Protein tyrosine phosphatases act in conjunction with protein kinases to regulate the tyrosine phosphorylation events that control cell activation and differentiation. We have isolated a previously undescribed human phosphatase, Lyp, that encodes an intracellular 105-kD protein containing a single tyrosine phosphatase catalytic domain. The noncatalytic domain contains four proline-rich potential SH3 domain binding sites and an NXXY motif that, if phosphorylated, may be recognized by phosphotyrosine binding (PTB) domains. Comparison of the Lyp amino acid sequence with other known proteins shows 70% identity with the murine phosphatase PEP. The human Lyp gene was localized to chromosome 1p13 by fluorescence in situ hybridization analysis. We also identified an alternative spliced form of Lyp RNA, Lyp2. This isoform encodes a smaller 85-kD protein with an alternative C-terminus. The lyp phosphatases are predominantly expressed in lymphoid tissues and cells, with Lyp1 being highly expressed in thymocytes and both mature B and T cells. Increased Lyp1 expression can be induced by activation of resting peripheral T lymphocytes with phytohemagglutinin or anti-CD3. Lyp1 was found to be constitutively associated with the proto-oncogene c-Cbl in thymocytes and T cells. Overexpression of lyp1 reduces Cbl tyrosine phosphorylation, suggesting that it may be a substrate of the phosphatase. Thus, Lyp may play a role in regulating the function of Cbl and its associated protein kinases.
PROTEIN TYROSINE phosphorylation, a key mechanism of cellular signal transduction, is regulated by the action of both protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPases). Originally, PTKs were believed to control the process of tyrosine phosphorylation, with a small number of PTPases playing largely housekeeping roles. Unexpectedly, the structural diversity of the growing number of PTPases has called this idea into question, and it has become apparent that PTPases have important roles in the regulation of growth and differentiation in both normal and neoplastic cells.1 2
All PTPases contain a catalytic domain of approximately 200 to 300 residues, including a subset of highly conserved amino acids that play a role in substrate recognition and tyrosine dephosphorylation.3 The PTPase family can be divided broadly into two major classes: membrane-bound receptors and intracellular phosphatases,4,5 both dividing into subfamilies based on their sequence similarities and noncatalytic domain structural motifs.6,7 The receptor PTPases may contain one or two intracellular phosphatase domains and often Ig-like and fibronectin-like extracellular regions6 that play a role in cell-cell or cell-matrix interactions.8 Some receptor PTPases appear to participate in homophilic and hetrophilic binding interactions, which is suggestive of a role in cell guidance and contact inhibition.7 8
The nonreceptor phosphatases display various intracellular localizations determined by amino acid sequences outside the catalytic domain.9-11 Some also contain conserved noncatalytic domains such as SH2 and SH3, allowing them to interact with tyrosine phosphorylated proteins and proteins containing proline-rich sequences, respectively.12,13 Cytoplasmic PTPases have been found associated with a variety of PTKs, including Csk and the Jak kinases and a number of cytokine and antigen receptors.7 13-15
Within the immune system several lines of evidence indicate that PTPases are essential for lymphocyte development and activation. CD45, a transmembrane phosphatase expressed exclusively in hematopoetic cells,16 is required for antigenic activation of B and T lymphocytes.17,18 CD45-deficient mice show that CD45 also plays a pivotal role in thymic development and T-cell apoptosis.19,20 Recent studies have shown that the hematopoetic-specific intracellular phosphatase SHP-1 negatively regulates signaling through the B-cell receptor, FcγRIIB1,21 and the inteleukin-3 (IL-3) receptor β chain.22 SHP1 also participates in T-cell signaling events through dephosphorylation of the T-cell receptor, p56lck, and Zap-70.23 Mutations in the murine motheaten locus, encoding the SHP1 protein, result in severe combined immunodeficiency and systemic autoimmunity, as well as many other hematopoietic abnormalities.24 Furthermore, expression of HePTP, a hematopoetic-specific cytoplasmic PTPase, is induced in lymphocytes stimulated by phytohemagglutinin (PHA), concavalin A, lipopolysaccharide, and anti-CD3,25 which is suggestive of a role in mature lymphocyte signaling pathways. All of these studies suggest a critical role for phosphatases not only in regulating signaling, but also in the development of the immune system.
Our search for PTPases involved in lymphocyte growth and development led to the isolation of a novel human cytoplasmatic phosphatase that is predominantly expressed in lymphoid cells, designated lymphoid phosphatase (Lyp). The Lyp gene was localized to human chromosome 1p13. The Lyp protein is closely related (70% homology) to the murine phosphatase PEP.12 Lyp is expressed in both immature and mature B and T cells. The level of Lyp protein expression was found to increase significantly upon stimulation of resting peripheral T lymphocytes with PHA or anti-CD3. We also describe an isoform of Lyp (Lyp2) that is the product of C-terminal alternative RNA splicing and that demonstrates a different pattern of RNA expression to Lyp1. In contrast to Lyp1, Lyp2 protein could only be detected in resting T cells.
Upon anti-CD3 stimulation of human thymocytes, a 116-kD phosphorylated protein was found to become associated with Lyp1. This protein was subsequently identified as the molecular adapter protein c-Cbl.26 Furthermore, expression of Lyp1 in COS cells results in a reduction in endogenous cbl phosphorylation, suggesting a role for Lyp in regulating the function of cbl in the TCR signaling pathway.
MATERIALS AND METHODS
Polymerase chain reaction (PCR) and subcloning of PTPases clones.
Total RNA was prepared from thymocytes using Trizol reagent (GIBCO-BRL, Gaithersburg, MA). First-strand cDNA synthesis was performed with oligo-dT primer and Superscript II RT (GIBCO-BRL). This was used as a template for PCR amplification with Taq DNA polymerase (Perkin Elmer Cetus, Norwalk, CT) and the following degenerate primers: PTP1, GCGGATCCTCIGA(C/T)TA(C/T)AT(A/C/T)AA(T/C)GC (sense); and PTP2, GCGAATTCCCIACICCIGC(A/G)CT(G/A)CA(G/A)TG (antisense). These degenerate primers are designed to match two highly conserved sequences within PTPase catalytic domains, XDYINA and HCSAGI/VG, respectively. PCR was performed as follows: five cycles of 60 seconds at 94°C, 30 seconds at 37°C, and 60 seconds at 72°C and a further 25 cycles with an annealing temperature of 45°C. The PCR products (∼400 bp) were isolated, cloned, and sequenced.
Isolation and sequencing of Lyp1 and Lyp2 cDNA clones.
An oligo-dT–derived λgt10 DNA library from human thymocytes was screened with a 32P-labeled 430-bp Lyp1 fragment obtained by PCR. λ Plaques were transferred to ICN Biotrans nylon filters (ICN, Costa Mesa, CA) and screened by hybridization at 65°C in 5× SSC, 5× Denhart’s solution, 0.1% sodium dodecyl sulfate (SDS).27 Phage DNA was prepared from positive plaques, and cDNA inserts were excised, subcloned into pUC19, and sequenced. To obtain the complete Lyp1 cDNA, secondary and tertiary library screenings were performed with 1.3-kb and 0.6-kb partial Lyp cDNA clones isolated in the first screening (Fig 1). One clone (P5) from the second screening was found to contain the carboxy-terminal sequence of the alternatively spliced form of Lyp1 (Lyp2).
Schematic diagram of Lyp1 and Lyp2 deduced from the cDNA clones. Boxes indicate the open reading frame, with thin lines representing the 5′ and 3′ untranslated regions. The PTPase catalytic domain is colored in gray. The six overlapping cDNA clones obtained from a human thymus cDNA library (bold black lines) are shown under the schematic structures of the cDNAs.
Schematic diagram of Lyp1 and Lyp2 deduced from the cDNA clones. Boxes indicate the open reading frame, with thin lines representing the 5′ and 3′ untranslated regions. The PTPase catalytic domain is colored in gray. The six overlapping cDNA clones obtained from a human thymus cDNA library (bold black lines) are shown under the schematic structures of the cDNAs.
Fluorescence in situ hybridization (FISH) detection and image analysis.
A 1.8-kb Lyp cDNA fragment was used as a probe to examine the chromosomal location of the human Lyp gene. Biotinylated Lyp probe was prepared by nick translation for FISH to normal human lymphocyte chromosomes (counterstained with propidium iodide and 4′,6-diamidin-3-phenylindol-dihydrochloride [DAPI] according to published methods28,29). The probe was detected with avidin-fluorescein isothiocyanate (FITC), followed by biotinylated antiavidin antibody and avidin-FITC. Images of metaphase preparations were captured by a thermoelectrically cooled charge coupled camera (Photometrics, Tucson, AZ). Separate images of DAPI-banded chromosomes30 and FITC-targeted chromosomes were obtained and merged electronically using image analysis software (courtesy of Tim Rand and David Ward, Yale University, New Haven, CT) and pseudo-colored blue (DAPI) and yellow (FITC) as described by Boyle et al.29 The band assignment was determined by measuring the fractional chromosome length and by analyzing the banding pattern generated by the DAPI counterstained image.31
Northern blot analysis.
Total RNA was extracted from thymocytes using Trizol reagent (GIBCO-BRL). Poly A+ RNA was isolated by two passages through an oligo(dT) column. Two micrograms of Poly A+ RNA per lane was electrophoresed in a 1% agarose formaldehyde gel and capillary blotted onto nitrocellulose filters. Filters and human multiple tissue poly A+ RNA membrane (Clontech, La Jolla, CA) were hybridized overnight at 42°C with32P-labeled Lyp cDNA probes in 50% formamide, 5× SSC, 5× Denhart’s solution, 0.1% SDS, 50 mmol/L Na2HPO4, pH 6.5, and denatured Salmon sperm DNA (100 μg/mL). After hybridization, the final wash was performed in 0.2% SSC, 0.1% SDS at 55°C.27
Lymphocyte isolation.
Thymuses were obtained from children undergoing open heart surgery. Mononuclear cells were isolated by Ficoll-Hypaque gradient centrifugation. Adherent cells were removed by incubation to plastic dishes for 60 minutes at 37°C. The resulting thymocytes are typically greater than 95% CD3+. Lymphocytes were isolated from tonsil tissue or from peripheral blood of healthy volunteers by Ficoll-Hypaque gradient centrifugation, followed by rosetting with neuraminidase-treated sheep red blood cells (RBCs) to isolate T lymphocytes. After isolating rosettes on Ficoll-Hypaque, T cells were released by ACT treatment (0.75% NH4Cl in 20 mmol/L Tris, pH 7.2) of the rosettes to lyse the RBCs. The buffy layer, containing the B cells, was washed three times with phosphate-buffered saline (PBS). The resultant T lymphocytes are typically 98% to 99% CD3+ and the B lymphocytes from tonsils are typically 85% to 90% CD19+. To induce activation and maturation of peripheral T lymphocytes, 25 × 106 T cells were stimulated with 2.5 μg/mL of anti-CD3 (Calbiochem) or 10 μg/mL of PHA (GIBCO-BRL) for 24 to 48 hours at 37°C in RPMI (10% fetal calf serum [FCS]).
Cell lines.
The OCI/AML3 cell line was kindly provided by Dr Mark D. Minden (Ontario Cancer Institute, Toronto, Ontario, Canada). The origin and properties of this myeloid cell line have been previously described.32 The G2 pre-pre-B–cell line was derived from a patient with acute lymphocytic leukemia.33 All of the other cell lines used for this study were obtained from the American Type Culture Collection (Rockville, MD). All cells were maintained in RPMI 1640 containing 10% FCS.
Antibodies.
Rabbit polyclonal antibodies were raised to a mixture of two peptides of LyP with the amino acid sequences RTKSTPFELIQQR and SKMSLDLPEKQDG. These peptides were chosen from a potentially exposed area, as predicted by Hopp and Woods,33a in the noncatalytic domain. A second polyclonal antibody was raised to a bacterial fusion protein of the catalytic domain of LyP (Pet vector; Novagen, Madison, WI). After careful testing, these antibodies were used for immunoprecipitation and Western blotting. T7 antibody was purchased from Novagen (WI); anti-cbl, anti-Jak3, and anti-p110 were purchased from Santa Cruz Biotech (Santa Cruz, CA); and antiphosphotyrosine was purchased from from UBI (Lake Placid, NY).
Transfection of COS-7 cells.
COS-7 cells (0.5 × 106) were transfected with 5 μg plasmid DNA in 50 μL of Lipofectamine (GIBCO-BRL) for 5 hours according to the manufacturer’s instructions. Forty-eight hours after transfection, the COS-7 cells were harvested and solubilized in cold lysis buffer (20 mmol/L Tris, pH 7.5, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP 40, and 1 mmol/L phenylmethyl sulfonyl fluoride [PMSF]).
Immunoprecipitation and Western blotting.
One percent NP-40 cell lysates were precleared by centrifugation. Immunoprecipitation of T7-tagged Lyp was performed by the addition of 1 μg of T7 antibody or by the addition of 5 μL of the lyp antiserum followed by the addition of 20 μL of a 50:50 suspension of protein G sepharose (Pharmacia, Uppsala, Sweden) and incubation overnight at 4°C. Immunoprecipitates were washed three times with lysis buffer and separated by 6% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were electrophortically transferred to Hybond C Super nitro-cellulose membrane (Amersham Life Science, Arlington Heights, IL). Membranes were blocked with 5% nonfat milk and blotted with anti-T7 (1:10,000) or with anti Lyp (1:800). Detection was performed with horseradish peroxidase-conjugated second antibodies from Amersham Life Science and chemiluminescence reagent from Kirkeggard & Perry Laboratories (Gaithersburg, MD).
Intracellular localization by indirect immunofluorescence.
Lyp1 and Lyp2 were inserted into the pCDNA3 eucaryotic expression vector (Invitrogen, San Diego, CA) and a T7 tag or HA epitope (YPYDVPDYA), as a three-tandem repeat, was inserted at the 5′ end of the coding sequences. Constructs were verified by sequencing. COS-7 cells were transfected with 2 μg DNA and 17 μL of lipofectamine for 5 hours, incubated on sterile cover slips in 6-well plates (0.3 × 106/plate) in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FCS, and stained 48 hours posttransfection. The COS-7 cells were then washed in PBS and fixed for 30 minutes at room temperature in 2% paraformaldehyde. Cell permeabilization was performed with 0.1% Triton X100 and, after blocking nonspecific sites with 5% donkey serum, the cells were incubated with monoclonal anti-HA (1:1,000) from Baco-Berkely (Richmond, CA) for 60 minutes at room temperature. The cells were washed and exposed for 45 minutes to cy3-conjugated affinipure Donkey antimouse IgG (1:1000 in PBS) from Jackson Immunoresearch Laboratories, Inc. (West Grove, PA). After 3 to 4 washes, immunoreactivity was detected by fluorescence microscopy.
Phosphatase assay.
The synthetic peptide Raytide was phosphorylated according to the method described by Guan et al34 on tyrosine by p60src (Oncogene Research Products, Cambridge, MA) as follows: 10 μg Raytide in 50 mmol/L HEPES, pH 7.5, 10 mmol/L MgCl2, 0.067% β-mercapto-ethanol, 0.05 mmol/L ATP was incubated with 300 μCi γ33P ATP/mL and 2 μg p60src in a final volume of 30 μL. The reaction was allowed to proceed for 30 minutes at 30°C and was stopped by the addition of 120 μL 10% phosphoric acid.
The sample was spotted onto two 1 × 1 cm sheets of P81 phosphocellulose paper and extensively washed with 0.5% phosphoric acid. Phosphorylated peptide was eluted twice with 1 mL 500 mmol/L (NH4)2CO3, lyophilized, and resuspended in 100 μL H2O.
The phosphorylated substrate was used in the phosphatase assay as described by Stueli et al.35 The phosphatase assay mixture (50 μL) contains 5 μL of ×10 phosphatase buffer (250 mmol/L HEPES, pH 7.3, 50 mmol/L EDTA, 100 mmol/L dithiothreitol), 5 μL of radioactive substrate (Raytide), and 5 μL of sample (Lyp immunoprecipitate) and H2O to final volume.
The assay was allowed to proceed at 30°C for the indicated time and the reaction was terminated by the addition of 750 μL of a charcoal mixture (0.9 mol/L HCl, 90 mmol/L sodium pyrophosphate, 2 mmol/L NaH2PO4, 4% vol/vol Norit A). After centrifugation, the free 33P in the supernatant was measured.
RESULTS
Isolation of novel human PTPases.
To identify novel members of the PTPase gene family that are expressed in thymocytes, we used a PCR-based approach with degenerate oligonucleotides directed at conserved regions of the PTPase catalytic domain. We amplified a fragment of approximately 400 bp from thymocyte cDNA and identified clones corresponding to seven different phosphatases. Six clones were identical to previously isolated human phosphatases: PTP-PEST,36 PTP1B,37TCPTP,38 HPTPδ,6 CD45, and PTPMEG2.39 A seventh clone had no known human homologue but was 90% homologous to a portion of the catalytic domain of the murine phosphatase PEP.12 This clone was used to screen a human thymocyte cDNA library. The first screening isolated two overlapping clones: P1 and P2 (Fig 1). Clone P2 was used to isolate a further three overlapping clones, P3, P4, and P5, from the cDNA library. Assembly of the five overlapping clones showed a single cDNA of 2,300 bp containing an open reading frame (ORF) of 2,076 bp, predicting a protein of 692 amino acids. The sequence surrounding the putative ATG/methionine start codon contained a purine (A) at position −3 and G at +4, both regarded as important criteria for an eucaryotic initiation site.40 The N-terminal region of the amino acid sequence (Fig 2) contained a single PTPase catalytic domain characterized by the conserved sequence motif (I/V)HCXXGXXRS/T. This sequence is thought to form the phosphate binding pocket for substrate, is found in all PTPases, and is essential for their enzymatic activity.3 In addition to the five overlapping clones, a single 1-kb clone was isolated (P6; Fig 1) with the 200 bp of its 5′-end overlapping nucleotides 1950 through 2055 of the complete cDNA previously isolated. However, this was followed by an alternative 700 bp, encoding an ORF totalling 2,424 bp (808 amino acids). The long and short forms share nucleotides 1 through 2097 but contain alternative C-terminal sequences. We have designated these forms Lyp1 (long) and Lyp2 (short). We suggest that LyP2 is an alternative spliced isoform of Lyp1.
Alignment of Lyp1 and PEP amino acid sequences. The PTPase domain is indicated by brackets. An arrow indicates the end of the amino acid sequence shared by Lyp1 and Lyp2 and the beginning of the unique C-terminal sequence of Lyp1. The NXXY motif is indicated by a line above the sequence. The four potential SH3 domain binding sites are also indicated (asterisks). A consensus sequence is shown below the alignment. The unique seven amino acids of Lyp2 are shown in the box below the alignment.
Alignment of Lyp1 and PEP amino acid sequences. The PTPase domain is indicated by brackets. An arrow indicates the end of the amino acid sequence shared by Lyp1 and Lyp2 and the beginning of the unique C-terminal sequence of Lyp1. The NXXY motif is indicated by a line above the sequence. The four potential SH3 domain binding sites are also indicated (asterisks). A consensus sequence is shown below the alignment. The unique seven amino acids of Lyp2 are shown in the box below the alignment.
Hydropathy analysis indicates that Lyp contains no obvious signal sequence or hydrophobic segments and is therefore likely to be an intracellular protein. The N-terminal regions of Lyp1 and Lyp2 contain a single phosphatase catalytic domain. Amino acid sequence analysis showed an overall identity of 70% between Lyp1 and the murine phosphatase PEP (89% within the catalytic domain and 61% within the noncatalytic portion), whereas the homology between the phosphatase domain of Lyp1 and other PTPases varies between 30% and 60%. On the basis of this analysis, Lyp1 and PEP certainly belong to the same phosphatase family and may possibly be homologues.
Chromosomal mapping of the Lyp gene.
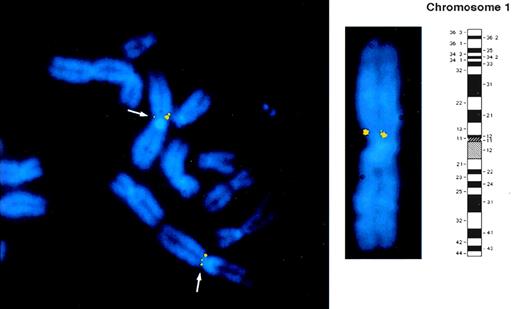
FISH hybridization was performed using a Lyp 1.8-kb cDNA probe to determine the chromosomal localization of the Lyp gene. The regional assignment of this cDNA probe was determined by the analysis of 40 well-spread metaphases. Positive hybridization signals at the short arm of human chromosome 1 in region p13 (shown schematically in Fig 3) were noted in approximately 10% of the cells. The band assignment was determined by measuring the fractional chromosome length and by analyzing the banding pattern generated by DAPI counterstained image. The low frequency of hybridization obtained with this probe is commonly seen with small cDNA probes of this size. Signals were visualized on both homologues in 90% of the positive spreads (Fig 3). No fluorescence signal was seen on any other chromosome, implying that the human LyP gene is located on chromosome 1 in the p13 region.
Regional mapping of the Lyp gene by FISH to normal human lymphocyte chromosomes counterstained with DAPI. Biotinylated cDNA probe was detected with avidin-FITC. Separate images of DAPI counterstained metaphase chromosomes and of LyP cDNA probe hybridization signals were captured and overlaid electronically as described in Materials and Methods. Part of a representative metaphase preparation is shown to indicate the position of the Lyp probe FISH signals that are visible as two yellow fluorescent spots on the p arm of chromosome 1. A DAPI banded chromosome 1 together with schematic ideogram is shown to indicate that the Lyp1 probe hybridizes to band 1p13.
Regional mapping of the Lyp gene by FISH to normal human lymphocyte chromosomes counterstained with DAPI. Biotinylated cDNA probe was detected with avidin-FITC. Separate images of DAPI counterstained metaphase chromosomes and of LyP cDNA probe hybridization signals were captured and overlaid electronically as described in Materials and Methods. Part of a representative metaphase preparation is shown to indicate the position of the Lyp probe FISH signals that are visible as two yellow fluorescent spots on the p arm of chromosome 1. A DAPI banded chromosome 1 together with schematic ideogram is shown to indicate that the Lyp1 probe hybridizes to band 1p13.
Lyp2 is produced by alternative RNA splicing of the Lyp1 message.
To confirm the hypothesis that LyP2 was produced by alternative splicing of Lyp1 RNA, three oligonucleotide-matching sequences around the putative splicing site were used in PCR amplifications on a genomic DNA template (Fig 4A). Oligonucleotide 1 corresponded to the common nucleotides 2076-2097 of Lyp1 and Lyp2 (Fig 4), olignucleotide 2 to Lyp2 untranslated area adjacent to the stop codon (nucleotides 2150-2168), and olignucleotide 3 to sequence immediately downstream of primer 1 in lyp1 (nucleotides 2098-2120). The resultant PCR products are shown in Fig 4B. PCR with primers 1 and 3 created an approximately 3.5-kb fragment, suggesting the presence of an intron between the primers. However, PCR with primers 1 and 2 resulted in a much smaller fragment of 100 bp, the size expected from Lyp2 cDNA sequence. Upon sequencing, the 5′ end of the 3.5-kb fragment was found to contain the alternative C-terminus, stop codon, and untranslated nucleotide sequence of Lyp2 (Fig 4C). This clearly demonstrated that LyP1 and LyP2 are the alternatively spliced transcripts of a single gene. Whereas the 3.5-kb intron is spliced out of the Lyp1 form, this does not occur in the Lyp2 isoform, and as a result only 7 amino acids are added and an alternative stop codon is used.
Lyp2 is a result of alternative splicing of the Lyp1 gene. (A) A schematic map of the PCR strategy used. Primer 1 corresponds to the last 20 nucleotides shared by both the Lyp1 and Lyp2 sequences, primer 2 to Lyp2 untranslated area, and primer 3 to the beginning of the unique Lyp1 sequence, immediately downstream of primer 1 (see also [C]). (B) The results of the PCR amplification on genomic DNA. Lane 1, DNA ladder; lane 2, a product of 3.5 kb was amplified with primers 1 and 3; lane 3, a product of 100 bp was amplified with primers 1 and 2. (C) Schematic map of Lyp1 splicing. The sequences before the vertical line represent the splice donor site, whereas the nucleotide sequences after it are the Lyp1 intronic sequence that code for the unique C-terminal seven amino acids, stop codon (asterisk), and untranslated sequence (lower case letters) of Lyp2. An open box represents the common cDNA sequence shared by Lyp1 and Lyp2; the solid and the light gray boxes represent the unique sequences of Lyp1 and Lyp2, respectively.
Lyp2 is a result of alternative splicing of the Lyp1 gene. (A) A schematic map of the PCR strategy used. Primer 1 corresponds to the last 20 nucleotides shared by both the Lyp1 and Lyp2 sequences, primer 2 to Lyp2 untranslated area, and primer 3 to the beginning of the unique Lyp1 sequence, immediately downstream of primer 1 (see also [C]). (B) The results of the PCR amplification on genomic DNA. Lane 1, DNA ladder; lane 2, a product of 3.5 kb was amplified with primers 1 and 3; lane 3, a product of 100 bp was amplified with primers 1 and 2. (C) Schematic map of Lyp1 splicing. The sequences before the vertical line represent the splice donor site, whereas the nucleotide sequences after it are the Lyp1 intronic sequence that code for the unique C-terminal seven amino acids, stop codon (asterisk), and untranslated sequence (lower case letters) of Lyp2. An open box represents the common cDNA sequence shared by Lyp1 and Lyp2; the solid and the light gray boxes represent the unique sequences of Lyp1 and Lyp2, respectively.
Predominantly lymphoid expression of Lyp RNA and differences between Lyp1 and Lyp2.
Northern blot analysis of mRNA from various human tissues using a Lyp cDNA probe common to both Lyp isoforms showed a major transcript of approximately 4.4 kb in all of the lymphoid tissues examined (Fig 5A). Substantial levels of Lyp mRNA were detected in spleen, thymus, tonsil, and B and T lymphocytes. In contrast, Lyp transcripts were not detected in prostate, ovary, testis, or colon tissues (or other human tissues, including heart, lung, brain, placenta, or liver; data not shown). However, a low level of Lyp expression could be detected in small intestine and appendix mucosa, probably due to the presence of contaminating lymphocytes. From its expression pattern in normal human tissues and cells, Lyp appears to be a predominantly lymphoid phosphatase, although a low level of expression could also be detected in the monocyte cell line, U937. Myeloid (OCI/AML3) and erythrolukemia (K562) cell lines displayed little to no expression.
Expression profile of Lyp1 and Lyp2 transcripts. (A) Two micrograms of poly A+ RNA from various human tissues and cell lines (OCI/AML3, acute myeloblastic leukemia cell line; K562, erythroleukemia cell line; and U937, monocytes cell line) were hybridized with a 1.3-kb cDNA probe common to both Lyp1 and Lyp2 (exposure time, 7 days) and with actin (exposure time, 24 hours). (B) RNA from immune relevant human tissues (Clontech) was blotted first with a cDNA probe from the unique 280-bp 3′ nucleotides sequence of Lyp2 (including the untranslated sequence) and then, after stripping, with a 600-bp cDNA probe from the unique 3′ nucleotide sequence of Lyp1 (exposure time, 7 days) and with actin (exposure time, 24 hours). The sizes of the RNA markers are indicated in kilobases.
Expression profile of Lyp1 and Lyp2 transcripts. (A) Two micrograms of poly A+ RNA from various human tissues and cell lines (OCI/AML3, acute myeloblastic leukemia cell line; K562, erythroleukemia cell line; and U937, monocytes cell line) were hybridized with a 1.3-kb cDNA probe common to both Lyp1 and Lyp2 (exposure time, 7 days) and with actin (exposure time, 24 hours). (B) RNA from immune relevant human tissues (Clontech) was blotted first with a cDNA probe from the unique 280-bp 3′ nucleotides sequence of Lyp2 (including the untranslated sequence) and then, after stripping, with a 600-bp cDNA probe from the unique 3′ nucleotide sequence of Lyp1 (exposure time, 7 days) and with actin (exposure time, 24 hours). The sizes of the RNA markers are indicated in kilobases.
To further characterize the expression of the Lyp isoforms, Northern blots were performed with a Lyp2-specific cDNA probe on human mRNA from lymphoid and hematopoietic tissues. This showed a single 5.2-kb transcript in all of the tissues examined, with the highest level of expression found in fetal liver (Fig 5B). Subsequent blotting of the same membrane with a Lyp1-specific probe showed the dominant 4.4-kb transcript previously observed. Lyp1 demonstrated a high level of expression not only in the mature lymphoid tissues, but also in the thymus. In contrast to Lyp2, Lyp1 mRNA could not be detected in fetal liver and only a low level of expression could be seen in bone marrow.
Characterization and intracellular localization of the Lyp1 and Lyp2 proteins.
To determine the actual size of the Lyp1 and Lyp2 proteins, the full-length cDNAs were cloned by PCR from oligo-dT selected mRNA, tagged with a T7 epitope, and transfected into COS-7 cells. The deduced amino acid sequences of Lyp1 and Lyp2 predict molecular weights of 92 and 78 kD, respectively. Immunoprecipitation of the transfected proteins with anti-T7 or anti-LyP antibodies and blotting with the T7 antibody showed the protein Lyp2 to have an apparent molecular weight of 85 kD, which is slightly higher than the predicted molecular weight. Two proteins with apparent molecular weights of 96 and 105 kD were observed in COS-7 cells transfected with the Lyp1 cDNA (Fig 6). Both of these proteins were recognized by the T7 and Lyp antibodies. The lower molecular weight product probably represents the result of proteolytic degradation, whereas the 105-kD protein is intact Lyp1. When immunoprecipitated from lymphoid cell lines, the native Lyp1 protein has an apparent molecular weight of 105 kD, in agreement with the size observed in transfected COS-7 cells (see Fig 8).
Transfection of Lyp1 and Lyp2 cDNA. T7-tagged Lyp1 (A) and Lyp2 (B) were transfected into COS-7 cells and immunoprecipitated with anti-LyP or anti-T7 antibody and blotted with anti-T7. (A) Lyp1 transfection results in a transfected protein of 105 kD and a probable degradation product of 96 kD, whereas (B) shows LyP2 as a protein of 85 kD.
Transfection of Lyp1 and Lyp2 cDNA. T7-tagged Lyp1 (A) and Lyp2 (B) were transfected into COS-7 cells and immunoprecipitated with anti-LyP or anti-T7 antibody and blotted with anti-T7. (A) Lyp1 transfection results in a transfected protein of 105 kD and a probable degradation product of 96 kD, whereas (B) shows LyP2 as a protein of 85 kD.
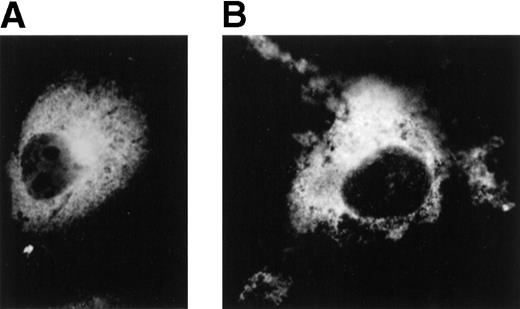
To determine the intracellular localization of the lyp phosphatases, the distribution of Lyp1 and Lyp2 was determined by indirect immunofluorescence in transiently transfected COS-7 cells. An HA epitope was attached as a three tandem repeat to the 5′ end of both the Lyp1 and Lyp2 cDNAs in the pCDNA3 eucaryotic expression vector. Forty-eight hours after transfection, cells were fixed and incubated with antibodies to the HA epitope. COS-7 cells transfected with either Lyp1 or Lyp2 displayed prominent perinuclear and cytoplasmatic staining, but no staining of the nucleus (Fig 7B). No fluorescence was observed in COS-7 cells transfected with vector alone (Fig 7A). The pattern of staining suggests that both of the phosphatases are predominantly cytoplasmatic.
Localization of Lyp1 and Lyp2 in transiently transfected COS-7 cells by immunofluorescence. COS-7 cells were transiently transfected with HA-tagged LyP1 and LyP2 cDNAs in pcDNA3 and immunofluorescent detection was performed with a monoclonal antibody against the HA tag (original magnification × 1,000). (A) Cells transfected with HA-Lyp2 cDNA. (B) Cells transfected with HA-LyP1 cDNA.
Localization of Lyp1 and Lyp2 in transiently transfected COS-7 cells by immunofluorescence. COS-7 cells were transiently transfected with HA-tagged LyP1 and LyP2 cDNAs in pcDNA3 and immunofluorescent detection was performed with a monoclonal antibody against the HA tag (original magnification × 1,000). (A) Cells transfected with HA-Lyp2 cDNA. (B) Cells transfected with HA-LyP1 cDNA.
Characterization of Lyp protein expression.
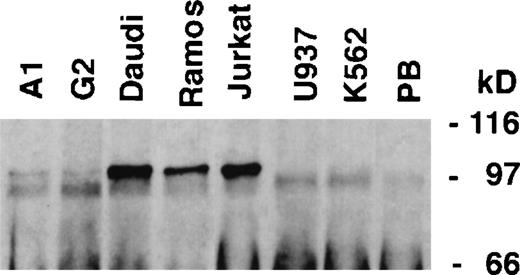
Two rabbit polyclonal antibodies were raised against the Lyp protein. After careful testing, an antibody raised against two peptides from a common C-terminal area of the Lyp isoforms was used for immunoprecipitation, because this antibody appeared to precipitate more efficiently than an antibody raised against a catalytic domain fusion protein. Both antibodies appeared to Western blot equally well. These polyclonal antibodies were first used to characterize the expression of Lyp proteins in human hematopoietic cell lines (Fig 8). A single band of 105 kD could be seen in both T-cell (Jurkat) and B-cell lines (Daudi and Ramos), the same size as observed upon transfection of Lyp1 cDNA into COS-7 cells (Fig 6). Lyp1 expression could not be detected in either the monocytic (U937) or myeloid (K562) cell lines, whereas low levels of expression could be seen in pre-B cells (G2 and A1). This pattern of protein expression correlates well with that of Lyp1 mRNA observed by Northern blotting. We could not detect a protein of the predicted size of Lyp2 (85kD) in any of the cell lines examined.
Lyp protein expression in lymphoid and myeloid cell lines. Lyp was immunoprecipitated from cell lines (107cells) and blotted with Lyp antibodies. A protein band of 105 kD corresponding to LyP1 could be detected in Jurkat, Daudi, Ramos, A1, and G2 cells, whereas U937 and K562 do not appear to have detectable amounts of Lyp. (PB), preimmune serum control.
Lyp protein expression in lymphoid and myeloid cell lines. Lyp was immunoprecipitated from cell lines (107cells) and blotted with Lyp antibodies. A protein band of 105 kD corresponding to LyP1 could be detected in Jurkat, Daudi, Ramos, A1, and G2 cells, whereas U937 and K562 do not appear to have detectable amounts of Lyp. (PB), preimmune serum control.
Having observed the predominantly lymphoid pattern of Lyp mRNA expression, we also examined expression of the lyp protein in primary lymphoid cells (Fig 9A). Both thymocytes and tonsil T lymphocytes expressed Lyp1, whereas resting T cells from peripheral blood, in addition to expressing low levels of Lyp1, also expressed an 85-kD protein recognized by both polyclonal Lyp antibodies. This is the predicted molecular weight of Lyp2, the shorter alternatively spliced form of Lyp1.
Expression of Lyp proteins in resting and activated T cells. (A) Lyp was precipitated from thymocytes (80 × 106cells), peripheral blood T cells (25 × 106 cells), and tonsil T cells (10 × 106 cells) and immunoblotted with anti-Lyp. Preimmune serum controls (PB) are presented in each case. A band of 105 kD is present in each sample and a band of 85 kD can be seen only in resting peripheral T cells. (B) Lyp was immunoprecipitated from peripheral blood T cells (25 × 106 cells) before and after stimulation with anti-CD3 (2.5 μg/mL) or PHA over a period of 48 hours. There is increased 105-kD Lyp1 expresssion, whereas the 85-kD protein appears to be downregulated.
Expression of Lyp proteins in resting and activated T cells. (A) Lyp was precipitated from thymocytes (80 × 106cells), peripheral blood T cells (25 × 106 cells), and tonsil T cells (10 × 106 cells) and immunoblotted with anti-Lyp. Preimmune serum controls (PB) are presented in each case. A band of 105 kD is present in each sample and a band of 85 kD can be seen only in resting peripheral T cells. (B) Lyp was immunoprecipitated from peripheral blood T cells (25 × 106 cells) before and after stimulation with anti-CD3 (2.5 μg/mL) or PHA over a period of 48 hours. There is increased 105-kD Lyp1 expresssion, whereas the 85-kD protein appears to be downregulated.
To determine whether expression of the lyp proteins may be regulated by activation in T cells, normal peripheral blood T lymphocytes were incubated with either PHA or anti-CD3 and harvested after 24 or 48 hours (Fig 9B). An increase in the level of Lyp1 protein expression was observed after 24 hours of either stimulus, with a further increase seen after 48 hours with anti-CD3. Interestingly, the 85-kD protein could no longer be detected after 24 hours of incubation with either PHA or anti-CD3.
Lyp phosphatase activity.
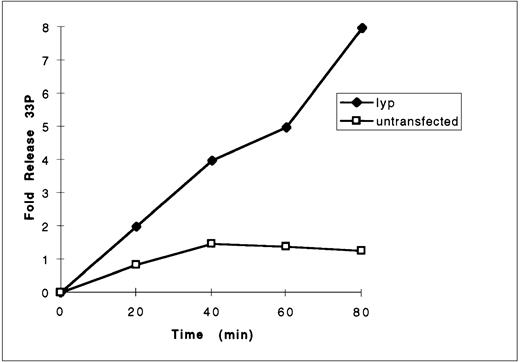
To determine whether Lyp1 possessed a catalytically active tyrosine phosphatase domain, COS cells were transfected with T7-Lyp cDNA, the protein immunoprecipitated with anti-T7 and used to dephosphorylate a labeled synthetic peptide, Raytide, in an in vitro phosphatase assay. Raytide peptide was 33P labeled on tyrosine residues in vitro using the tyrosine kinase p60src and purified on phosphocellulose paper. Release of 33P over time was measured in the phosphatase assay and compared with controls from untransfected cells. The results showed a sevenfold increase in 33P release from the substrate incubated with Lyp immunoprecipitates compared with control immunoprecipitates (Fig 10), demonstrating that Lyp does indeed possess tyrosine phosphatase activity. This activity could be completely inhibited by pervanadate (not shown).
Measurement of lyp1 phosphatase activity. Anti-lyp immunoprecipitates from untransfected and pcDNA3-lyp1–transfected cells were prepared in pervanadate-free lysis buffer and incubated with33P-labeled substrate Raytide. At the indicated time points, reactions were stopped by the addition of charcoal and the free33P released from the peptide and now present in the supernatant was measured by liquid scintillation counting.
Measurement of lyp1 phosphatase activity. Anti-lyp immunoprecipitates from untransfected and pcDNA3-lyp1–transfected cells were prepared in pervanadate-free lysis buffer and incubated with33P-labeled substrate Raytide. At the indicated time points, reactions were stopped by the addition of charcoal and the free33P released from the peptide and now present in the supernatant was measured by liquid scintillation counting.
Involvement of Lyp1 in TCR signaling.
One of the earliest events after TCR stimulation of T cells is the induction of tyrosine phosphorylation. To determine whether Lyp played a role in TCR signaling, human thymocytes were stimulated with anti-CD3 for various periods of time, Lyp immunoprecipitated, and blotted with antiphosphotyrosine. This showed that, whereas Lyp itself is not detectably tyrosine phosphorylated, a heavily phosphorylated protein of 116 to 120 kD coprecipitates with lyp, appearing within 1 minute of stimulation (Fig 11A). Once activated, the phosphorylation level of this protein remained constant over a period of 20 minutes. We attempted to identify the 116-kD phosphorylated protein by Western blotting of Lyp immunoprecipitates from CD3-stimulated thymocytes with antibodies to various candidate proteins. The 116-kD protein associated with LyP1 was found to be c-Cbl (Fig 11B), but not p125Fak, p116 Jak3, or p110 PI3-kinase. No alteration in the amount of Cbl coimmunoprecipitating with lyp could be detected upon anti-CD3 stimulation, suggesting that Lyp1 and Cbl are constitutively associated, although Cbl can be inducibly phosphorylated. This interaction was also observed in the mature T-cell line Jurkat (not shown) and further confirmed by transfection of Lyp1 into COS-7 cells and examining its association with the endogenous Cbl protein (Fig 11C). Lyp1 was found not only to coprecipitate with cbl in COS cells, but also to significantly reduce the basal level of cbl tyrosine phosphorylation (Fig 11D). This suggests that lyp1 may serve to regulate cbl function in lymphoid cells and possibly that of Cbl-associated proteins.
Involvement of Lyp1 in TCR signaling. (A) Lyp immunoprecipitates from thymocytes (80 × 106cells) stimulated with anti-CD3 were blotted with antiphophotyrosine. A single phosphorylated band of 116 kD was detected coimmunoprecipitating with Lyp. Lyp protein loading was quantitated by anti-Lyp Western blot after stripping. (B) Immunoblotting with anti-Cbl identified the 116-kD phosphorylated protein as Cbl, whereas immunoblotting with anti-FAK or anti-p110 (subunit of PI-3 kinase) showed them not to be associated with Lyp. (C) Lyp1 was transfected into COS-7 cells and Cbl immunoprecipitates prepared from these and untransfected cells. Western blotting was performed with Lyp antibodies. The position of lyp is indicated by an arrow. Cbl immunoprecipitates were also prepared and blotted with antiphosphotyrosine (D) and then anti-cbl after stripping.
Involvement of Lyp1 in TCR signaling. (A) Lyp immunoprecipitates from thymocytes (80 × 106cells) stimulated with anti-CD3 were blotted with antiphophotyrosine. A single phosphorylated band of 116 kD was detected coimmunoprecipitating with Lyp. Lyp protein loading was quantitated by anti-Lyp Western blot after stripping. (B) Immunoblotting with anti-Cbl identified the 116-kD phosphorylated protein as Cbl, whereas immunoblotting with anti-FAK or anti-p110 (subunit of PI-3 kinase) showed them not to be associated with Lyp. (C) Lyp1 was transfected into COS-7 cells and Cbl immunoprecipitates prepared from these and untransfected cells. Western blotting was performed with Lyp antibodies. The position of lyp is indicated by an arrow. Cbl immunoprecipitates were also prepared and blotted with antiphosphotyrosine (D) and then anti-cbl after stripping.
DISCUSSION
We have isolated two novel intracellular phosphatase cDNA sequences from a human thymus cDNA library: Lyp1 and its splice variant, Lyp2. Sequence analysis of Lyp1 showed significant homology with the murine phosphatase PEP, an intracellular PTPase widely expressed in hematopoietic tissues.12 Lyp1 shares an overall 70% amino acid identity with PEP (Fig 2). Although there is 89% identity between the catalytic domain of Lyp1 and PEP, significantly less homology is observed within the noncatalytic portions (61%). This degree of homology is lower than any other currently described pair of murine-human phosphatase homologues. Within this low homology area Lyp1 contains four proline-rich sequences, which are also present in PEP (see Fig 2), forming putative PXXP and class II (XPPLPXR) SH3 domain binding motifs.12,41 A recent study has demonstrated association between one of the proline-rich motifs of PEP (PPPLPERTP, also present in Lyp) and the SH3 domain of the protein tyrosine kinase p50csk.15 The same motif occurs in the phosphatase PTP-PEST, where it is also responsible for binding to csk.16 Preliminary experiments also show Lyp1 to associate with csk in T cells (not shown). The Lyp1 noncatalytic domain also contains a large area of unique sequence, including an NXXY motif (see Fig 2). When tyrosine phosphorylated, this motif may be recognized by a phosphotyrosine binding (PTB) domain42 found in adaptor proteins such as IRS, Shc, and cbl.
The lyp-related murine phosphatase PEP also has several PEST consensus sequences.12 These contain an unusually high percentage of proline (P), glutamic/aspartic acid (E/D), serine (S), and threonine (T) residues; have been identified in many proteins; and were proposed to confer a high susceptibility to rapid degradation.43Early studies on the rapidly degraded enzyme ornithine decarboxylase provided support for this hypothesis,44 but such a claim could not be substantiated for PEP11 or for other PEST-containing proteins.45 46 Analysis of the Lyp1 protein sequence using the program PEST-FIND (PC analysis software; Oxford Molecular Group, Oxford, UK) indicated the presence of only a single PEST motif (amino acids 702-736), whereas five were confirmed in PEP.
The relatively high degree of homology between the catalytic domains of Lyp1 and PEP suggests that Lyp1 may be the human homologue of PEP. However, there are significant differences between the other domains and, although the function of both phosphatases remains unclear, it is difficult to determine whether they are in fact homologues.
Using FISH analysis, the Lyp gene was localized to chromosome 1p13 (Fig3). This region is of particular interest, because it is a common site of chromosomal rearrangement in both solid and hematopoietic cancers.47-52 Several lines of evidence already suggest that PTPases may act as tumor suppression genes.2 53Knowledge of the precise chromosomal locus, along with comparative studies in normal and malignant cells, will help to establish whether Lyp may play a role in tumorigenesis.
The shorter form of Lyp mRNA isolated by library screening, Lyp2, was found to be derived from alternative splicing of a Lyp gene intron. A 3.5-kb intronic sequence of Lyp1 contains an alternative exon coding for the C-terminal 7 amino acids of Lyp2 and at least part of its 3′ untranslated sequence (Fig 4). Consequently, the 116 amino acid C-terminal of Lyp1 is replaced by 7 alternative amino acids in Lyp2. Precisely the same mechanism of generating an alternative C-terminal has been observed in the receptor phosphatase PTP-P1.53 An intronic sequence of PTP-P1 was found to contain an alternative exon coding for the C-terminal 26 amino acid of PTP-PS and its 3′-untranslated region. Several other studies have also shown that diversity can be generated by alternative splicing in cytoplasmic PTPases.54-57 Thus, the creation of C-terminal diversity by alternative RNA splicing appears to be a general mechanism of generating functional diversity in phosphatases.
The significance of the alternative C-terminal sequences of Lyp1 and Lyp2 remains unclear, but there are several differences that may be key to showing functional divergence. The C-terminus of Lyp1, but not Lyp2, contains a consensus sequence XS/TPXK/R (741KTPGK745) recognized by the p34cdc2 kinase,58a cell cycle regulatory kinase,59 suggesting that Lyp1 may be phosphorylated in a cell cycle-dependent manner. Lyp1 also contains four potential SH3 domain binding sites, compared with a single motif in Lyp2, suggesting that the isoforms may interact with different sets of SH3 domains.
The pattern of Lyp1 expression observed by Northern blot showed it to be preferentially expressed in lymphoid cells (Fig 5), particularly in thymocytes and mature B and T cells. A low level of Lyp1 expression was also seen in tissues rich in lymphoid infiltrates, such as the small intestine and appendix. The pattern of Lyp1 protein expression detected by antibodies in human hematopoietic cell lines correlated well with Lyp1 mRNA expression (Fig 8). This pattern suggests that Lyp1 may play a role in aspects of both the early and late stages of T-cell differentiation. The lack of expression in fetal liver tissue, which contains a large population of pre-B cells, suggests a different role in the biology of B-cell development. However, it should be noted that Northern blot analysis indicated significant expression of Lyp2 in fetal liver tissue.
We have differentiated between the mRNA expression of Lyp1 and its isoform, Lyp2, by the use of specific cDNA probes (Fig 5B). The two isoforms demonstrated different patterns of RNA expression. Although Lyp2 was present at lower levels then LyP1 in all the lymphoid tissue examined, its expression was higher in fetal liver tissue. This clearly indicates that there are differences in the regulation of expression between the two isoforms. None of the cell lines examined (Fig 8) expressed a protein of the expected size of Lyp2 (85 kD), as observed in lysate from Lyp2 COS-7 cell transfectants (Fig 7). Only resting peripheral T lymphocytes demonstrated expression of an 85-kD protein recognized by the LyP-specific antibodies. Interestingly, stimulation of T lymphocytes with PHA or anti-CD3 resulted in the induction of the Lyp1 protein, with a simultaneous downregulation of the 85-kD protein (Fig 9B). The 85-kD protein is believed to be Lyp2 on the basis of its apparent molecular weight and the fact both Lyp antibodies can recognize it. This finding suggests that Lyp2 may play an important role in resting cells, because thymocytes, tonsil T cells, and lymphoid cell lines, which are activated cells, do not express the protein. Further investigation of its role will require the generation of Lyp2-specific antisera directed against its unique C terminal sequence. However, these observations also suggest that the two isoforms may play significantly different roles in T-cell development.
While examining antigen receptor- and cytokine-mediated activation of T cells, anti-CD3 stimulation of thymocytes was found to induce the association of a 116-kD phosphorylated protein with Lyp1. Western blotting of Lyp immunoprecipitates identified the phosphorylated band as the proto-oncogene c-Cbl. Although inducibly phosphorylated, cbl was found to be constitutively associated with Lyp1. From previous studies it is known that Cbl is heavily tyrosine phosphorylated after TCR stimulation60 and can associate with the Syk and ZAP tyrosine kinases, negatively regulating their activities.61-65 Treatment of Jurkat cells with the phosphatase inhibitor pervanadate leads to a marked increase in the phosphorylation of Cbl,63 suggesting that tyrosine phosphatases keep Cbl in a basally dephosphorylated state. We have shown that Lyp1 is basally associated with Cbl in thymocytes and confirmed this interaction in Jurkat (not shown) and in COS cells by transfection, where Cbl phosphorylation is also reduced by Lyp1 overexpression (Fig 11). This strongly suggests that Lyp may play a role in regulating Cbl activity through modulation of its tyrosine phosphorylation status. Because Cbl is an adaptor protein that associates with numerous protein tyrosine kinases, it is also conceivable that Lyp may play a role in the regulation of these proteins.64 Although we could not detect tyrosine phosphorylation of Lyp1, a minor variant (EPNY) of the Cbl PTB domain consensus binding motif (D(N/D)XpY) is present in the noncatalytic domain that could form the basis for interaction. Alternatively, in the absence of other identifiable interactive domains in either protein, a multiple SH3 domain adaptor protein such as Grb2 may serve to link Lyp and Cbl.
ACKNOWLEDGMENT
The authors are indebted to Sergio Grinsten for helping with the cellular localization of Lyp, to Barbara Beatty for assisting with chromosomal localization of Lyp, and to Michal Shahar for excellent technical assistance.
Supported by the Medical Research Council of Canada, the National Cancer Institute, and The Lotte and John Hecht Memorial Foundation.
The GenBank accession number for Lyp1 cDNA is AF 001846 and for Lyp2 isAF001847.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Chaim M. Roifman, MD, Division of Immunology/Allergy, Infection, Immunity, Injury and Repair Programme, The Hospital for Sick Children, 555 University Ave, Toronto, Ontario, Canada M5G 1X8.



![Fig. 4. Lyp2 is a result of alternative splicing of the Lyp1 gene. (A) A schematic map of the PCR strategy used. Primer 1 corresponds to the last 20 nucleotides shared by both the Lyp1 and Lyp2 sequences, primer 2 to Lyp2 untranslated area, and primer 3 to the beginning of the unique Lyp1 sequence, immediately downstream of primer 1 (see also [C]). (B) The results of the PCR amplification on genomic DNA. Lane 1, DNA ladder; lane 2, a product of 3.5 kb was amplified with primers 1 and 3; lane 3, a product of 100 bp was amplified with primers 1 and 2. (C) Schematic map of Lyp1 splicing. The sequences before the vertical line represent the splice donor site, whereas the nucleotide sequences after it are the Lyp1 intronic sequence that code for the unique C-terminal seven amino acids, stop codon (asterisk), and untranslated sequence (lower case letters) of Lyp2. An open box represents the common cDNA sequence shared by Lyp1 and Lyp2; the solid and the light gray boxes represent the unique sequences of Lyp1 and Lyp2, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/6/10.1182_blood.v93.6.2013.406k25_2013_2024/5/m_blod40625004w.jpeg?Expires=1769358619&Signature=xGTwwAtxtTdQ5wfoxQpmasPBSuZDK8IRc1dzaGZflSiMnniDY~n65N~IX6pU-4QaAFI~797qBO0f2lRNMIpe~2WAcOSUPfl6gG07rke9B~K55IXOTzWKigo5Z5JVO-x00sagsGr4-6YD62d3Y6nArUGtg4ljs~03oxQj3g00pawOW8Qyw9bwpbjFuXifwd4H3Lr8sg43VwHvIcVBTkiv775s4IRDGE-5I5s5r5iMiPixXbvyaAFsvLqVV4EIOOZ9rY8hNvqwzaLBerXOAVvh1d1MjHGM6gzjhWYvAhYOw0TCV~~cVDX~XuGjjmvAIw72yTefyK09WYSZi04MDlzrYg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal