.9% for light (VL) chains and intraclonal diversity based on somatic mutation. These findings identify germinal center (GC) B cells as the precursors of the transformed B cells in TCRBCL. The study also exemplifies various means how Ig gene rearrangements can be modified by GC B cells or their malignant counterparts in TCRBCL: In one case, the tumor precursor may have switched from κ to λ light chain expression after acquiring a crippling mutation within the initially functional κ light chain gene. In another case, the tumor cells harbor two in-frame VH gene rearrangements, one of which was rendered nonfunctional by somatic mutation. Either the tumor cell precursor entered the GC with two potentially functional in-frame rearrangements or the second VHDHJHrearrangement occurred in the GC after the initial in-frame rearrangement was inactivated by somatic mutation. Finally, in each of the six cases, at least one cell contained two (or more) copies of a clonal Ig gene rearrangement with sequence variations between these copies. The presence of sequence variants for V region genes within single B cells has so far not been observed in any other normal or transformed B lymphocyte. Fluorescence in situ hybridization (FISH) points to a generalized polyploidy of the tumor cells.
IN THE REVISED European-American Lymphoma (REAL) Classification, T-cell–rich B-cell lymphoma (TCRBCL) belongs to the diffuse large cell lymphomas (DLL).1 TCRBCL is characterized by few putative malignant B cells dispersed as single cells in an infiltrate consisting mainly of T cells, histiocytes, and plasma cells. The frequency of T cells can vary from 50% to more than 90% of cells in the tissue. The histology is usually diffuse, but some cases may also contain nodular areas.2-4 The size of the tumor B cells can be variable ranging from centroblast-like to lymphocytic and histiocytic (L&H)-like cells, as they are found in lymphocyte predominant Hodgkin’s disease (LP HD).5 The tumor cells are CD20+ and CD15- and CD30-.6 In a few cases, they harbor the Epstein-Barr virus (EBV).7 8 Besides the large neoplastic B cells, varying amounts of small B lymphocytes (up to 50% of all cells) are found in the tumor tissue.
TCRBCLs were first recognized as a separate entity by Ramsey et al4 in 1988, demonstrating polyclonal T-cell receptor gene (TCR) rearrangements by Southern Blot analysis and clonal B-cell populations by monotypic immunoglobulin (Ig) light chain expression in five lymphomas (previously diagnosed as peripheral T-cell lymphomas (PTCL)) with T-cell contents of more than 90%. By applying immunohistochemistry and analysis of Ig and TCR gene rearrangements, also other groups identified TCRBCLs among cases diagnosed previously as PTCLs.6,7,9 10
Furthermore, the distinction of TCRBCL from the diffuse form of LP HD can be difficult,11,12 but is important for adequate therapy, as TCRBCL is much more aggressive than LP HD.13Useful markers may be epithelial membrane antigen (EMA) and CD80, for which the L&H cells in some cases of LP HD, but not the large neoplastic B cells in TCRBCL, are positive.6,14,15 In addition, in LP HD, significant numbers of CD57-positive T cells, which can form rosettes around the L&H cells, and follicular dendritic cells (FDCs) are found, whereas in TCRBCL, FDCs are absent and CD57+ T cells are rare.11 16
The differentiation stage of B cells can be studied by analysis of rearranged V region genes. Naive IgM+IgD+ B cells carry unmutated Ig gene rearrangements. In a germinal center (GC) reaction, antigen-specific B cells clonally expand and diversify their rearranged V genes by somatic hypermutation.17-19 Thus, somatic mutations in rearranged V genes are found in GC B cells and their descendents, ie, memory B cells and plasma cells. On this basis, B-cell lymphoma precursors have been assigned to distinct stages of normal B-cell differentiation (see Klein et al18 for review).
We recently analyzed V gene rearrangements in five subtypes of DLL, including one case of TCRBCL, using DNA isolated from paraffin-embedded tissues.20 Tumors of all five subtypes harbored mutated V region genes and are thus derived from antigen experienced B cells. However, as the template DNA was extracted from whole tissue sections and polymerase chain reaction (PCR) products were directly sequenced, we could not analyze the DLLs for intraclonal diversity, which may allow a discrimination between a GC or memory B-cell derivation of the tumor cells in TCRBCL. Here, we characterize the differentiation stage of TCRBCL precursors by the analysis of Ig gene rearrangements in single large CD20+ cells isolated by micromanipulation from seven cases of TCRBCL.
MATERIALS AND METHODS
Tissues and clinical data.
Clinical data for the patients are given in Table 1. All biopsies were performed for diagnostic purposes.
Immunostaining and EBV-encoded RNA (EBER) in situ hybridization.
Immunohistochemistry with antibodies against CD20, CD15, CD30, CD3 (DAKO, Hamburg, Germany), and CD57 (Becton Dickinson, Heidelberg, Germany) was performed with the avidin-biotin-complex technique, alkaline phosphatase and Fast Red as substrate. The anti-FDC antibody DR53 was a gift from Dr G. Delsol (Toulouse, France). EBER in situ hybridization was performed with paraffin-embedded tissue sections as described.21 The EBER probes were kindly provided by Dr G. Niedobitek (Erlangen, Germany)22 and digoxigenin-labeled using the DIG RNA Labeling kit (Boehringer Mannheim, Mannheim, Germany).
Micromanipulation of single cells.
For micromanipulation, 5 to 10-μm sections of frozen biopsies were stained with anti-CD20 or anti-CD3 antibody and alkaline phosphatase/Fast Red. Single cells were mobilized and aspirated with micropipettes and the help of a hydraulic micromanipulator under a microscope as described.23 In cases 1 to 4, L&H-like cells not surrounded by smaller CD20+ cells were isolated. In cases 5 to 7, the micromanipulated centroblast-like cells were often surrounded by other CD20+ cells. Aspirated cells were transferred to PCR tubes containing 20 μL of Expand High Fidelity PCR buffer (Boehringer Mannheim) and stored at −20°C.
Single cell PCR.
Rearranged VH, Vκ, and Vλ genes were amplified in a seminested approach, using family-specific primers for the VH leader region and primers binding to sequences within framework region (FR) I of human VH, Vκ, and Vλ genes together with two sets of nested primers for the joining (J) domains of each locus, as previously described (the VHFRI 1 and 6 primers were not used in this study).23-25 The primers used in the first round of PCR for the amplification of Vλ rearrangements were: Vλ1 5′-GGTCCTGGGCCCAGTCTGTG-3′, Vλ2 5′-CAGTCTGCCCTGACTCAGCCT-3′, Vλ3a 5′-CTCAGCCACCCTCAGTGTCCGT-3′, Vλ3b 5′-CTCAGCCACCCTCGGTGTCAGT-3′, Vλ4 5′-TTTCTTCTGAGCTGACTCAGGAC-3′, Vλ6 5′-GAGTCTCCGGGGAAGACGGTA-3′, Vλ7 5′-GTGGTGACTCAGGAGCCCTCAC-3′, Vλ8 5′-ACTGTGGTGACCCAGGAGCCA-3′, Vλ9 5′-CCTGTGCTGACTCAGCCACCT-3′, Jλ1 5′-GCCACTTACCTAGGACGGTGAC-3′, Jλ23 5′-GAAGAGACTCACCTAGGACGGTC-3′ and Jλ67 5′-GGAGACT(C/T)ACCGAGGACGGTC-3′. The same Vλ primers and the following nested Jλ primers were used in the second round of amplification: Jλ1i 5′-GGACGGTGACCTTGGTCCCAGT-3′, Jλ237i 5′-GACGGTCAGCTTGGT(G/C)CCTCC-3′ and Jλ6i 5′-GACGGTCACCTTGGTGCCACT-3′. Cells were digested with 0.25 mg/mL Proteinase K (Boehringer Mannheim) in 20 μL PCR buffer for 1 hour at 50°C followed by a 10-minute incubation at 95°C. The first round of amplification consisted of a 2-minute denaturation step at 95°C, 4 minutes at 65°C, and 45 seconds at 72°C followed by 34 cycles of 30 seconds at 95°C, 30 seconds at 61°C, and 45 seconds at 72°C. The first round was performed in 1x Expand buffer (Boehringer Mannheim) with 2 mmol/L MgCl2, 200 μmol/L of each deoxynucleoside triphosphate (dNTP), 0.04 μmol/L of each primer and 1 U of Expand enzyme mix (Boehringer Mannheim). One-microliter aliquots of the first round were used in the second amplification round, where a separate PCR was performed for each V gene family with the family-specific leader or FRI primer and a nested J primer set. After a 2-minute denaturation at 95°C and 4 minutes at 65°C (VHFRI 2 and 5, Vκ 1-6) or 67°C (VHL 1-5, Vλ 1-4, and 6-9) or 69°C (VHFRI 3 and 4) and 45 seconds at 72°C, 39 cycles were performed at 95°C for 30 seconds, 61°C (VHFRI 2 and 5, Vκ 1-6) or 63°C (VHL 1-5, Vλ 1-4, and 6-9) or 65°C (VHFRI 3 and 4) for 30 seconds and 72°C for 45 seconds, in 1x PCR buffer (Boehringer Mannheim) with 1.5 (VHL 1 and 5, VHFRI 2-5, Vλ 1-4, and 6-9) or 2.0 mmol/L MgCl2 (VHL 2-4, Vκ 1-6), 200 μmol/L each dNTP, 0.125 μmol/L of each primer with the exception of the JH primer set, which was used at a concentration of 0.031 μmol/L and 0.7 U of Taq DNA polymerase (Boehringer Mannheim).
To test whether the L&H-like cells contain several alleles of rearranged Ig loci, 12 cells from case 4 were each digested in 60 μL PCR buffer with 0.25 mg/mL Proteinase K and after extensive mixing split into three 20-μL aliquots. After 10 minutes at 95°C, a standard seminested PCR was performed for each aliquot.
Analysis of PCR products.
After agarose gel electrophoresis, PCR products were excised from the gels, the DNA extracted with the QiaExII gel extraction kit (Qiagen, Hilden, Germany) and directly sequenced using the dRhodamine or BigDye Terminator cycle sequencing kits and an automated sequencer (ABI377, Applied Biosystems, Weiterstadt, Germany). PCR products were sequenced from both sides. Sequences were compared with the EMBL IMGT database (http://www.genetik.uni-koeln.de/dnaplot/) and the GenBank data library (release 98) using the GeneWorks software (Intelligenetics, Oxford, UK).
Fluorescence in situ hybridization (FISH).
Cytospin slides of isolated nuclei were prepared by mechanic disaggregation of cyropreserved tumor tissue and subsequent pepsin digest as described recently.26 For chromosomes 7, 12, X and Y, the indirectly-labeled centromeric probes D7Z1 (biotin-labeled) and D12Z3 (digoxigenin-labeled) (Oncor, Heidelberg, Germany) and the directly-labeled probes CEPX and CEPY (Vysis, Stuttgart, Germany) were used, respectively. Denaturation, hybridization, detection of indirectly-labeled probes, and counterstaining was performed as described.26 For each cytospin slide two probes were used for hybridization (D7Z1 together with D12Z3 and CEPX together with CEPY). Hybridization signals were analyzed with a fluorescence microscope (Zeiss, Jena, Germany) and documented using the ISIS imaging system (MetaSystems, Altrussheim, Germany).
RESULTS
Micromanipulation of single cells from TCRBCLs.
Single cells were micromanipulated from CD20-stained frozen tissue sections of seven cases of TCRBCL (20 to 63 cells per patient, Tables 1 and2). Criteria for selection of cells were CD20 expression and size. In cases 1 to 4, the large micromanipulated cells resembled L&H cells with few small CD20+ cells in the background. In cases 5, 6, and 7, centroblast-like CD20+cells were micromanipulated (Table 1). Here, the numbers of small CD20+ cells in the background were higher than in cases 1 to 4, and L&H-like cells were not seen. In three cases, two micromanipulation experiments were performed (Table 2). Aliquots of the buffer covering the sections, and in case 2 also CD3+ cells from an adjacent anti-CD3–stained section, were aspirated as negative controls.
Case Description of Patients With T-Cell–Rich B-Cell Lymphoma
| Patient . | Age/Sex . | Presentation . | Biopsy Site . | CD20+ Cells Analyzed . |
|---|---|---|---|---|
| 1 | 65/M | First | Axillary LN | L&H-like |
| 2 | 32/F | First | Axillary LN | L&H-like |
| 3 | 84/M | First | Axillary LN | L&H-like |
| 4 | 33/F | Second, first also TCRBCL, CT | Bone marrow trephine | L&H-like |
| 5 | 54/M | Second, first follicular mixed lymphoma, CT | Axillary LN | Centroblast-like |
| 6 | 54/F | First | Femur | Centroblast-like |
| 7 | 85/F | First | Cervical LN | Centroblast-like |
| Patient . | Age/Sex . | Presentation . | Biopsy Site . | CD20+ Cells Analyzed . |
|---|---|---|---|---|
| 1 | 65/M | First | Axillary LN | L&H-like |
| 2 | 32/F | First | Axillary LN | L&H-like |
| 3 | 84/M | First | Axillary LN | L&H-like |
| 4 | 33/F | Second, first also TCRBCL, CT | Bone marrow trephine | L&H-like |
| 5 | 54/M | Second, first follicular mixed lymphoma, CT | Axillary LN | Centroblast-like |
| 6 | 54/F | First | Femur | Centroblast-like |
| 7 | 85/F | First | Cervical LN | Centroblast-like |
Abbreviations: CT, chemotherapy; LN, lymph node.
Summary of Single Cell Analysis of Six Cases of T-Cell–Rich B-Cell Lymphoma
| Patient . | Experiment . | Cells Positive . | PCR Products . | Rearrangements . | Controls Positive . | ||
|---|---|---|---|---|---|---|---|
| Total . | Sequenced . | Repeated . | Unique . | ||||
| 1 | a | 25/32 | 16VH4L* | 16 | 16VH4 | 0/13 buffer | |
| 24Vκ3 | 23 | 16Vκ3(a) | |||||
| 5Vκ3(b) | |||||||
| 2Vκ3(a and b) | |||||||
| 2 | a | 16/33 | 10VH3L | 10 | 10VH3 | 0/6 T cells | |
| 14Vκ1 | 9 | 8Vκ1(a) | 0/13 buffer | ||||
| 1Vκ1(a and b) | |||||||
| b | 13/30 | 5VH3L | 5 | 5VH3 | 0/12 buffer | ||
| 13Vκ1 | 12 | 9Vκ1(a) | |||||
| 3Vκ1(b) | |||||||
| 3 | a | 36/60 | 19VH4L | 16 | 16VH4 | 0/24 buffer | |
| 33Vκ2 | 21 | 21Vκ2 | |||||
| 4 | a | 13/20 | 10Vκ1 | 9 | 9Vκ1 | 0/8 buffer | |
| 6Vκ2 | 6 | 6Vκ2 | |||||
| 0VHL | |||||||
| b | 29/39 | 23VH3 | 23 | 23VH3 | 0/17 buffer | ||
| 28Vλ1 | 20 | 20Vλ1 | |||||
| 28Vλ3 | 6 | 6Vλ3 | |||||
| 5† | a | 34/63 | 0VHL | 0/24 buffer | |||
| 10VH4 | 10 | 10VH4 | |||||
| 10Vκ4 | 4 | 4Vκ4 | |||||
| 20Vλ2 | 20 | 20Vλ2 | |||||
| 6‡ | a | 15/30 | 4VH1L | 4 | 4VH1 | 0/12 buffer | |
| 12VH4L | 12 | 11VH4 | |||||
| 1VH4 | |||||||
| 1VH3L | 1 | 1VH3 | |||||
| 3Vκ1 | 3 | 3Vκ12-153 | |||||
| 1Vκ2 | 1 | 1Vκ2 | |||||
| b | 18/29 | 10VH1L | |||||
| 7VH4L | |||||||
| 11Vλ1 | 11 | 11Vλ1 | 0/14 buffer | ||||
| 18Vλ3 | 8 | 8Vλ3 | |||||
| Patient . | Experiment . | Cells Positive . | PCR Products . | Rearrangements . | Controls Positive . | ||
|---|---|---|---|---|---|---|---|
| Total . | Sequenced . | Repeated . | Unique . | ||||
| 1 | a | 25/32 | 16VH4L* | 16 | 16VH4 | 0/13 buffer | |
| 24Vκ3 | 23 | 16Vκ3(a) | |||||
| 5Vκ3(b) | |||||||
| 2Vκ3(a and b) | |||||||
| 2 | a | 16/33 | 10VH3L | 10 | 10VH3 | 0/6 T cells | |
| 14Vκ1 | 9 | 8Vκ1(a) | 0/13 buffer | ||||
| 1Vκ1(a and b) | |||||||
| b | 13/30 | 5VH3L | 5 | 5VH3 | 0/12 buffer | ||
| 13Vκ1 | 12 | 9Vκ1(a) | |||||
| 3Vκ1(b) | |||||||
| 3 | a | 36/60 | 19VH4L | 16 | 16VH4 | 0/24 buffer | |
| 33Vκ2 | 21 | 21Vκ2 | |||||
| 4 | a | 13/20 | 10Vκ1 | 9 | 9Vκ1 | 0/8 buffer | |
| 6Vκ2 | 6 | 6Vκ2 | |||||
| 0VHL | |||||||
| b | 29/39 | 23VH3 | 23 | 23VH3 | 0/17 buffer | ||
| 28Vλ1 | 20 | 20Vλ1 | |||||
| 28Vλ3 | 6 | 6Vλ3 | |||||
| 5† | a | 34/63 | 0VHL | 0/24 buffer | |||
| 10VH4 | 10 | 10VH4 | |||||
| 10Vκ4 | 4 | 4Vκ4 | |||||
| 20Vλ2 | 20 | 20Vλ2 | |||||
| 6‡ | a | 15/30 | 4VH1L | 4 | 4VH1 | 0/12 buffer | |
| 12VH4L | 12 | 11VH4 | |||||
| 1VH4 | |||||||
| 1VH3L | 1 | 1VH3 | |||||
| 3Vκ1 | 3 | 3Vκ12-153 | |||||
| 1Vκ2 | 1 | 1Vκ2 | |||||
| b | 18/29 | 10VH1L | |||||
| 7VH4L | |||||||
| 11Vλ1 | 11 | 11Vλ1 | 0/14 buffer | ||||
| 18Vλ3 | 8 | 8Vλ3 | |||||
The micromanipulated cells were analyzed together with buffer or T-cell controls in a blinded fashion. The data summarized as one experiment contain all data obtained from cells and controls micromanipulated in one micromanipulation session. Two different Vκ rearrangements using Vκ genes of the same V gene family were amplified from cells of patients 1 and 2. The different rearrangements are labeled with the suffixes (a) and (b) in Tables 2 and 3.
Ig heavy chain rearrangements with the suffix ‘L’ were amplified with the VH leader primers.
For 20 cells the VHL and Vκ primer sets were used, for 43 cells the VHFRI and the Vλset.
In experiment b of case 6 the VHL and Vλprimer sets were used, but nested PCR for VHL was only performed for Vλ positive cells.
An unmutated VκL12a in-frame rearrangement was coamplified with the clonal VH4 rearrangement from cell BKL1. As no Vκ rearrangements were amplified from the 10 other cells from which clonal VH rearrangements were obtained, the amplification of this unique Vκ1 rearrangement is most likely due to contamination with fragments from other cells during the micromanipulation procedure.
All large irregular shaped cells were CD3−, CD15−, and CD30− in immunhistochemistry and EBV-negative applying EBER-ISH (not shown). In all cases, only a few scattered CD57+ T cells were found and FDCs (DR53+ cells) were not seen.
PCR analysis of IgV gene rearrangements in the micromanipulated cells.
V gene rearrangements for heavy (VH) and light chains (VL) were amplified from single cells using sets of family-specific V gene leader and/or FR I primers with two nested sets of primers for the J regions, as described.23-25 In cases in which amplifications with the VH leader (VHL) and Vκ primer sets did not yield products or only out-of-frame Vκ rearrangements were amplified, additional cells were analyzed using VH FRI (VHFRI) and Vλ primer sets. Amplification efficiencies ranged from 39% to 78% (cells were counted as positive when at least one V gene rearrangement was amplified). The negative controls from the micromanipulations were included in a blinded fashion; all were negative for V gene rearrangements (Table 2).
Clonality of the tumor cells in TCRBCL.
For cases 1 to 5, only clonal heavy and light chain rearrangements were amplified (Table 2). In case 6, 34 of 40 amplified V gene rearrangements for heavy and light chains belonged to four clonal rearrangements (two VH and two Vλ region genes). Two VH and four Vκ rearrangements were unique. This indicates that in this case a small fraction of B cells not belonging to the tumor clone was present in the lymph node. Indeed, this case contained a considerable number of small B cells besides the centroblast-like putative tumor cells (Table 1).
In case 7, clonal Ig gene rearrangements were neither detected by single cell PCR nor by Southern blot analysis (data not shown). Therefore, this case is not further considered here.
The large CD20+ cells in TCRBCLs harbor mutated, clonal Ig heavy and light chain rearrangements.
All clonal VH genes and in-frame VLrearrangements, as well as two VL out-of-frame rearrangements amplified from single B cells of cases 1 to 6 were mutated, with mutation frequencies ranging from 7.2% to 24% (average frequency, 15.5%) for VH and 0.4% to 11.6% (average frequency, 5.9%) for VL genes (Table 3). From each of the cases, a potentially functional VH and VL rearrangement was amplified. Three out-of-frame Vκ and one out-of-frame Vλ gene rearrangements were unmutated. The lack of somatic mutation in the out-of-frame Vκ gene rearrangements of tumor cells that carry somatically mutated VH and Vλ genes is likely due to inactivation of the loci by rearrangements using the Kappa deleting element (KDE), which is frequently seen in human B cells.27-31 Because a deleting element has not been described for the λ locus, it is unclear why the out-of-frame Vλ rearrangement in case 6 is unmutated despite the high load of mutations in the other rearrangements from the same tumor clone.
Sequence Analysis of Clonal Ig Heavy and Light Chain Gene Rearrangements
| Patient . | V Gene . | In-frame3-150 . | % Mutation . | Intraclonal Diversity . | No. of Variants . | Mixed3-151 Sequences . | Remarks . |
|---|---|---|---|---|---|---|---|
| 1 | VH 4-34 | + | 7.2 | Yes | 3 | 2x 308G/C | |
| Vκ A27(a) | + | 7 | No | ||||
| Vκ A27(b) | − | 0 | |||||
| 2 | VH 3-33 | + | 7.8 | Yes | 3 | 1x 303G/A | 7 sequences with 19 bp deletion |
| Vκ L12a(a) | + | 2.7 | Yes | 2 | |||
| Vκ O12(b) | − | 9.4 | No | 1 nonsense mutation in codon 353-152; duplication of 109 bp | |||
| 3 | VH 4-31 | + | 18.7 | Yes | 3 | 7 sequences with 21 bp duplication | |
| Vκ A19 | + | 6.9 | Yes | 2 | 1x 23T/C | ||
| 12x 23G/C | |||||||
| 4 | VH 3-30 | + | 8.9 | Yes | 3 | 1x 240A/G | |
| 241A/G | |||||||
| 2x 158C/G | |||||||
| 274T/A | |||||||
| 5x 158 C/G | |||||||
| 273A/G | |||||||
| 274T/A | |||||||
| Vκ L1 | − | 0 | |||||
| Vκ A2 | + | 6.9 | No | 1 nonsense mutation in codon 93; deletion of 33 bp; Cys88-Ser | |||
| Vλ 1g | + | 1.4-4.0 | Yes | 6 | 1x 108A/G 109T/C 223A/G 275A/G | 2 sequences with a 1 and a 6 bp deletion and Cys23-Ser | |
| 6x variants with and without deletion | |||||||
| Vλ 3m | − | 0.4-1.2 | Yes | 2 | |||
| 5 | VH 4-39 | + | 23.6 | Yes | 2 | 1x 114T/C | Cys92-Tyr |
| Vκ B3 | − | 0 | |||||
| Vλ 2b2 | + | 11.6 | No | 1x 94T/C | |||
| 178C/T | |||||||
| 6 | VH 1-8 | + | 24.0 | Yes | 2 | 3 nonsense mutations in codons 1, 3 and 91; 1 and 3 bp insertion; Cys22-His | |
| VH4-34 | + | 18.3 | Yes | 3 | |||
| Vλ 1b | + | 7.1 | Yes | 2 | 1x 15T/C | ||
| 52T/C | |||||||
| 1x 181T/C | |||||||
| Vλ 3r | − | 0 |
| Patient . | V Gene . | In-frame3-150 . | % Mutation . | Intraclonal Diversity . | No. of Variants . | Mixed3-151 Sequences . | Remarks . |
|---|---|---|---|---|---|---|---|
| 1 | VH 4-34 | + | 7.2 | Yes | 3 | 2x 308G/C | |
| Vκ A27(a) | + | 7 | No | ||||
| Vκ A27(b) | − | 0 | |||||
| 2 | VH 3-33 | + | 7.8 | Yes | 3 | 1x 303G/A | 7 sequences with 19 bp deletion |
| Vκ L12a(a) | + | 2.7 | Yes | 2 | |||
| Vκ O12(b) | − | 9.4 | No | 1 nonsense mutation in codon 353-152; duplication of 109 bp | |||
| 3 | VH 4-31 | + | 18.7 | Yes | 3 | 7 sequences with 21 bp duplication | |
| Vκ A19 | + | 6.9 | Yes | 2 | 1x 23T/C | ||
| 12x 23G/C | |||||||
| 4 | VH 3-30 | + | 8.9 | Yes | 3 | 1x 240A/G | |
| 241A/G | |||||||
| 2x 158C/G | |||||||
| 274T/A | |||||||
| 5x 158 C/G | |||||||
| 273A/G | |||||||
| 274T/A | |||||||
| Vκ L1 | − | 0 | |||||
| Vκ A2 | + | 6.9 | No | 1 nonsense mutation in codon 93; deletion of 33 bp; Cys88-Ser | |||
| Vλ 1g | + | 1.4-4.0 | Yes | 6 | 1x 108A/G 109T/C 223A/G 275A/G | 2 sequences with a 1 and a 6 bp deletion and Cys23-Ser | |
| 6x variants with and without deletion | |||||||
| Vλ 3m | − | 0.4-1.2 | Yes | 2 | |||
| 5 | VH 4-39 | + | 23.6 | Yes | 2 | 1x 114T/C | Cys92-Tyr |
| Vκ B3 | − | 0 | |||||
| Vλ 2b2 | + | 11.6 | No | 1x 94T/C | |||
| 178C/T | |||||||
| 6 | VH 1-8 | + | 24.0 | Yes | 2 | 3 nonsense mutations in codons 1, 3 and 91; 1 and 3 bp insertion; Cys22-His | |
| VH4-34 | + | 18.3 | Yes | 3 | |||
| Vλ 1b | + | 7.1 | Yes | 2 | 1x 15T/C | ||
| 52T/C | |||||||
| 1x 181T/C | |||||||
| Vλ 3r | − | 0 |
Mutation frequencies refer to the germline sequences from Schäble,52 Matsuda,53 and Williams.54 In cases with intraclonal diversity, mutation frequencies refer to the sequence variant with greatest homology to the germline sequence. The suffixes (a) and (b) for the Vκrearrangements of patients 1 and 2 refer to Table 2, where they are used to label different Vκ rearrangements using Vκ gene segments of the same V gene family. All sequences have been deposited in the EMBL database (AJ130895 through AJ130914). For rearrangements with intraclonal diversity, the sequence with the greatest homology to the germline sequence has been deposited.
All in-frame rearrangements are potentially functional when somatic mutations are disregarded.
Mixed sequences contain at one or more positions two nucleotides with approximately equal fluorescense intensities on sequencing gels (confirmed by sequencing both strands) or consisted of partly double sequences, when a mixture of variants with and without a deletion was sequenced. Behind the number indicating how often a mixed sequence was found, the positions and the nucleotides found at the corresponding positions are given.
Codons are numbered according to Kabat et al.55
In three cases, V gene rearrangements that had most likely originally been potentially functional were rendered nonfunctional by somatic mutations. In the in-frame VH rearrangement of case 5, the cysteine at position 92, which is thought to be important for proper folding of the protein,32 is mutated to a tyrosine. In case 4, the mutated clonal in-frame Vκ rearrangement is rendered nonfunctional by a stop codon. In addition, nearly the complete complementarity determining region (CDR) I is missing due to a 33-bp deletion, and the cysteine at position 88, which is important for an intrachain disulfide bridge,32 is mutated to a serine. However, case 4 also harbors a clonal in-frame Vλrearrangement, which is potentially functional and somatically mutated.
In case 6, two clonal in-frame VH rearrangements were amplified. These two rearrangements were repeatedly amplified from the same single cells. While the rearrangement using VH4-34 is potentially functional, the VH1-8 rearrangement contains three stop codons and two insertions in FRIV, one of which resulted in loss of the correct reading frame. The original rearrangement may have been functional and inactivated by somatic hypermutation.
Intraclonal diversity in TCRBCLs.
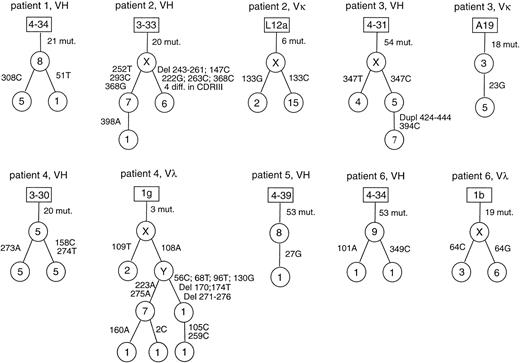
In all six cases with clonal rearrangements, intraclonal diversity was observed among the heavy chain rearrangements, and in four cases also among the light chain sequences (Table 3, Fig 1). Usually, 2 to 3 sequence variants of a clonal rearrangement were found. The greatest diversity was observed for the potentially functional Vλ rearrangement in case 4 with six sequence variants. Most of the sequence variants are due to single nucleotide exchanges and were amplified several times. None of these diversifying point mutations in the potentially functional rearrangements rendered the genes nonfunctional. In cases 2 and 4, besides nucleotide exchanges, also deletions, and in case 3, a duplication were observed in several cells. Whereas in cases 2 and 4, the deletions abolished the coding capacity of these rearrangements, the 21-bp duplication in the VH CDRIII of case 3 preserves the correct reading frame.
Intraclonal diversity in TCRBCL. Genealogical trees for the potentially functional rearrangements with intraclonal diversity. Letters and figures in the rectangles are the designations of the germline genes with the greatest homologies. Assumed intermediates in the genealogical trees are designated with X and Y. Figures in the circles indicate how often a specific sequence variant was found. Beside the lines connecting the sequence variants, positions of nucleotide exchanges and the new nucleotides are given. Mixed clonal sequences are excluded from this figure.
Intraclonal diversity in TCRBCL. Genealogical trees for the potentially functional rearrangements with intraclonal diversity. Letters and figures in the rectangles are the designations of the germline genes with the greatest homologies. Assumed intermediates in the genealogical trees are designated with X and Y. Figures in the circles indicate how often a specific sequence variant was found. Beside the lines connecting the sequence variants, positions of nucleotide exchanges and the new nucleotides are given. Mixed clonal sequences are excluded from this figure.
Analysis of mutation pattern.
The ratio of replacement (R) to silent (S) mutations in the FRs of functional V gene rearrangements can give hints whether cells have been under selective pressure for expression of an antigen receptor. In that case, R mutations are counterselected in the FRs to preserve the evolutionary optimized structure of the antibody V domain. In the absence of selection, like in out-of-frame rearrangements, R mutations in the FRs are not counterselected.
Indeed, the R/S ratios of the mutations within the FRs in productive rearrangements of various B-cell subsets with mutated V gene rearrangements have been shown to be in the range between 1.0 and 1.6, whereas for a collection of nonfunctional out-of-frame rearrangements, the ratio is 3.0, the value expected assuming random mutagenesis.18 20 For the 190 mutations in the FRs of the 12 potentially functional rearrangements in the six cases of TCRBCL analyzed, a R/S ratio of 1.5 was found, which is within the range typical for antigen-selected cells.
A R/S analysis was also performed for two other in-frame rearrangements (VκA2 of case 4 and VH1-8 of case 6), which might have been functional before they accumulated inactivating mutations. Indeed, these rearrangements show R/S ratios of 1.5 and 2.0, respectively, indicating that they had been positively selected before inactivation.
The large CD20+ cells in TCRBCL are polyploid and contain different sequence variants of the same clonal V gene rearrangement within a single cell.
From several single tumor B cells, mixed sequences representing two variants of the same clonal rearrangement were obtained, indicating the presence of two or more copies of the clonal V gene rearrangement in the respective cells. These mixed sequences were mainly due to single nucleotide differences, and in one case, to two deletions (Vλ1g rearrangement of case 4). Such mixed sequences were found at least once in each case. In half of the cases in which a mixed sequence was detected, it was seen in several single cells (Table 3).
To verify the existence of multiple copies of rearranged V genes in large CD20+ cells, the DNA of single micromanipulated cells from one case (case 4) was split into three aliquots and the standard seminested amplification protocol was performed for each of the aliquots (see Materials and Methods). Five clonal VH and five clonal Vλ rearrangements were amplified from 12 cells. From two aliquots of one cell, an identical clonal Vλ gene was amplified. From two aliquots of another cell, clonal VH rearrangements differing at two nucleotide positions were obtained. This confirms that at least in case 4 multiple copies of the same clonal rearrangement, which may show sequence variation, are present within some or all tumor cells.
The existence of multiple copies of clonal rearrangements could be due to duplications of the rearrangements, gain of single chromosomes 2, 14, or 22 carrying the V gene loci or a gain of a whole chromosome set leading to polyploidy. The latter two possibilities can be distinguished by FISH using probes for chromosomes other than those carrying the V genes. Thus, FISH was performed on nuclei isolated from frozen tissues of cases 1 to 3 using probes for chromosomes 7, 12, X and Y. In all three cases, additional signals for chromosomes 7, 12, and the gonosomes were seen in a significant percentage (5% to 10%) of interphase nuclei evaluated. Thus, the results of the FISH study suggest polyploidy and render the presence of isolated gains of the chromosomes carrying the V genes unlikely. As the proportion of the large CD20+ cells in the three tissues analyzed is about 10% to 15%, it is likley that the large CD20+ cells represent the polyploid cells. Thus, the large CD20+ cells in TCRBCL contain abnormal numbers of some (and potentially all) chromosomes.
DISCUSSION
Clonality of the L&H- or centroblast-like cells in TCRBCL.
From six cases of TCRBCL, clonal Ig gene rearrangements were amplified from single micromanipulated CD20+ cells of irregular size. This confirms that in TCRBCL the irregular L&H-like cells (cases 1 to 4) or the centroblast-like cells (cases 5 and 6), assumed to be the neoplastic cells, indeed represent clonal B-cell populations. The amplification of five unique rearrangements in case 6 shows that also nonclonal infiltrating B cells are present at least in this TCRBCL and sometimes may be difficult to distinguish from the neoplastic cells.
TCRBCL are derived from GC B cells.
All clonal VH and in-frame VL gene rearrangements were mutated with mutation frequencies ranging from 0.4% to 24%. These data are in agreement with those obtained for the one case analyzed previously20 and show that the tumor cells are derived from mature B cells that had participated in a GC reaction (ie, GC or post-GC B cells). Moreover, because the process of somatic hypermutation appears to be restricted to GC B cells, the finding of intraclonal diversity in all six cases identifies GC B cells as the precursors of the tumor clone in TCRBCL. However, it is unclear whether the somatic hypermutation machinery was still active in the tumor cells when the biopsies were taken. It is likewise possible that the observed intraclonal diversity resulted from ongoing mutation at a very early stage of tumor clone expansion and that subclones generated in this way were then stably propagated. A GC B cell derivation of TCRBCL is further supported by the expression of the GC B cell-specific transcription factor bcl-6 by 10% to 90% of the lymphoma cells33 34 (indeed, bcl-6 was expressed by the tumor cells in all six cases analyzed in the present study; data not shown) and the centroblast-like morphology of the large irregular cells in some cases.
The somatic mutations in the rearranged V genes were not restricted to nucleotide exchanges, but also included duplications, insertions, and deletions (Table 3 and Fig 1). The occurrence of duplications, insertions, and deletions in somatically mutated V region genes is in accord with the work of Goossens et al35 and Wilson et al36 who recently showed that such events occur during the process of somatic hypermutation.
For each of the six cases, potentially functional Ig heavy, as well as light chain, genes were amplified. The average R/S ratio of 1.5 for the FRs of those rearrangements is in the range that is typical for normal antigen-selected B cells.18 This indicates that, in general, the precursors of the tumor clones were selected for a functional antigen receptor.
Crippling mutations in TCRBCL?
Despite those clear signs for antigenic selection in the tumor precursors, in several of the cases, potentially crippling mutations were observed in clonal V gene rearrangements. In case 2, seven of 15 in-frame VH gene sequences were rendered nonfunctional by a 19-bp deletion. In case 4, a fraction of the sequences harbored two deletions, one of which resulted in loss of the correct reading frame. In addition, it is conceivable that nearly half of the sequences of the VH4-31 rearrangement in case 3 are no more able to bind to the initial antigen recognized by the B-cell receptor, because of a 21-bp duplication in CDRIII. In case 5, the cysteine at codon 92 is mutated to tyrosine in all clonal VH rearrangements amplified. This cysteine is thought to be important for correct folding of the protein.32 Thus, it is possible that in some cases of TCRBCL, the tumor cells or subpopulations of them lost dependence on expression of a functional antigen receptor. However, the following aspects are important in evaluating the “crippling” mutations: (1) it has been described that a hybridoma expressed a functional antigen receptor despite a cysteine 92 to tyrosine mutation.37 Therefore, mutations of the conserved cysteine 92 are not necessarily crippling. (2) In case 4, a mixed sequence of the Vλ gene rearrangement with and without the two deletions was amplified from six cells (Table 3). Thus, these tumor cells most likely contained two copies of the clonal Vλgene rearrangement, one of which was still functional. This may well be true for all cells of this tumor, given the limited efficiencies of V gene amplification from single, micromanipulated cells. That the tumor cells in TCRBCL often contain more than two copies of several (or even all) chromosomes is supported by the FISH analysis, which showed additional copies of the chromosomes investigated, ie, chromosomes 7, 12, and the gonosomes, in the three cases analyzed. Moreover, it has been reported that 91% of DLLs show numerical chromosomal aberrations.38
Thus, in TCRBCL, single tumor cells may contain two or more copies of a V gene rearrangement and between these copies there may be sequence variation. In case 2, the functional and crippled variants of the VH gene rearrangement were never amplified together from a single cell. Thus, in this case, a subclone of the tumor cells might have lost the capacity to express a functional antigen receptor. However, because the PCR efficiency and the total number of sequences was lower in this case than in case 4, it is possible that the tumor cells harbor two copies of the VH gene rearrangement (one functional and one crippled) also in this case.
Indications for V gene editing in human GCs.
In two instances we observed crippling mutations in tumor cell-derived V gene rearrangements, which are indicative of secondary V gene rearrangements (receptor editing) in the course of the GC reaction.39 40 In case 4, an in-frame Vκ gene rearrangement was rendered nonfunctional by a stop codon within the Vκ gene segment and a 33 bp deletion in CDRI. Interestingly, the tumor cells also harbor a potentially functional Vλ gene rearrangement. Conceivably, the tumor precursor, a GC B cell, initially expressed a κ light chain gene and when this rearrangement was crippled by the stop codon and/or the deletion, it successfully performed a productive Vλ gene rearrangement. The considerably lower mutation load in the Vλ gene (1.4% to 4%) compared with the VH, as well as Vκ gene (8.9% and 6.9%, respectively) is in line with the interpretation that the Vλ gene rearrangement occurred in the course of the GC reaction, so that it went through less rounds of somatic mutation than the two other V genes. In addition, the R/S ratio of 1.5 for mutations in the FRs of the Vκ rearrangement indicates that the gene was initially selected and hence expressed.
In case 6, two clonal in-frame VH gene rearrangements were amplified. One of the rearrangements had been inactivated by somatic mutation. These two in-frame VH rearrangements could have been generated during B-cell development, and somatic mutation could then have inactivated one rearrangement of a potential double producer. The expression of two functional heavy chain genes has indeed been described for a fraction of cases of B-cell chronic lymphocytic leukemia (B-CLL).41 However, it is also possible that the tumor precursor contained initially only one functional VH rearrangement, which was inactivated in the GC by somatic mutation and the second VH in-frame rearrangement occurred thereafter. The R/S ratio of 2 for the mutations within the FRs of the inactivated rearrangement may reflect such a partial selection for functionality.
The relationship between TCRBCL, other B-cell non-Hodgkin’s lymphoma (NHL) and HD.
Other types of DLLs also harbor mutated Ig gene rearrangements and, based on the analysis of a collection of cases from different types of DLLs (10 centroblastic lymphomas, 5 mediastinal sclerosing lymphomas, 2 immunoblastic lymphomas, one large cell anaplastic lymphoma, and one TCRBCL), the tumor precursors appear to be selected for expression of a functional antigen receptor also in these other entities.20However, it is unclear whether the other DLLs are also derived from mutating GC B cells. Whereas ongoing mutation was not observed in 17 cases analyzed, in one study,42 ongoing mutation has been described in a centroblastic lymphoma20 and four DLLs not further specified.43 44
In terms of the differentiation stage of the tumor precursors, TCRBCL appears to be closely related to follicular lymphoma45 and LP HD25,46,47 in which the consistent finding of ongoing mutation indicates a derivation from mutating GC B cells as well. Moreover, intraclonal diversity of rearranged V genes is occasionally also seen in sporadic and endemic Burkitt’s lymphoma48,49and mucosa-associated lymphoid tissue (MALT) lymphomas.50 Thus, although the precursors of all these lymphomas derive from the same B-cell compartment, they give rise to different types of lymphomas. It is likely that GC B-cell–derived lymphomas differ in their dependence on antigenic triggering, in the transforming events, and the subsets of GC B cells that are subject to the initial transforming events.18
The diagnostic distinction of TCRBCL from the diffuse variants of LP HD is often difficult, but important for adequate therapy, as both entities show a strikingly different clinical behavior, TCRBCL being much more aggressive than the clinical indolent LP HD.13 In LP HD, the tumor cells seem to be dependent on a GC-like microenvironment with FDCs and CD57+ T cells, while in TCRBCL, no GC remnants are found.11,16 Comparison of the V gene rearrangements and their mutation pattern in TCRBCL and LP HD25 does not show a clear distinction between the two diseases: (1) both types of tumor cells harbor highly mutated clonal Ig gene rearrangements (with average VH mutation frequencies of 10.1% for LP HD and TCRBCL) and in all cases analyzed, potentially functional in-frame rearrangements were amplified for Ig heavy and light chains.18,25 (2) The mutation pattern indicates that in both entities the tumor precursors are generally selected for the expression of a functional antigen receptor, in distinction to classical HD, where the tumor as a rule appears to be derived from crippled GC B cells.24 (3) Although the interpretation of the crippled rearrangements in two cases of TCRBCL of the present study is ambigious, it appears that in LP HD, as well as in TCRBCL, subclones of the tumor may occasionally lose dependence on antigen receptor expression, resulting in the outgrowth of subclones harboring crippling mutations.46 51
The only molecular distinction between TCRBCL and LP HD that became evident from the present study is the consistent finding of two or more copies of clonal rearrangements showing mixed sequences within single cells in each of the six cases of TCRBCL, while this was only rarely observed in LP HD (two of 27 sequences in one of five cases, unpublished observation). The consistent finding of such mixed sequences in TCRBCL indicates that chromosomal aneuploidy is a frequent feature of the tumor cells in this disease and that the hypermutation machinery is still active when or after chromosomes have been duplicated. The low frequency of mixed sequences in the cases of LP HD so far analyzed25 suggests that numerical chromosomal abnormalities involving the chromosomes harboring the Ig loci are either rare in LP HD or that the hypermutation machinery was no longer active when such hypothetical chromosomal duplications happened.
Taken together, the present study identifies mutating GC B cells as the precursors of the tumor clones in TCRBCL, adding this disease to the large collection of GC B-cell–derived malignancies and showing a close relationship between this lymphoma and LP HD at the level of the tumor precursors.
ACKNOWLEDGMENT
We thank Christiane Gerhard and Tanja Schaffer for excellent technical assistance. We are grateful to Holger Kanzler for critically reading the manuscript.
Supported by the Deutsche Forschungsgemeinschaft through SFB 502, the Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung, and the Wilhelm-Sanders-Stiftung. B.S. holds a Hermann and Lilly Schilling professorship.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Andreas Bräuninger, PhD, Department of Pathology, University of Frankfurt, Theodor Stern Kai 7, 60590 Frankfurt, Germany; e-mail: Braeuninger@em.uni-frankfurt.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal