Abstract
Combined factor V-factor VIII deficiency (F5F8D) is a rare, autosomal recessive coagulation disorder in which the levels of both coagulation factors V and VIII are diminished. The F5F8D locus was previously mapped to a 1-cM interval on chromosome 18q21. Mutations in a candidate gene in this region, ERGIC-53, were recently found to be associated with the coagulation defect in nine Jewish families. We performed single-strand conformation and sequence analysis of the ERGIC-53 gene in 35 F5F8D families of different ethnic origins. We identified 13 distinct mutations accounting for 52 of 70 mutant alleles. These were 3 splice site mutations, 6 insertions and deletions resulting in translational frameshifts, 3 nonsense codons, and elimination of the translation initiation codon. These mutations are predicted to result in synthesis of either a truncated protein product or no protein at all. This study revealed that F5F8D shows extensive allelic heterogeneity and all ERGIC-53 mutations resulting in F5F8D are “null.” Approximately 26% of the mutations have not been identified, suggesting that lesions in regulatory elements or severe abnormalities within the introns may be responsible for the disease in these individuals. In two such families, ERGIC-53 protein was detectable at normal levels in patients’ lymphocytes, raising the further possibility of defects at other genetic loci.
COMBINED FACTOR V-factor VIII deficiency (F5F8D) (MIM 227310) is a rare, recessive coagulation disorder characterized by reduction in levels of both factor V and factor VIII, to less than 20 U/dL.1-4 The severity ranges from mild (factor V and VIII levels of 10 to 20 U/dL) to moderate (levels 5 to 10 U/dL).5 Linkage of the F5F8D locus to human chromosome 18q was reported in nine nonAshkenazi Jewish families by homozygosity mapping6 and by classical linkage analysis in 19 families of Irani, Pakistani, and Algerian origin.7 The same locus was, therefore, implicated in different ethnic groups. The difference in clinical severity and the lack of a specific haplotype in the second group of families,7 in contrast to two distinct founder haplotypes reported by Nichols et al,6 suggested the existence of allelic heterogeneity, ie, that more than one mutation was responsible for the disease in this sample of families. Critical recombination events localized the F5F8D gene to an interval of approximately 1 cM, between markers D18S849 and D18S1103.7 ERGIC-53, which had been previously mapped to this region8 and codes for a 53-kD transmembrane protein resident in the endoplasmic reticulum-Golgi intermediate compartment,9 was shown by mutation analysis to be the gene responsible for F5F8D.1 Two mutations were identified. All Sephardic Jewish patients from five families tested were homozygous for a donor splice site mutation leading to premature protein truncation, and all patients from four Middle Eastern Jewish families had a single base pair insertion at codon 30. In Epstein-Barr virus (EBV) transformed cell lines from these patients, Western blotting and immunofluorescence analysis indicated complete absence of ERGIC-53 expression.1 To investigate the molecular origin of this coagulation disorder we have studied 35 additional F5F8D families from different ethnic backgrounds by single-strand conformational analysis (SSCA) and sequencing of polymerease chain reaction (PCR) products of exons 1-13 and flanking intronic sequences of the ERGIC-53 gene. As expected from our previous haplotype analysis,7 numerous mutations in the ERGIC-53 gene were found to be responsible for the disease. All 13 identified discrete mutations were predicted to lead to a deficiency or absence of functional protein. These mutations accounted for a total of 52 of 70 mutant alleles (74%). In addition, a number of polymorphisms were identified, some of which result in amino acid changes.
MATERIALS AND METHODS
Description of families.
Blood was collected from 16 families from Iran, 6 families of Pakistani origin, 8 from Italy, 2 of Chinese origin, and 1 each from Algeria, Britain, and South Africa (Table1). Informed consent was obtained from all families. The majority of Iranian and Pakistani families were consanguineous (19 of 22) involving first cousin marriages. Factor V and factor VIII assays were performed by a one-stage procedure by using congenitally deficient plasma substrates and normal pooled plasma as reference.10 The levels of factor V:C and factor VIII:C detected in the plasma of affected individuals are shown in Table 1.
Identified Mutations, Geographical Origin, and Clinical Characteristics of F5F8D Patients
| Patient . | Origin (Consanguinity) . | Phenotype Factor V/VIII . | Mutation . | ERGIC-53 Protein . |
|---|---|---|---|---|
| A1 | Iran (C) | 13/14 | 912-913insA (cd 305) | ND |
| 912-913insA (cd 305) | ||||
| A2 | Iran (C) | 7/7 | 822G>A (IVS7 DS −1) | −* |
| 822G>A (IVS7 DS −1) | ||||
| A3 | Iran (C) | 14/13 | 912-913insA (cd 305) | ND |
| 89-90insG (cd 30) | ||||
| A4 | Iran (C) | 10/11 | IVS9 +2 T>G | ND |
| IVS9 +2 T>G | ||||
| A5 | Iran (C) | 5/10 | ? | ND |
| ? | ||||
| A6 | Iran (C) | 14/13 | 822G>A (IVS7 DS −1) | ND |
| 822G>A (IVS7 DS −1) | ||||
| A7 | Iran (C) | 7/7 | ? | ND |
| ? | ||||
| A8 | Iran (C) | 5/6 | 1214-1218delAAATG (cd 406) | ND |
| 1214-1218delAAATG (cd 406) | ||||
| A9 | Iran (C) | 18/8 | 822G>A (IVS7 DS −1) | ND |
| 822G>A (IVS7 DS −1) | ||||
| A10 | Iran (C) | 12/15 | 23delG (cd 8) | ND |
| 23delG (cd 8) | ||||
| A11 | Iran (C) | 18/17 | 89-90insG (cd 30) | ND |
| 89-90insG (cd 30) | ||||
| A12 | Iran (C) | 2.5/2.2 | 822G>A (IVS7 DS −1) | −* |
| 822G>A (IVS7 DS −1) | ||||
| A13 | Iran (C) | 4.6/5.5 | ? | ND |
| ? | ||||
| A14 | Iran | 10/6 | ? | ND |
| ? | ||||
| A16 | Iran | 9/13.5 | R202X (CGA>TGA) | ND |
| R202X (CGA>TGA) | ||||
| A17 | Iran (C) | 5/7.5 | R202X (CGA>TGA) | ND |
| R202X (CGA>TGA) | ||||
| A18 | Pakistan (C) | 10/15 | K302X (AAA>TAA) | −* |
| K302X (AAA>TAA) | ||||
| A19 | Pakistan (C) | 14/14 | R456X (CGA>TGA) | −* |
| R456X (CGA>TGA) | ||||
| A20 | Pakistan (C) | 8/6 | K302X (AAA>TAA) | −* |
| K302X (AAA>TAA) | ||||
| A21 | Italy | 10/11 | ? | +† |
| ? | ||||
| A22 | Italy | 8/24 | ? | −* |
| ? | ||||
| A24 | Italy | 17/26 | M1T (ATG>ACG) | ND |
| M1T (ATG>ACG) | ||||
| A25 | Italy | 9/10 | M1T (ATG>ACG) | −* |
| M1T (ATG>ACG) | ||||
| A26 | Pakistan (?) | 18/18 | K302X (AAA>TAA) | ND |
| K302X (AAA>TAA) | ||||
| A27 | China | 7/9 | R456X (CGA>TGA) | −* |
| R456X (CGA>TGA) | ||||
| A28 | UK | ? | ND | |
| ? | ||||
| A29 | South Africa | 11/22 | ? | +† |
| ? | ||||
| A30 | Pakistan (C) | 6/3 | K302X (AAA>TAA) | ND |
| K302X (AAA>TAA) | ||||
| A31 | Pakistan (C) | 14/18 | K302X (AAA>TAA) | ND |
| K302X (AAA>TAA) | ||||
| A33 | Italy | ? | ND | |
| ? | ||||
| A34 | Italy | 9/27 | IVS5 +1 G>T | ND |
| IVS5 +1 G>T | ||||
| IVS5 +1 G>T | ||||
| A36 | Italy | 1.4/2.7 | 1208-1209insT (cd 403) | ND |
| 1208-1209insT (cd 403) | ||||
| A37 | China (?) | 17/19 | R456X (CGA>TGA) | ND |
| R456X (CGA>TGA) | ||||
| B1 | Algeria C) | 31delG (cd 11) | ND | |
| 31delG (cd 11) |
| Patient . | Origin (Consanguinity) . | Phenotype Factor V/VIII . | Mutation . | ERGIC-53 Protein . |
|---|---|---|---|---|
| A1 | Iran (C) | 13/14 | 912-913insA (cd 305) | ND |
| 912-913insA (cd 305) | ||||
| A2 | Iran (C) | 7/7 | 822G>A (IVS7 DS −1) | −* |
| 822G>A (IVS7 DS −1) | ||||
| A3 | Iran (C) | 14/13 | 912-913insA (cd 305) | ND |
| 89-90insG (cd 30) | ||||
| A4 | Iran (C) | 10/11 | IVS9 +2 T>G | ND |
| IVS9 +2 T>G | ||||
| A5 | Iran (C) | 5/10 | ? | ND |
| ? | ||||
| A6 | Iran (C) | 14/13 | 822G>A (IVS7 DS −1) | ND |
| 822G>A (IVS7 DS −1) | ||||
| A7 | Iran (C) | 7/7 | ? | ND |
| ? | ||||
| A8 | Iran (C) | 5/6 | 1214-1218delAAATG (cd 406) | ND |
| 1214-1218delAAATG (cd 406) | ||||
| A9 | Iran (C) | 18/8 | 822G>A (IVS7 DS −1) | ND |
| 822G>A (IVS7 DS −1) | ||||
| A10 | Iran (C) | 12/15 | 23delG (cd 8) | ND |
| 23delG (cd 8) | ||||
| A11 | Iran (C) | 18/17 | 89-90insG (cd 30) | ND |
| 89-90insG (cd 30) | ||||
| A12 | Iran (C) | 2.5/2.2 | 822G>A (IVS7 DS −1) | −* |
| 822G>A (IVS7 DS −1) | ||||
| A13 | Iran (C) | 4.6/5.5 | ? | ND |
| ? | ||||
| A14 | Iran | 10/6 | ? | ND |
| ? | ||||
| A16 | Iran | 9/13.5 | R202X (CGA>TGA) | ND |
| R202X (CGA>TGA) | ||||
| A17 | Iran (C) | 5/7.5 | R202X (CGA>TGA) | ND |
| R202X (CGA>TGA) | ||||
| A18 | Pakistan (C) | 10/15 | K302X (AAA>TAA) | −* |
| K302X (AAA>TAA) | ||||
| A19 | Pakistan (C) | 14/14 | R456X (CGA>TGA) | −* |
| R456X (CGA>TGA) | ||||
| A20 | Pakistan (C) | 8/6 | K302X (AAA>TAA) | −* |
| K302X (AAA>TAA) | ||||
| A21 | Italy | 10/11 | ? | +† |
| ? | ||||
| A22 | Italy | 8/24 | ? | −* |
| ? | ||||
| A24 | Italy | 17/26 | M1T (ATG>ACG) | ND |
| M1T (ATG>ACG) | ||||
| A25 | Italy | 9/10 | M1T (ATG>ACG) | −* |
| M1T (ATG>ACG) | ||||
| A26 | Pakistan (?) | 18/18 | K302X (AAA>TAA) | ND |
| K302X (AAA>TAA) | ||||
| A27 | China | 7/9 | R456X (CGA>TGA) | −* |
| R456X (CGA>TGA) | ||||
| A28 | UK | ? | ND | |
| ? | ||||
| A29 | South Africa | 11/22 | ? | +† |
| ? | ||||
| A30 | Pakistan (C) | 6/3 | K302X (AAA>TAA) | ND |
| K302X (AAA>TAA) | ||||
| A31 | Pakistan (C) | 14/18 | K302X (AAA>TAA) | ND |
| K302X (AAA>TAA) | ||||
| A33 | Italy | ? | ND | |
| ? | ||||
| A34 | Italy | 9/27 | IVS5 +1 G>T | ND |
| IVS5 +1 G>T | ||||
| IVS5 +1 G>T | ||||
| A36 | Italy | 1.4/2.7 | 1208-1209insT (cd 403) | ND |
| 1208-1209insT (cd 403) | ||||
| A37 | China (?) | 17/19 | R456X (CGA>TGA) | ND |
| R456X (CGA>TGA) | ||||
| B1 | Algeria C) | 31delG (cd 11) | ND | |
| 31delG (cd 11) |
F5F8D levels were measured as described in Materials and Methods. Western blot analysis of ERGIC-53 protein levels was as described (see Materials and Methods and Fig 3).
Abbreviation: ND, not determined.
Indicates no ERGIC-53 protein detected.
Indicates normal ERGIC-53 levels.
DNA isolation and PCR amplification.
Genomic DNA was purified from blood leukocytes, according to standard protocols. The ERGIC-53 gene structure was determined by PCR and sequence analysis as described in another report (Nichols et al,11 this issue). For the mutation screening, one affected patient from each family (35 individuals) and four normal individuals were used. The ERGIC-53 gene was analyzed by PCR amplification of all 13 exons, including intron-exon junctions (with the exception of the intron 7-exon 8 junction). The primers used for the PCR amplifications (5′ > 3′) are shown in Table2.
Primers Used for PCR Amplifications (5′ > 3′)
| Exon . | Forward . | Reverse . |
|---|---|---|
| 1 | TCG CGT TCC AGA ATC CAA G | TCA GCA CAC CAG GGT AGC |
| 2 | CAG TTT GGA AAT GTA CAT TGA G | GGG AAC AGT TAG AGG CTA G |
| 3 | CTA GCC TCT AAC TGT TCC C | CTC ACA GCC TAA CTC TGT TG |
| 4 | TGT AAG TCA CTT CAT AGT AC | CAA TGT ATT TCA TAA GGA TTC C |
| 5 | TGA AAA GCT GAG TGT CTT GT | GAA AAG TGA TAC TGT AAC ATT G |
| 6 | GAA ACA AAA CTG AAT AGT AGT C | ACA AGT CTA CAT ATC CCT AA |
| 7 | AGA GTG CCA TTG CCT TTA CC | CAA ACC TAA GTT AGT CTT CC |
| 8 | CAC CAG ATA AAG AAA TTT CG | AGG CAA CAC AGA GAC TCA AG |
| 9 | CAC TTT GGT CAC TTA CGT TA | TCT ATG AGC ACA TAG TAC AG |
| 10 | GGG AAG TAA AGA AGA AGG GC | AAT CAC ATA ACA CAC AAA CG |
| 11 | GTG ATT TTA TTG TAT CAA GAG | AGT ATG AGT TCT TCC TTT CC |
| 12 | GGG GAT AGA AGG TTT TCT GG | GAA CAT AGA TAA CTT AGT TG |
| 13 | CTG TTC ATT TCA GTT CAC AT | AAT TCC CTC AAA ACG ACA TC |
| Exon . | Forward . | Reverse . |
|---|---|---|
| 1 | TCG CGT TCC AGA ATC CAA G | TCA GCA CAC CAG GGT AGC |
| 2 | CAG TTT GGA AAT GTA CAT TGA G | GGG AAC AGT TAG AGG CTA G |
| 3 | CTA GCC TCT AAC TGT TCC C | CTC ACA GCC TAA CTC TGT TG |
| 4 | TGT AAG TCA CTT CAT AGT AC | CAA TGT ATT TCA TAA GGA TTC C |
| 5 | TGA AAA GCT GAG TGT CTT GT | GAA AAG TGA TAC TGT AAC ATT G |
| 6 | GAA ACA AAA CTG AAT AGT AGT C | ACA AGT CTA CAT ATC CCT AA |
| 7 | AGA GTG CCA TTG CCT TTA CC | CAA ACC TAA GTT AGT CTT CC |
| 8 | CAC CAG ATA AAG AAA TTT CG | AGG CAA CAC AGA GAC TCA AG |
| 9 | CAC TTT GGT CAC TTA CGT TA | TCT ATG AGC ACA TAG TAC AG |
| 10 | GGG AAG TAA AGA AGA AGG GC | AAT CAC ATA ACA CAC AAA CG |
| 11 | GTG ATT TTA TTG TAT CAA GAG | AGT ATG AGT TCT TCC TTT CC |
| 12 | GGG GAT AGA AGG TTT TCT GG | GAA CAT AGA TAA CTT AGT TG |
| 13 | CTG TTC ATT TCA GTT CAC AT | AAT TCC CTC AAA ACG ACA TC |
Single-strand conformational analysis.
Each sample was analyzed independently with either small- or large-format polyacrylamide gels:
(1) PCR was performed from genomic DNA in a total volume of 15 μL, containing 2.6 pmol of each primer, 1.3 μmol/L of each dNTP, and 0.25 U Taq polymerase. After denaturation at 94°C, the amplification program consisted of 10 touchdown cycles of 30 seconds denaturation at 94°C, 30 seconds annealing between 60 and 50°C, and 30 seconds elongation at 72°C followed by 20 cycles (30 seconds 94°C, 30 seconds 50°C, 30 seconds 72°C). Ten additional cycles were then performed in a new reaction on an aliquot (2 μL) of the first PCR to reduce the amount of genomic DNA versus amplified product before SSCA. PCR products were denatured with an equal volume of denaturation buffer (95% formamide, 0.05% xylene cyanol, 0.05% bromophenol blue) for 10 minutes at 94°C. A 6-μL sample of each was then loaded on a 12.5% GeneGel Excel (Pharmacia Biotech, St Albans, UK). Electrophoresis was at 600 V at 12°C for 3 hours (for fragments of approximately 300 bp: ie, exons 1, 2, 6, 7, 9, 11, 12, 13) or 2 hours (for fragments of approximately 200 bp: ie, exons 3, 4, 5, 8, 10). The gels were stained by DNA silver staining (Pharmacia Biotech). SSCA variants were purified and directly sequenced with the primers used for the amplification with a semiautomated sequencer (ABI 377; Perkin-Elmer Applied Biosystems, Foster City, CA), using standard protocols. Mutant alleles of heterozygous patients were cloned by using TA cloning (Invitrogen, La Jolla, CA) and purified and sequenced as previously described.
(2) PCR was performed from genomic DNA in a total volume of 50 μL of 50 mmol/L KCl, 10 mmol/L Tris-HCl pH 8.8, 1.5 mmol/L MgCl2, 0.1% Triton X-100 containing 150 ng of each primer, 200 μmol/L of each dNTP and 1 U of Red Hot DNA polymerase (Advanced Biotechnologies, Epsom, UK). After denaturation at 94°C for 7 minutes, 30 cycles of denaturation: 94°C, 1 minute; annealing: either 50°C or 55°C, 1 minute; extension: 72°C, 3 minutes were performed followed by a final extension at 72°C for 10 minutes. PCR products were labeled by the incorporation of α-33Pd-ATP (56 kBq/50 μL reaction, 377-110 Tbq/mmol; Amersham Life Science, Little Chalfont, UK). SSCA was performed as described by Michaelides et al12 on a 40-cm polyacrylamide gel run at 4°C. PCR products were purified by filtration by using a Microcon 100 spin column (Amicon, Stonehouse, UK) before direct sequencing with the thermosequenase dye terminator cycle sequencing kit (Amersham Life Science) according to the manufacturer’s instructions and analyzed on an Applied Biosystems 373A DNA automated sequencer (Perkin-Elmer Applied Biosystems, Warrington, UK). The primers used for PCR and direct sequence analysis were as in Table 2, with the exception of (5′ > 3′) exon 1, F:TCGCGTTCCAGAATCCAAG, R:AGCACACCAGGGTAGCCG; exon 6, F:AGTCATAAAATGGATCGATTG, R:TTCCCAATAAAACACACCTC; and exon 8, F:TGTTAACCTTTCCGTAGTGG, R:GCTAGGCAACACAGACTCAA.
Allele-specific PCR.
The 822G > A mutation was analyzed by allele-specific PCR amplification using the following primers: (5′ > 3′): GTAATCTCCTATGGAACTTTT and either the wild-type TTGAAAATATGTAAAATTACT or mutant TTGAAAATATGTTTGTAAAATTACC. After denaturation at 94°C for 7 minutes, 30 cycles of denaturation: 94°C, 1 minute; annealing: 53°C, 1 minute; extension: 72°C, 3 minutes were performed followed by a final extension at 72°C for 10 min. The products were analyzed by electrophoresis on 2% agarose gels.
Analysis of intragenic polymorphic markers.
The G3R polymorphism was analyzed by restriction digestion of the exon 1 PCR product with BamH1 (G3, BamH1 site; R3, noBamH1 site). Similarly, the R14Q polymorphism was analyzed by restriction digestion of the exon 1 PCR product with EagI (R14EagI site; Q14, no EagI site). The R117 polymorphism was studied by SSCA of the exon 2 PCR product.
RT-PCR analysis.
Polyadenylated RNA was isolated from EBV transformed lymphocytes and first-strand cDNA synthesis performed according to standard techniques (Amersham Pharmacia Biotech, St Albans, UK) with 200 ng of an ERGIC-53 exon 8 specific antisense primer: 5′-TTTATCCAATTCTTGTTGAAAG-3′. Five microliters of the reaction was used for PCR amplification with an ERGIC-53 exon 6 specific oligonucleotide primer: 5′-AATGATCAATAATGGCTTTACA-3′ and the exon 8 specific primer used for the cDNA synthesis under the following conditions: (1) Initial denaturation 95°C, 5 minutes then 25 cycles of denaturation, 94°C, 30 seconds; (2) Annealing, 57°C, 30 seconds; (3) Elongation, 72°C, 30 seconds, and (4) A final elongation step at 72°C, 10 minutes. A further 5 μL of the reverse transcription (RT) was used for PCR amplification of GAPDH by using the following oligonucleotides: 5′-TGAGTACGTCGTGGAGTCCAC-3′ and 5′-ACCAGGAAATGAGCTTGACA-3′. The products were analyzed by 2% agarose gel electrophoresis.
Western blot analysis.
ERGIC-53 protein was detected by Western blot analysis as previously described1 on EBV transformed cell lines from affected and carrier individuals in families A18, A19, A20, A21, A22, A25, A27, and A29.
RESULTS
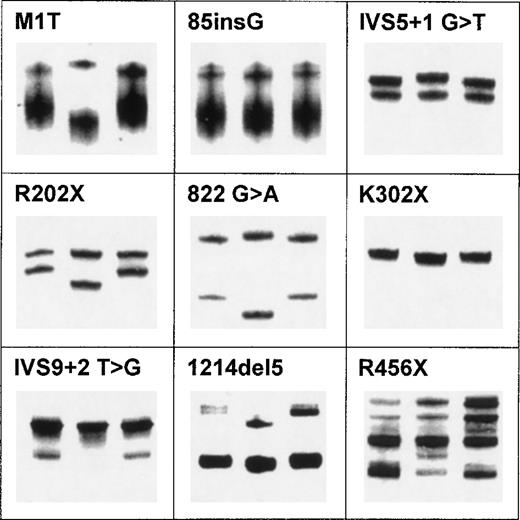
A total of 35 patients, ie, one affected individual per family, were analyzed for mutations in the ERGIC-53 gene. Each of the 13 exons and the adjacent intron-exon boundaries of the ERGIC-53 gene were amplified by PCR and analyzed in two laboratories by SSCA with both small and large format polyacrylamide gels. Representative SSCA patterns of nine mutations are shown in Fig 1. In families for whom no mutation was identified by SSCA, each exon (complete with the flanking intronic sequences) was sequenced.
Examples of SSCA for nine discrete mutations in ERGIC-53. SSCA was performed according to protocol 1 in Materials and Methods. In each panel, the abnormal pattern caused by the mutation is in the middle lane surrounded by two controls.
Examples of SSCA for nine discrete mutations in ERGIC-53. SSCA was performed according to protocol 1 in Materials and Methods. In each panel, the abnormal pattern caused by the mutation is in the middle lane surrounded by two controls.
Many variant SSCA patterns were identified. For each discrete pattern, the appropriate fragment was subjected to nucleotide sequencing. In all cases, a particular SSCA pattern was associated with a unique sequence difference. All heterozygous sequence variants were characterized by direct DNA sequencing of both strands of the PCR products and, in some cases, the PCR products were cloned and sequenced.
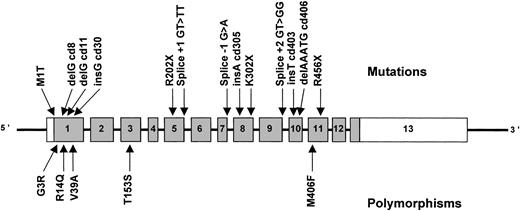
A total of 13 definite mutations, representing 52 of 70 F5F8D alleles, were identified (Table 1, Fig 2). We have confirmed the high degree of allelic heterogeneity suggested by our previous haplotype analysis,7 which is in marked contrast to the founder effect observed in Jewish families by Nichols et al.1 6 The majority of the patients in whom mutations were identified are true homozygotes rather than compound heterozygotes (25 of 26), largely because of the high degree of consanguinity found in the parents of patients with this rare, autosomal recessive disease (Table 1).
Schematic representation of the ERGIC-53 gene showing mutations causing combined F5F8D (above the gene) and normal polymorphisms (below). Exons, indicated by rectangles, are numbered from 1 to 13 and are drawn to scale. The coding portion of the gene is shaded, with the white portion of exon 1 representing the 5′ UTR and the white portion of exon 13 the 3′ UTR, the exact size of which is unknown. The introns are indicated by narrow lines and are not to scale.
Schematic representation of the ERGIC-53 gene showing mutations causing combined F5F8D (above the gene) and normal polymorphisms (below). Exons, indicated by rectangles, are numbered from 1 to 13 and are drawn to scale. The coding portion of the gene is shaded, with the white portion of exon 1 representing the 5′ UTR and the white portion of exon 13 the 3′ UTR, the exact size of which is unknown. The introns are indicated by narrow lines and are not to scale.
Nonsense mutations.
Three different nonsense mutations were found, in exons 5, 8, and 11, accounting for a total of 20 F5F8D alleles (27% of all mutant alleles) (Table 1, Fig 2). One of these, K302X (AAA > TAA), was homozygous in five of the six Pakistani families studied. The other two, R202X and R456X, were CGA to TGA mutations in accordance with the known CpG to TpG hypermutability.13 The R456X was found in both Chinese and Pakistani families on a different haplotype (A19 homozygous for R117R cgg; A27 homozygous for R117R cga) showing recurrence of this mutation.
Deletions and insertions.
A total of six small deletions and insertions leading to disruption of the reading frame and premature termination of translation were identified, on 14 different mutant alleles (Table 1, Fig 2). These were in exon 1, 23delG (in a G4 tract), 31delG (in a G3 tract), 89-90insG (in a G4 tract); in exon 8, 912-913insA (in an A8 tract); in exon 10, 1208-1209insT, and 1214-1218delAAATG (deletion of one of two copies of a repeated pentanucleotide). The 89-90insG mutation, which we found in two Iranian families, is the common mutation of Middle Eastern Jewish families.1
Missense mutation in the initiator ATG.
A mutation of particular interest was found in the translation initiation codon in two Italian families from our study (Table 1, Fig2), and four Italian families in the report by Nichols et al.11 This ATG > ACG mutation is predicted to abolish translation since the next in frame ATG in the coding sequence is found in exon 6, thus leading to the complete absence of ERGIC-53, as confirmed by Western blotting.
Splice site mutations.
Two different splice site mutations predicted to result in severe abnormality of RNA processing and one putative splice site mutation were identified in three different exons accounting for a total of 14 F5F8D alleles (Table 1, Fig 2). In intron 5, a G > T substitution at the invariant GT dinucleotide of the donor splice site (GT > TT) was found in homozygosity in families A34 and A35, both from Italy. In intron 9, the donor splice site invariant dinucleotide GT was mutated to GG in both alleles of the affected members of family A4 from Iran. Both of these mutations are expected to result in abnormal splicing of the ERGIC-53 mRNA.14 Indeed, a different mutation in the same donor splice site of intron 9 (GT > GC) was previously identified in five Sephardic Jewish families, resulting in an apparently complete block of splicing of this intron.1
Another modification, a G > A change in the last nucleotide of exon 7 (822G > A), ie, in the −1 position of the donor splice site consensus sequence of intron 7 was found in homozygosity in four Iranian families: A2, A6, A9, and A12 (Table 1, Fig 2). This mutation does not change the amino acid corresponding to the modified codon as both CCG and CCA code for proline. However, it is possible that abnormal splicing occurs at least in some mRNA molecules because the sequence surrounding the splice site is modified. We screened the DNA of 24 unaffected and the remaining 13 affected Iranians by allele-specific PCR; none carried the mutation.
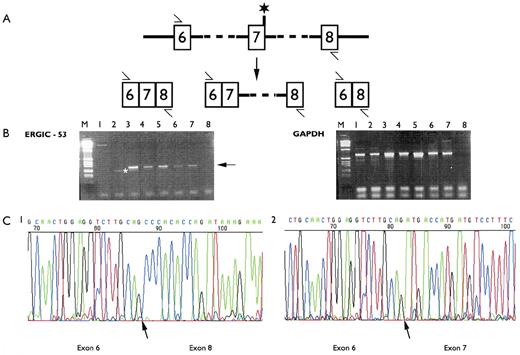
To determine the consequences of this mutation on ERGIC-53 mRNA we isolated polyadenylated RNA from lymphoblastoid cell lines from patients: A2, A12 (both homozygous 822G > A), A24 and A25 (homozygous M1T), and one unaffected individual. cDNA synthesis was primed by using an oligonucleotide corresponding to the antisense sequence of exon 8. RT-PCR between exons 6 and 8 followed by sequencing of the products confirmed that in A2 and A12, exons 6 and 8 were directly contiguous after the skipping of exon 7; no normal cDNA was present. No skipping of exon 7 was observed in the other tested individuals (Fig 3). Skipping of exon 7 leads to absence of ERGIC-53 protein in lymphoblasts (Table 1, A2 and A12).
(A) Cartoon representation of intron 6-intron 8 of the ERGIC-53 gene. The primers used for the RT-PCR analysis are indicated by an arrow and the asterisk indicates the position of the IVS7-1 mutation. Potential RT-PCR products resulting from normal splicing, inclusion of intron 7 or skipping of exon 7 are shown. (B) Agarose gel electrophoresis of the RT-PCR products after amplification of polyadenylated RNA with ERGIC-53 exon 6-exon 8 specific primers and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) specific primers. The GAPDH amplification was a control for mRNA integrity. The cDNA synthesis was primed with the exon 8 specific primer and oligo dT. Lane M, markers; lane 1, individual A2 (homozygous 822G > A); lane 2, individual A12 (homozygous 822G > A); lane 3, human endothelial cell line; lane 4, unaffected individual; lanes 5, 6, and 7, affected individuals with the M1T mutation; lane 8, no DNA control. The arrow marks the normally spliced product. The asterisk marks the incorrectly spliced product. (C) DNA sequence chromatograms of: 1, the incorrectly spliced product, resulting from skipping of exon 7, observed in RT-PCR of polyadenylated lymphoblastoid RNA from individual A12; 2, the major product observed in RT-PCR of polyadenylated RNA from a human endothelial cell line. The exon boundaries are indicated by an arrow.
(A) Cartoon representation of intron 6-intron 8 of the ERGIC-53 gene. The primers used for the RT-PCR analysis are indicated by an arrow and the asterisk indicates the position of the IVS7-1 mutation. Potential RT-PCR products resulting from normal splicing, inclusion of intron 7 or skipping of exon 7 are shown. (B) Agarose gel electrophoresis of the RT-PCR products after amplification of polyadenylated RNA with ERGIC-53 exon 6-exon 8 specific primers and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) specific primers. The GAPDH amplification was a control for mRNA integrity. The cDNA synthesis was primed with the exon 8 specific primer and oligo dT. Lane M, markers; lane 1, individual A2 (homozygous 822G > A); lane 2, individual A12 (homozygous 822G > A); lane 3, human endothelial cell line; lane 4, unaffected individual; lanes 5, 6, and 7, affected individuals with the M1T mutation; lane 8, no DNA control. The arrow marks the normally spliced product. The asterisk marks the incorrectly spliced product. (C) DNA sequence chromatograms of: 1, the incorrectly spliced product, resulting from skipping of exon 7, observed in RT-PCR of polyadenylated lymphoblastoid RNA from individual A12; 2, the major product observed in RT-PCR of polyadenylated RNA from a human endothelial cell line. The exon boundaries are indicated by an arrow.
Polymorphisms.
The SSCA and nucleotide sequence analysis showed several common polymorphisms in the ERGIC-53 gene that result in amino acid substitutions. These substitutions are not associated with nor do they cause F5F8D because they were found in homozygosity in normal individuals, or in affected alleles in which another, deleterious, mutation was also present. These polymorphisms are R14Q (CGG to CAG), V39A (GTC to GCC), T153S (ACT to TCT), and M410L (ATG to TTG) (Fig 2). In addition there is a common CGA to CGG polymorphism at the Arg codon 117 that does not result in an amino acid substitution. Furthermore, a deletion of 2 Ts at nucleotides 16-17 of IVS4 was also commonly observed. These polymorphisms may be useful in linkage studies involving chromosome 18q21. In our study, these polymorphisms allowed us to confirm the compound heterozygosity (ie, two different mutations on the two ERGIC-53 alleles) observed in the Iranian patient A3 (Table1) because this patient was also heterozygous for the R14Q and R117 polymorphisms.
In Iranian family A14, there was a nucleotide substitution G to A at codon 3 resulting in an amino acid substitution glycine to arginine. This substitution is within the hydrophobic 30–amino acid signal sequence. This substitution was not present in the DNA of any other affected individual from the Iranian sample and was not present in 24 unaffected Iranian individuals tested.
Western blot analysis of ERGIC-53 protein levels.
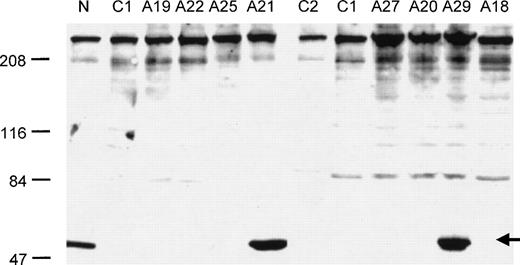
Western blot hybridization confirmed the absence of detectable ERGIC-53 protein for two nonsense mutations (K302X, R456X) and one missense mutation (M1T), (Table 1 and Fig4). It also showed the presence of detectable ERGIC-53 protein in individuals A21 and A29, two patients with no detectable mutation in the ERGIC-53 coding sequence and flanking intron-exon boundaries.
Western blot analysis of ERGIC-53 protein levels in selected families with F5F8D (see Table 1 for the nature of the ERGIC-53 mutation present in each affected individual). N indicates a normal control, lanes C1 and C2 are two different mutations from reference 1 (C1: 86-89insG; C2 IVS9 + 2 T > C). The arrow indicates the position of the ERGIC-53 protein band. Normal levels of ERGIC-53 protein were also found in two unaffected family members from family A19.
Western blot analysis of ERGIC-53 protein levels in selected families with F5F8D (see Table 1 for the nature of the ERGIC-53 mutation present in each affected individual). N indicates a normal control, lanes C1 and C2 are two different mutations from reference 1 (C1: 86-89insG; C2 IVS9 + 2 T > C). The arrow indicates the position of the ERGIC-53 protein band. Normal levels of ERGIC-53 protein were also found in two unaffected family members from family A19.
DISCUSSION
In this study we have analyzed the ERGIC-53 gene in 35 families with F5F8D. We have confirmed that mutations in the ERGIC-53 gene are definitely responsible for the deficiency in 74% of the families analyzed. In contrast to the two distinct founder mutations found to be responsible for the disorder in Sephardic and Middle Eastern Jewish families, we identified 13 distinct mutations accounting for 52 mutant alleles in F5F8D families of multiple ethnic origins. There were 3 different splice site mutations, 6 insertions and deletions resulting in translational frameshifts, 3 nonsense mutations, and a missense mutation in the initiator methionine. In addition we also identified several amino acid polymorphisms.
One of the splice site mutations, found in homozygosity in four Iranian families but absent in 74 other Iranian alleles tested, was situated at position −1 of the donor splice site consensus sequence of intron 7 (822G > A). At this position in the consensus donor splice site, G is found in 78% and A in only 10% of mammalian genes; therefore, we confirmed by RT-PCR analysis that this modification leads to exon skipping and loss of the open reading frame. There are 56 known G to A mutations in the −1 nucleotide of the donor splice site in human genes associated with disorders14 (Human Gene Mutation Database; http://www.uwcm.ac.uk/uwcm/mg/hgmd0.html), including factor V, factor VII, factor VIII, and factor IX. Abnormal splicing was studied in several of these cases and the resulting exon skipping ranged from 30% to 100%. In addition, use of cryptic donor splice sites has been reported as resulting in translational frameshift and abnormal protein.14 16
One of the sequence differences, G3R, which was found in homozygosity in one Iranian family (A14) but was absent in 80 other Iranian alleles tested, is within the 30-amino acid signal sequence and may be either a polymorphism or a causative mutation. This position is not highly conserved. For example, the rat sequence contains valine at position 3 (Genbank no. U44129). The G3R substitution does not change the probability that the first 30 amino acids is a signal peptide (http://psort.nibb.ac.jp; http://www.cbs.dtu.dk/services/SignalP). However, it is now recognized that signal sequences have a more complex structure than previously anticipated, allowing for multiple and independent interactions with the translocation machinery17(see Note Added in Proof).
There appears to be little or no correlation between the precise mutation and the severity of the F5F8D phenotype because (1) all identified mutations are predicted to lead to an absence of mature protein and (2) some recurrent mutations were found in association with strikingly different levels of factor V and factor VIII (compare, for example, families A6 and A12 or families A26 and A30).
The human ERGIC-53 is a 53-kD transmembrane resident protein of the ER-Golgi intermediate compartment, a distinctive vesicular organelle in the secretory pathway.18 The protein is homologous to leguminous lectins, presenting mannose-selective and calcium-dependent binding.19-21 Additional ERGIC-53 homologues have been identified in the rat, Xenopus laevis, andCaenorhabditis elegans22,23 (Genbank accession no. Z81097). The importance of ERGIC-53 protein in the efficient secretion of the coagulation factors V and VIII has clearly been established by its causative role in F5F8D (data presented in this report, the accompanying report by Nichols et al,11 and reference 1). Factor V and factor VIII are homologous proteins that share a conserved domain structure, having derived from a common ancestor molecule, with the A and C domains of the two factors showing 40% sequence identity.24 25 Both factor V and factor VIII are subject to extensive posttranslational modification, which includes the addition of multiple oligosaccharide residues, predominantly in the B-domain. Therefore, ERGIC-53 most probably interacts with the B-domains of factor V and factor VIII via a lectin like linkage. All of the mutations in ERGIC-53 described to date are null mutations. However, there is still some factor V and factor VIII activity in the plasma of the affected individuals, suggesting that there may be several bypass mechanisms for the transport of factor V and factor VIII from the ER to the Golgi. ERGIC-53 may also be required for the secretion of many other glycoproteins whose loss is not sufficient to cause a clinically recognizable phenotype.
Although we have identified approximately 74% of the mutations responsible for F5F8D in our patient sample, 26% of the mutations have not been identified despite screening by SSCA with two different experimental conditions and sequencing the entire coding sequence and intron-exon boundaries of the ERGIC-53 gene. These results are similar to those reported by Nichols et al11 in the accompanying report, with no mutations found in the ERGIC-53 gene in 8 of 19 families. A number of explanations are possible for this incomplete detection. The missing mutations may be located within intronic regions that were not analyzed in the current study, leading, for example, to aberrant splicing or other RNA anomalies, or in regulatory regions situated up to several hundred kilobases away from the ERGIC-53 gene. Such mutations could only be identified by detailed investigation of ERGIC-53 transcripts in the remaining families.
However, a number of observations strongly support an alternative explanation, which is the existence of mutations at other, currently unidentified, loci. First, in all but one of the consanguineous families in whom we have identified ERGIC-53 mutations, as expected the patients are true homozygotes. In contrast, in two of the three consanguineous (first-cousin marriage) families in whom no mutations were identified, the affected individuals are heterozygous for the ERGIC-53 gene according to intragenic and flanking polymorphisms7 (families A5: heterozygous for R14Q and R117, and A13: heterozygous for R14Q and M410L). Secondly, affected individuals from another two families with no identified ERGIC-53 mutations (A21 and A29) have normal levels of ERGIC-53 according to Western blotting. The existence of further loci responsible for F5F8D is currently under investigation.
ACKNOWLEDGMENT
We thank all members of these families for their cooperation and for donating blood samples for our study. We are grateful to Dr L. Tengborn, Salgrenska University Hospital, Göteborg, Sweden; Dr E.A.C. Chalmers, Royal Hospital for Sick Children, Glasgow, UK; Dr S.D. Wright, Watford General Hospital, Watford, UK; Dr P. Giangrande, Oxford Haemophilia Centre, Oxford, UK; Dr M. Makris, Sheffield Haemophilia and Thrombosis Centre, Sheffield, UK; Dr L.A. Parapia, Bradford Royal Infirmary, Bradford, UK; Dr J. Wilde, Haemophilia Unit, University Hospital, Birmingham, UK; Dr M.J. Strevens, Walsgrave Hospital, Coventry, UK; Dr Steven P. Field, South African Blood Transfusion Service, Johannesburg, South Africa. Prof L.C. Chan, University of Hong Kong, Hong Kong; Dr M. Morfini, Haemophilia Centre, Florence, Italy; Dr A. Gringeri, Haemophilia and Thrombosis Centre, Milan, Italy; Dr J. Reynaud, Hopitaux de St-Etienne, France, for the collection of families, and to Drs S. Zeinali, M. Akhtari, M. Lak, and R. Sharifan, Haemophilia Center and Haematology Department, Imam Khomeini Hospital, Tehran, Iran for their generous assistance to EGDT and FP. We also thank Dr H-P. Hauri (Biozentrum, Basel, Switzerland) for the gift of the anti–ERGIC-53 antibody and ERGIC-53 cDNA. We thank Dr Philip Chandler for his help in establishing lymphoblastoid cell lines and Dr Anni Schönbörner for DNA extractions.
M.N.-A. and K.M.J. contributed equally to this study.
M.N.-A. is a recipient of a Marie Heim-Voegtlin grant from the Swiss National Science Foundation. Supported by National Institutes of Health (NIH) Grant No. HL38165, funds from the University and Cantonal Hospital of Geneva and grant FN 31-49815.96 from the Swiss National Science Foundation (M.A.M.). Haemostasis Research Group is supported by the UK Medical Research Council. W.C.N. and D.G. are supported by NIH Grants No. R01-HL39693 and P01-HL57346. D.G. is a Howard Hughes Medical Institute Investigator.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
NOTE ADDED IN PROOF
Immunofluorescence analysis of cells expressing introduced genes for ERGIC-53 with wild-type or mutated (G3R) leader sequences showed no difference in signal peptide cleavage or distribution of protein. Hence, G3R cannot be the cause of disease in family A14. (Hans-Peter Hauri, personal communication, Basel, Switzerland.)
Author notes
Address reprint requests to E.G.D. Tuddenham, MD, Haemostasis Research Group, MRC Clinical Sciences Centre, Imperial College School of Medicine, DuCane Rd, London W12 ONN, e-mail: etuddenh@hgmp.mrc.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal