The mi locus encodes a member of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) protein family of transcription factors (hereafter called MITF). We reported that expression of the mouse mast cell protease 5 (MMCP-5) and MMCP-6 genes were deficient in cultured mast cells (CMC) derived from mutant mice ofmi/mi genotype. Despite the reduced expression of both MMCP-5 and MMCP-6, their regulation mechanisms were different. Because MMCP-5 is a chymase and MMCP-6 a tryptase, there was a possibility that the difference in regulation mechanisms was associated with their different characteristics as proteases. We compared the regulation mechanisms of another chymase, MMCP-4, with those of MMCP-5 and MMCP-6. The expression of the MMCP-4 gene was also deficient in mi/mi CMC. The overexpression of the normal (+) MITF but not of mi-MITF normalized the poor expression of the MMCP-4 gene in mi/mi CMC, indicating the involvement of +-MITF in transactivation of the MMCP-4 gene. Although MMCP-4 is chymase as MMCP-5, the regulation of MMCP-4 expression was more similar to MMCP-6 than to MMCP-5. We also showed the deficient expression of granzyme B and cathepsin G genes inmi/mi CMC. Genes encoding granzyme B, cathepsin G, MMCP-4, and MMCP-5 are located on chromosome 14. Because all these genes showed deficient expression in mi/mi CMC, there is a possibility that MITF might regulate the expression of these genes through a locus control region.
THE mi LOCUS OF MICE encodes a member of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) protein family of transcription factors (mi-transcription factor, MITF).1,2 The MITF encoded by the mutant mi locus deletes one of four consecutive arginines in the basic domain (hereafter called mi-MITF).1,3,4 The mi/mimice show microphthalmia, depletion of pigment in both hair and eyes, osteopetrosis, and decrease in the number of mast cells.5-9In addition to the decrease in number, the phenotype of mast cells was abnormal in mi/mi mice.9-13 We have shown that normal (+) MITF (hereafter called +-MITF) is involved in the expression of the mouse mast cell protease 6 (MMCP-6),14c-kit,15 p75 nerve growth factor (NGF) receptor,16 and MMCP-5 genes.17 We have also shown that the deficient transactivation ability of the mi-MITF is caused by two different mechanisms: the loss of DNA binding and the impaired nuclear localization.18 19
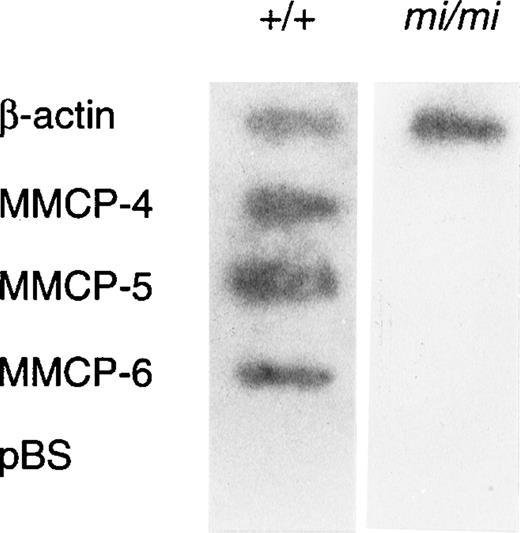
Mast cells of mice contain various proteases. The complementary (c) DNA and genes that encode mast cell carboxypeptidase A (MC-CPA) and six of the seven mouse mast cell–specific serine proteases have been cloned and sequenced.20-27 The MC-CPA is an exopeptidase that prefers C-terminal aliphatic amino acids. The MMCP-1, MMCP-2, MMCP-4, and MMCP-5 are predicted to be chymases from the deduced amino acid sequences, whereas MMCP-6 and MMCP-7 to be tryptases. The MMCP-1, MMCP-2, MMCP-4, and MMCP-5 genes reside on the chromosome 14 and link with a gene complex encoding blood cell proteases, such as the cathepsin G and the granzymes specific to cytotoxic T lymphocytes.28-31 On the other hand, the MC-CPA gene resides on the chromosome 3, and the MMCP-6 and MMCP-7 genes reside on the chromosome 17.28 Despite the significant reduction in expression of both MMCP-5 and MMCP-6 in mi/mi cultured mast cells (CMC), their regulation mechanisms appeared to be different.17 Although +-MITF directly bound CANNTG motifs in the promoter region of the MMCP-6 gene and transactivated it, the binding of +-MITF to the CANNTG motif in the promoter region of the MMCP-5 gene was not detectable. The +-MITF appeared to regulate the transactivation of the MMCP-5 gene indirectly.17 Moreover, addition of stem cell factor (SCF) to the medium normalized the expression of MMCP-5 but not of the MMCP-6 gene in mi/miCMC.17
In the present study, we investigated whether this difference of the regulation mechanisms between MMCP-5 and MMCP-6 were associated with the difference between chymase and tryptase. For this purpose, we examined the regulation of MMCP-4, whose expression was also poor inmi/mi CMC. Although MMCP-4 is chymase as MMCP-5, the regulation mechanisms of MMCP-4 expression was more similar to MMCP-6 than to MMCP-5.
MATERIALS AND METHODS
Mice.
The original stock of C57BL/6-mi/+ (mi/+) mice was purchased from the Jackson Laboratory (Bar Harbor, ME) and maintained in our laboratory by consecutive backcrosses to our own inbred C57BL/6 colony (more than 18 generations at the time of the present experiment). Female and male mi/+ mice were crossed together, and the resulting mi/mi mice were selected by their white coat color.5,6 The original stock of VGA-9-tg/tg mice, in which the mouse vasopressin-Escherichia coliβ-galactosidase transgene was integrated at the 5′ flanking region of the mi (MITF) gene, were kindly given by H. Arnheiter (National Institutes of Health, Bethesda, MD). The integrated transgene was maintained by repeated backcrosses to our own inbred C57BL/6 colony (more than eight generations at the time of the present experiment). Female and male tg/+ mice were crossed together, and the resulting tg/tg mice were selected by their white coat color.1
Cells.
Pokeweed mitogen-stimulated spleen cell–conditioned medium (PWM–SCM) was prepared according to the method described by Nakahata et al.32 Mice of mi/mi or tg/tg genotype and control C57BL/6-+/+ (+/+) mice were used at 2 to 3 weeks of age to obtain CMC. Mice were killed by decapitation after ether anesthesia and spleens were removed. Spleen cells derived from mi/mi, tg/tg,or +/+ mice were cultured in α-minimal essential medium ( α-MEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% PWM–SCM and 10% fetal calf serum (FCS; Nippon Bio-supp Center, Tokyo, Japan). Half of the medium was replaced every 7 days. More than 95% of cells were CMC 4-weeks after initiation of the culture.33, 34 The helper virus-free packaging cell line (ψ 2) was maintained in Dulbecco’s modified Eagle’s medium (DMEM; ICN Biomedicals) supplemented with 10% FCS.35 The NIH/3T3 fibroblast cell line was generously provided by Dr S.A. Aaronson (National Cancer Institute, Bethesda, MD) and maintained in DMEM supplemented with 10% FCS. The IC-2 cell line was provided by Dr I. Yahara (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan)36 and maintained in α-MEM supplemented with 10% PWM–SCM and 10% FCS. When various amounts of recombinant mouse SCF (rmSCF, a generous gift of Kirin Brewery Co Ltd, Tokyo, Japan) were added, mi/mi and +/+ CMC were cultured in α-MEM containing 10% PWM–SCM and 10% FCS.
In situ hybridization.
Skin pieces were removed from the back of 20-day-old mice and smoothed onto a piece of the filter paper to keep them flat. The skin pieces were fixed in 4% paraformaldehyde in 0.1 mol/L phosphate buffer (PB; pH 7.4), and embedded in paraffin. CMC were collected, washed with phosphate-buffered saline (PBS), and immobilized by agarose.37 The pieces of agarose containing CMC were fixed with 4% paraformaldehyde in 0.1 mol/L PB overnight, and then embedded in paraffin. To obtain the MMCP-4, MMCP-5, and MMCP-6 probes, total RNA was extracted from CMC of C57BL/6-+/+ mouse origin by lithium chloride-urea method.38 Single-stranded cDNA was generated from the RNA. A polymerase chain reaction (PCR) product was then amplified from the cDNA with specific primers for each protease.13 39 The products were subcloned into theEcoRV site of Bluescript KS (-) plasmid (pBS; Stratagene, La Jolla, CA) that contains T3 and T7 promoters to generate probes. The technique of in situ hybridization was described previously.37, 40 The number of MMCP-4 messenger RNA (mRNA)-positive cells and that of alcian blue–positive cells were counted in the serial sections, and the proportion of MMCP-4 mRNA-positive cells to alcian blue–positive cells was calculated.
Construction of retrovirus vector and its infection.
Plasmid pBS containing the whole coding region of +-MITF ormi-MITF (pBS-+-MITF and pBS-mi-MITF, respectively) had been constructed in our laboratory.18,19 A retroviral vector, pM5Gneo,41 a derivative of myeloproliferative sarcoma virus vector, was a kind gift from Dr W. Ostertag (Universität Hamburg, Hamburg, Germany). The purifiedSmaI-HincII fragment from pBS-+-MITF or pBS-mi-MITF was introduced into the blunted EcoRI site of pM5Gneo. The resulting pM5Gneo-+-MITF and pM5Gneo-mi-MITF were transfected into the packaging cell line (ψ2)35 by the calcium phosphate precipitation method,42 and neomycin-resistant ψ2 cell clones were selected by culturing in DMEM supplemented with 10% FCS and G418 (0.8 mg/mL; GIBCO-BRL, Grand Island, NY). For gene transfer, spleen cells obtained frommi/mi mice were incubated on irradiated (30 Gy) subconfluent monolayer of virus-producing ψ2 cells for 72 hours in α-MEM supplemented with 10% PWM–SCM and 10% FCS. Neomycin-resistant CMC were obtained by continuing the culture in α-MEM containing 10% PWM–SCM, 10% FCS, and G418 (0.8 mg/mL) for 4 weeks. In our previous study, Northern blotting analysis showed the apparent expression of the introduced +-MITF or mi-MITF cDNA in mi/mi CMC, whereas the mRNA expression of the endogenous mi gene was hardly detectable in either +/+ CMC or mi/mi CMC.14, 15
Cathepsin G cDNA probe.
Sense (5′-CATTGCTTGGGAAGCTCCATA-3′, nucleotide [nt] 328 to 348) and antisense (5′-ACCAGAATCACCCCTGAAGG-3′, nt 725 to 744) oligonucleotide primers were synthesized by conventional means. Nucleotide numbers were based on the report of Aveskogh et al43 Total RNA (5 μg) obtained from +/+ CMC was used as a template, and the single-strand cDNA was synthesized with a random primer by avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim GmbH Biochemica, Mannheim, Germany). The cDNA was amplified by PCR using sense and antisense primers. The products were subcloned into the plasmid pBS for further analysis. Nucleotide sequence was determined by Model 373A DNA sequencer (Applied Biosystems, Foster City, CA) using Taq dye deoxyterminater cycles sequencing kit (Applied Biosystems).44
Northern blot analysis.
Total RNA from CMC or IC-2 cells was prepared by the lithium chloride-urea method.38 Northern blot analysis was performed using MMCP-4,11,13 mouse β-actin,45 and mouse cathepsin G cDNA labeled with α-[32P]-dCTP (DuPont/NEN Research Products, Boston, MA) by random oligonucleotide priming as probes. After hybridization at 42°C, blots were washed to a final stringency of 0.2 × standard saline citrate (SSC; 1 × SSC is 150 mmol/L NaCl and 15 mmol/L trisodium citrate, pH 7.4) at 50°C and subjected to autoradiography.
Structure of MMCP-4 transcript.
Five micrograms of total RNA obtained from +/+ or mi/mi CMC were reverse transcribed in 20 μL of the reaction mixture containing 20 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim GmbH Biochemica) and random hexamer. One microliter of each reaction product was amplified in 25 μL of PCR mixture containing 0.125 U of Taq DNA polymerase (Takara Shuzou, Kyoto, Japan) and 12.5 pmol each of sense (5′-AGAATCTCTCTCCAAGCTGTGACCG-3′, nt 1 to 25) and antisense (5′-GGAGGTTAGGTCTTTACTGAGGTGCA-3′, nt 974 to 999)22 primers for the MMCP-4 gene by 30 cycles of denaturation at 94°C (30 seconds), annealing at 57°C (30 seconds), and synthesis at 72°C (1 minute). Inserts were subcloned into the plasmid pBS for further analysis. Nucleotide sequence was determined as described previously.44
Nuclear run-on assays.
CMC of C57BL/6-+/+ or -mi/mi mice were cultured in α-MEM supplemented with 10% PWM–SCM and 10% FCS. CMC (2 × 107) were washed with ice-cold PBS and lysed in 5 mL of ice-cold lysis buffer (10 mmol/L NaCl, 3 mmol/L MgCl2, 0.5% Nonidet P-40, and 10 mmol/L Tris-HCl, pH 7.4). After centrifugation of the lysates at 800g for 5 minutes at 4°C, the nuclei recovered in each bottom fraction were resuspended in 200 μL of storage buffer (40% glycerol, 5 mmol/L MgCl2, 0.1 mmol/L EDTA, and 50 mmol/L Tris-HCl, pH 7.5).46 Each nuclear run-on reaction was initiated in 400 μL of reaction buffer containing 30 mmol/L Tris-HCl, pH 8.0, 20% glycerol, 5 mmol/L MgCl2, 150 mmol/L KCl, 1 mmol/L dithiothreitol, 10 units of RNase inhibitor, 0.5 mmol/L adenosine triphosphate (ATP), 0.5 mmol/L cytidine triphosphate (CTP), 0.5 mmol/L guanosine triphosphate (GTP), and 100 μCi of α-[32P]-UTP (Amersham, Arlington Heights, IL). After a 30-minute incubation at 30°C, the reaction mixture was digested with DNase I (25 μg/mL, RNase-free) for 5 minutes and the radiolabeled transcripts were purified. Each slot blot, containing 5 μg of pBS DNA and gene-specific probes for MMCP-4, MMCP-5, MMCP-6, and β-actin11,13,17 45 immobilized onto separate regions of the nitrocellulose membrane, was placed in a small vial containing 1 to 2 mL of hybridization buffer and 1 to 5 × 106 cpm/mL 32P-labeled RNA. After a 36- to 48-hour incubation at 65°C, the slot blot was treated with RNase A for 20 minutes at 37°C, washed three times (20 minutes each) with 2 × SSC at 65°C, and subjected to autoradiography.
Construction of effector and reporter plasmids.
pEF-BOS expression vector47 was kindly provided by Dr S. Nagata (Osaka University Medical School, Osaka, Japan). TheSmaI-HincII fragment of pBS-+-MITF or pBS-mi-MITF was introduced into the blunted XbaI site of pEF-BOS. The resulting pEF-+-MITF and pEF-mi-MITF expression vectors were used as effectors. The luciferase gene subcloned into pSP72 (pSPLuc)48 was generously provided by Dr K. Nakajima (Osaka University Medical School, Osaka, Japan). To construct reporter plasmids, a DNA fragment containing a promoter region and the first exon (noncoding region) of the MMCP-4 gene (nt-1556 to +35, +1 is the transcription initiation site)22 obtained from genomic DNA of C57BL/6-+/+ mouse was cloned into the upstream region of the luciferase gene in pSPLuc. The deletion of the MMCP-4 promoter was produced by using the appropriate restriction enzyme. The mutations were introduced by PCR with mismatched primers. Deleted or mutated products were verified by sequencing.
Transient assay.
NIH/3T3 cells (5 × 105) were plated in a 10 cm dish 1 day before the procedure. Cotransfection with 10 μg of a reporter, 500 ng of an effector, and 5 μg of an expression vector containing the β-galactosidase gene was performed by the calcium phosphate precipitation method.42 The expression vector containing the β-galactosidase gene was used as an internal control. Because IC-2 cells expressed effector gene by themselves,16 the reporter and the expression vector containing the β-galactosidase gene were added to cell suspension (1 × 107) in 0.7 mL PBS, mixed gently, and incubated on ice for 10 minutes. For gene transfer, cells were electroporated by a single pulse (975 microfarads at 350 V) from a Gene Pulser II (Bio-Rad Laboratories, Richmond, CA). After incubation on ice for 10 minutes, the cells were suspended in 10 mL complete culture medium. NIH/3T3 cells were harvested 48-hours after transfection; IC-2 cells were harvested 8-hours after transfection. Cells were lysed with 0.1 mol/L potassium phosphate buffer (pH 7.4) containing 1% Triton X-100 (Sigma). Extracts were then used to assay luciferase activity with luminometer model LB96P (Berthold, Wildbad, Germany) and β-galactosidase activity. Luciferase activity was normalized by β-galactosidase activity and total protein concentration according to the method described by Yasumoto et al.49 The normalized value was divided by the value obtained with the cotransfection of the reporter and pEF-BOS, and was expressed as relative luciferase activity.
Electrophoretic gel mobility shift assay (EGMSA).
The production and purification of glutathione-S-transferase (GST)-+-MITF and GST-mi-MITF fusion proteins were described previously.18 19 Oligonucleotides were labeled with α-[32P]-dCTP by filling 5′-overhangs, and were used as probes for EGMSA. DNA binding assays were performed in a 20-μL reaction mixture containing 10 mmol/L Tris-HCl (pH 8.0), 1 mmol/L EDTA, 75 mmol/L KCl, 1 mmol/L dithiothreitol, 4% Ficoll type 400, 50 ng poly (dI-dC), 25 ng of the labeled DNA probe, and 3.5 μg of GST-+-MITF or GST-mi-MITF fusion protein. After incubation at room temperature for 15 minutes, the reaction mixture was subjected to electrophoresis at 14 V/cm at 4°C on a 5% polyacrylamide gel in 0.25 × tris hydroxymethyl aminomethane-borate, ethylenediaminetetraacetic acid (TBE) buffer (1 × TBE is 90 mmol/L Tris-HCl, 64.6 mmol/L boric acid, and 2.5 mmol/L EDTA, pH 8.3). The polyacrylamide gels were dried on Whatman 3MM chromatography paper (Whatman, Maidstone, UK) and subjected to autoradiography. Competitive DNA binding assays were performed as described above, except that the unlabeled competitive DNA was added to the reaction mixture before addition of GST-+-MITF fusion protein.
RESULTS
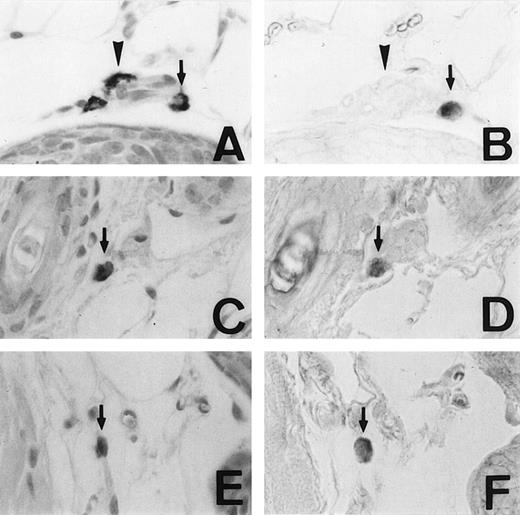
We examined the expression of the MMCP-4 gene in skin mast cells of +/+, mi/mi, and tg/tg mice by in situ hybridization. The criterion of the positive expression was arbitrarily determined, and we assessed that approximately 20% of the skin mast cells expressed the MMCP-4 mRNA in +/+ mice (Fig1A and B). Although approximately 2% of the skin mast cells showed the positive signals of the comparable degree in mi/mi andtg/tg mice (Fig 1C to F), most skin mast cells in these mutant mice did not show such significant signals. Then we calculated the proportion of MMCP-4 mRNA-positive mast cells by counting 180 to 210 alcian blue-positive mast cells in each mice (Table 1). The proportion was significantly greater in +/+ mice than in mi/mi and tg/tg mice.
Mast cells expressing MMCP-4 mRNA in the skin of +/+,mi/mi, and tg/tg mice. Serial sections from the skin of a +/+ mouse (A and B), a mi/mi mouse (C and D), and atg/tg mouse (E and F). A, C, and E, stained with alcian blue and nuclear fast red; B, D, and F, in situ hybridization with MMCP-4 probe. In each set of serial section, arrows show identical cells that expressed MMCP-4 gene and arrowheads show identical cells that did not express MMCP-4 gene. Original magnification ×400.
Mast cells expressing MMCP-4 mRNA in the skin of +/+,mi/mi, and tg/tg mice. Serial sections from the skin of a +/+ mouse (A and B), a mi/mi mouse (C and D), and atg/tg mouse (E and F). A, C, and E, stained with alcian blue and nuclear fast red; B, D, and F, in situ hybridization with MMCP-4 probe. In each set of serial section, arrows show identical cells that expressed MMCP-4 gene and arrowheads show identical cells that did not express MMCP-4 gene. Original magnification ×400.
Proportion of MMCP-4 mRNA Expressing Mast Cells in the Skin of +/+, mi/mi, and tg/tg Mice Shown by In Situ Hybridization
| Genotype of Mice . | No. of Mice . | No. of Alcian Blue+ Cells per cm of Skin* . | No. of MMCP-4 mRNA+ Cells per cm of Skin* . | Proportion of MMCP-4 mRNA+ to Alcian Blue+ Cells (%)* . |
|---|---|---|---|---|
| +/+ | 5 | 211 ± 10 | 43 ± 4 | 20 ± 1 |
| mi/mi | 5 | 51 ± 8† | 1 ± 1† | 2 ± 1† |
| tg/tg | 5 | 42 ± 9† | 1 ± 1† | 2 ± 1† |
| Genotype of Mice . | No. of Mice . | No. of Alcian Blue+ Cells per cm of Skin* . | No. of MMCP-4 mRNA+ Cells per cm of Skin* . | Proportion of MMCP-4 mRNA+ to Alcian Blue+ Cells (%)* . |
|---|---|---|---|---|
| +/+ | 5 | 211 ± 10 | 43 ± 4 | 20 ± 1 |
| mi/mi | 5 | 51 ± 8† | 1 ± 1† | 2 ± 1† |
| tg/tg | 5 | 42 ± 9† | 1 ± 1† | 2 ± 1† |
Mean ± SE.
P < .01 by t-test when compared with the value of +/+ mice.
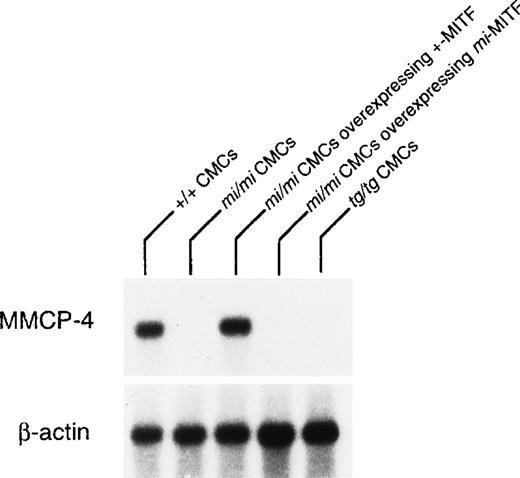
The expression of the MMCP-4 gene was also examined in +/+,mi/mi, and tg/tg CMC by Northern blot. The mRNA expression of the MMCP-4 gene was easily detectable in +/+ CMC but hardly in mi/mi and tg/tg CMC (Fig 2). To examine the involvement of +-MITF in the expression of the MMCP-4 gene, we used mi/mi CMC, to which cDNA encoding +-MITF or mi-MITF had been introduced. Overexpression of either +-MITF or mi-MITF in mi/mi CMC had been confirmed in previous studies.14 15 The poor mRNA expression of the MMCP-4 gene was normalized in mi/mi CMCs overexpressing +-MITF but not in mi/mi CMC overexpressing ofmi-MITF (Fig 2).
Reduced expression of MMCP-4 mRNA in mi/mi andtg/tg CMC and normalization of the MMCP-4 expression inmi/mi CMC by the introduction of the +-MITF cDNA but not ofmi-MITF cDNA. The blot was hybridized with32P-labeled cDNA probe of MMCP-4 or of β-actin.
Reduced expression of MMCP-4 mRNA in mi/mi andtg/tg CMC and normalization of the MMCP-4 expression inmi/mi CMC by the introduction of the +-MITF cDNA but not ofmi-MITF cDNA. The blot was hybridized with32P-labeled cDNA probe of MMCP-4 or of β-actin.
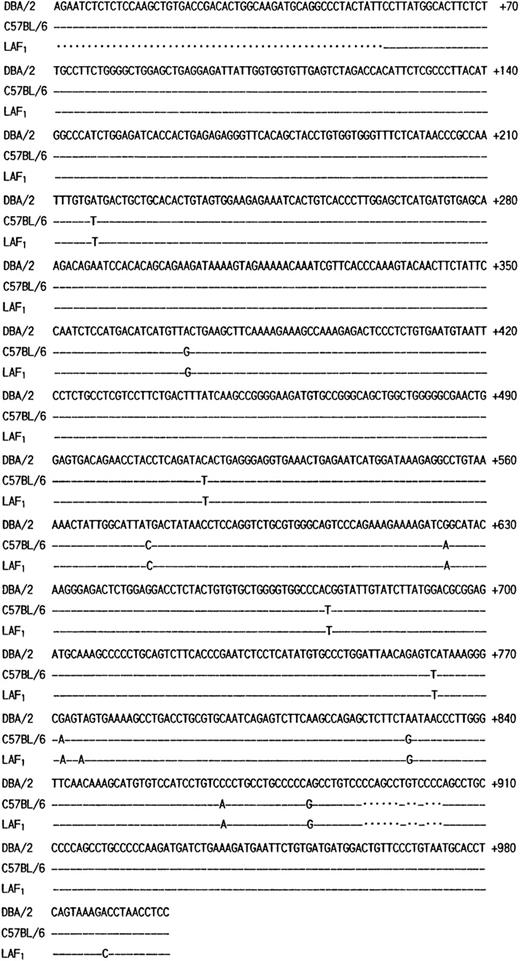
The poor expression of the MMCP-7 mRNA in the C57BL/6 strain is due to a splice-site mutation causing instability of its transcript.50 Both +/+ and mi/mi mice used in the present experiment were of C57BL/6 background. First the nucleotide sequence of MMCP-4 cDNA was compared between +/+ and mi/mi CMC. No difference was detectable (data not shown). Then we compared the sequence to that of the MMCP-4 cDNA obtained from the Kirsten sarcoma virus-immortalized mast cells (KiSV-MC1) derived from a DBA/2 mouse22 and also to that of the cDNA obtained from the mastocytoma cell line derived from a (Leaden × A1) F1(hereafter LAF1) mouse.25 Eight nucleotides were different between C57BL/6 and DBA/2 strains, but 5 of 8 nucleotide changes did not result in the alternation of amino acids (Fig 3). The remaining three changes caused the amino acid changes: an ATG codon at nt 217 to 219 changed to a TTG codon, leading to the change from Met to Leu at codon 41; an ACA codon at nt 514 to 516 changed to an ATA codon, leading to the change from Thr to Ile at codon 140; a GAG codon at nt 772 to 774 changed to an AAG codon, leading to the change from Glu to Lys at codon 226. Complete cDNA sequence of the mastocytoma cell line from LAF1 mouse has not been reported. The reported sequence started from codon 18, nt 53. Between nt 53 and 780, two nucleotide alternations leading to changes of an amino acid were observed between C57BL/6 and LAF1 mice: a TAG codon at nt 775 to 777 changed to an AAG codon, abolishing the stop codon observed in C57BL/6 mice (Fig 3). As a result, MMCP-4 of LAF1 mice appeared to contain one more amino acid (Lys) than that of C57BL/6 mice. These results suggested that the poor expression of the MMCP-4 mRNA in mast cells of C57BL/6-mi/mi mice was not attributable to the specific gene structure of the C57BL/6 strain.
Comparison of the structure of MMCP-4 transcript among KiSV-MC1 cells derived from a DBA/2 mouse, CMC derived from C57BL/6-+/+ mice and the mastocytoma cells derived from a LAF1 mouse. Numbering of the nucleotides begins at the transcription initiation site of the MMCP-4 transcript of KiSV-MC1 cells. Dashes indicate identical nucleotides. Dots indicate deletion of nucleotides in the sequence.
Comparison of the structure of MMCP-4 transcript among KiSV-MC1 cells derived from a DBA/2 mouse, CMC derived from C57BL/6-+/+ mice and the mastocytoma cells derived from a LAF1 mouse. Numbering of the nucleotides begins at the transcription initiation site of the MMCP-4 transcript of KiSV-MC1 cells. Dashes indicate identical nucleotides. Dots indicate deletion of nucleotides in the sequence.
Because amounts of mRNA of mast cell–specific chymases were reported to be regulated by a posttranscriptional mechanism,51 we examined whether such a mechanism was responsible to the decreased expression of MMCP-4 mRNA in mi/mi CMC. The nuclear run-on assay showed that MMCP-4, MMCP-5, and MMCP-6 genes were transcribed in +/+ CMC but not in mi/mi CMC (Fig4). In contrast, the transcription of the β-actin gene in +/+ CMC was comparable to that of mi/mi CMC. Because the MMCP-4, MMCP-5, MMCP-6, and β-actin probes were inserted in pBS, the pBS-derived DNA was used as a negative control. Hybridization of radiolabeled transcripts to pBS-derived DNA was not detected in the nuclear RNA preparation.
Nuclear run-on analysis of +/+ and mi/mi CMC.32P-labeled nuclear transcripts obtained from +/+ CMC (left lane) and mi/mi CMC (right lane) were examined for their ability to hybridize to DNA probes for β-actin, MMCP-4, MMCP-5, and MMCP-6. pBS-derived DNA was used as a negative control.
Nuclear run-on analysis of +/+ and mi/mi CMC.32P-labeled nuclear transcripts obtained from +/+ CMC (left lane) and mi/mi CMC (right lane) were examined for their ability to hybridize to DNA probes for β-actin, MMCP-4, MMCP-5, and MMCP-6. pBS-derived DNA was used as a negative control.
The transcription of MMCP-4 gene was poor in both mi/mi skin mast cells and mi/mi CMC. Skin mast cells of mice were considered to be supported by SCF52 and CMC by T-cell–derived growth factors.53 We compared the effect of SCF on the expression of MMCP-4, MMCP-5, and MMCP-6 between +/+ andmi/mi CMC. The poor expression of MMCP-5 in mi/mi CMC was normalized by the addition of SCF but the poor expression of MMCP-4 and MMCP-6 remained unchanged (Table 2). The expression of MMCP-4, MMCP-5, and MMCP-6 in +/+ CMC was not influenced by the addition of SCF, either (Table 2).
Effect of rmSCF on Proportions of CMCs Expressing MMCP-4, MMCP-5, or MMCP-6 mRNA in +/+ and mi/mi CMCs Shown by In Situ Hybridization
| Genotype of CMCs . | Concentration of rmSCF (ng/mL)* . | Proportion of MMCP-4 mRNA+ to Alcian Blue+ Cells (%)† . | Proportion of MMCP-5 mRNA+ to Alcian Blue+ Cells (%)† . | Proportion of MMCP-6 mRNA+ to Alcian Blue+ Cells (%)† . |
|---|---|---|---|---|
| +/+ | 0 | 23 ± 4 | 47 ± 2 | 46 ± 1 |
| mi/mi | 0 | 5 ± 1‡ | 4 ± 3‡ | 6 ± 1‡ |
| +/+ | 10 | 29 ± 2 | 55 ± 6 | 51 ± 3 |
| mi/mi | 10 | 6 ± 1‡ | 45 ± 42-153 | 5 ± 2‡ |
| +/+ | 50 | 30 ± 3 | 50 ± 5 | 52 ± 2 |
| mi/mi | 50 | 7 ± 2‡ | 40 ± 42-153 | 6 ± 1‡ |
| Genotype of CMCs . | Concentration of rmSCF (ng/mL)* . | Proportion of MMCP-4 mRNA+ to Alcian Blue+ Cells (%)† . | Proportion of MMCP-5 mRNA+ to Alcian Blue+ Cells (%)† . | Proportion of MMCP-6 mRNA+ to Alcian Blue+ Cells (%)† . |
|---|---|---|---|---|
| +/+ | 0 | 23 ± 4 | 47 ± 2 | 46 ± 1 |
| mi/mi | 0 | 5 ± 1‡ | 4 ± 3‡ | 6 ± 1‡ |
| +/+ | 10 | 29 ± 2 | 55 ± 6 | 51 ± 3 |
| mi/mi | 10 | 6 ± 1‡ | 45 ± 42-153 | 5 ± 2‡ |
| +/+ | 50 | 30 ± 3 | 50 ± 5 | 52 ± 2 |
| mi/mi | 50 | 7 ± 2‡ | 40 ± 42-153 | 6 ± 1‡ |
+/+ or mi/mi CMCs were incubated with 10% PWM-SCM and various concentrations of rmSCF for 2 weeks.
Mean ± SE of five independent experiments.
P < .01 by t-test when compared with the value of +/+ CMCs, which were cultured without rmSCF.
P < .01 by t-test when compared with the value ofmi/mi CMCs, which were cultured without rmSCF.
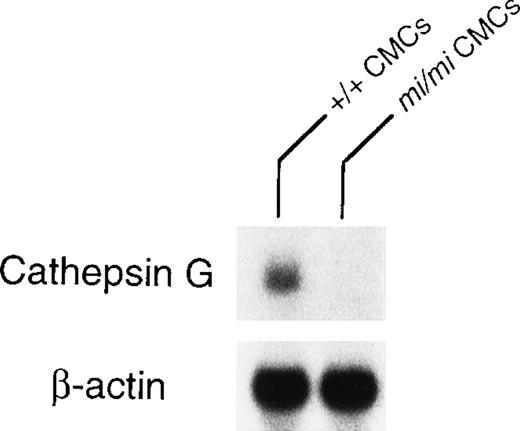
Granzyme B is located at the 5′ most end of the granzyme gene cluster, which includes (from 5′ to 3′) genes encoding granzymes B, C, F, G, D, E, cathepsin G, and mast cell chymase (MMCP-1, MMCP-2, MMCP-4, and MMCP-5).28 Pham et al54showed that the knock out of granzyme B gene affected the expression of granzymes C, D, and F genes but not the expression of cathepsin G gene. The knock out of cathepsin G did not affect the expression of the upstream granzymes. Pham et al54 suggested that there was probably a locus control region (LCR) for the granzyme genes and another LCR for cathepsin G and the downstream chymase genes. Because the expression of granzyme B, MMCP-4, and MMCP-5 genes are deficient inmi/mi CMC,17, 55 we investigated the involvement of the LCR in the expression of these genes. The MITF might have an effect on LCR. Because cathepsin G gene is located between the granzyme gene cluster and the genes encoding mast cell chymases, we compared the expression of cathepsin G gene between mi/mi and +/+ CMC. In fact, the mRNA expression of the cathepsin G gene was significantly lower in mi/mi CMC than that in +/+ CMC (Fig 5). This suggested a possibility that MITF may regulate the expression of granzyme B, cathepsin G, MMCP-4, and MMCP-5 through a single LCR or LCRs.
Expression of cathepsin G mRNA in +/+ and mi/miCMCs. The blot was hybridized with 32P-labeled cDNA probe of cathepsin G or of β-actin.
Expression of cathepsin G mRNA in +/+ and mi/miCMCs. The blot was hybridized with 32P-labeled cDNA probe of cathepsin G or of β-actin.
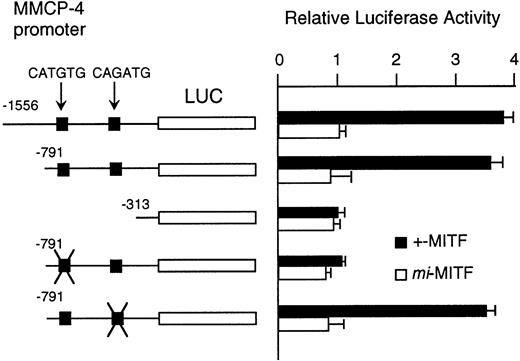
Although MITF may have an effect on LCR that might regulate the expression of the genes encoding granzyme B, cathepsin G, MMCP-4, and MMCP-5, we could not exclude another possibility that MITF could activate these genes in their different and appropriate elements. We have reported the direct and functional binding of +-MITF with 5′-upstream region of granzyme B.55 Thus, we attempted to study the interaction of MITF with 5′-upstream region of MMCP-4. We cloned 1556 bases of the 5′-upstream region of the MMCP-4 gene (Fig 6). The reporter plasmid that contained the luciferase gene under the control of the MMCP-4 promoter starting from nt-1556 was constructed. We also constructed the deleted reporter plasmids containing the MMCP-4 promoter starting from nt-791, -313 to examine the elements that mediate the transactivation. The expression plasmid containing +-ormi-MITF cDNA was cotransfected into NIH/3T3 fibroblasts with the luciferase construct under the control of the MMCP-4 promoter. Coexpression of +-MITF but not mi-MITF significantly increased the luciferase activity when we used the reporter plasmids containing the MMCP-4 promoter starting from both nt-1556 and -791 (Fig 7). Coexpression of +-MITF did not increase the luciferase activity when we used the reporter plasmid containing the MMCP-4 promoter starting from nt-313. Between nt-791 and -313, there were two CANNTG motifs, CATGTG and CAGATG. Mutation of the CATGTG motif (nt-383 to -378) to CTTGAG abolished the transactivation ability of the +-MITF when we used the reporter plasmids containing the MMCP-4 promoter starting from nt-791 (Fig 7). However, mutation of the CAGATG motif (nt-366 to -361) to CTGAAG did not abolish the transactivation of the +-MITF.
The nucleotide sequence of 5′ flanking region of the MMCP-4 gene. The CANNTG motif was boxed. A part of first exon was shown by capitals, and the 5′ flanking region is shown by lower case. The transcription initiation site was numbered as +1.
The nucleotide sequence of 5′ flanking region of the MMCP-4 gene. The CANNTG motif was boxed. A part of first exon was shown by capitals, and the 5′ flanking region is shown by lower case. The transcription initiation site was numbered as +1.
Effect of coexpression of cDNA encoding +-MITF ormi-MITF on luciferase activity. The luciferase gene under control of the normal, deleted, or mutated MMCP-4 promoter was cotransfected to NIH/3T3 fibroblasts. Bars indicate the standard error of these assays.
Effect of coexpression of cDNA encoding +-MITF ormi-MITF on luciferase activity. The luciferase gene under control of the normal, deleted, or mutated MMCP-4 promoter was cotransfected to NIH/3T3 fibroblasts. Bars indicate the standard error of these assays.
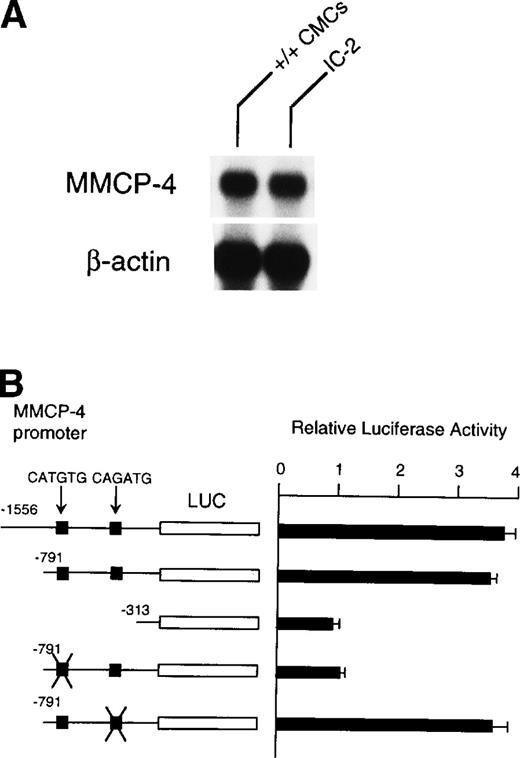
Because NIH/3T3 cells did not express +-MITF, we examined whether endogenous MITF can drive expression of the MMCP-4 reporter construct by using the IC-2 mast cell line that expressed +-MITF.16IC-2 cells also expressed the MMCP-4 mRNA (Fig 8A). We introduced the luciferase gene under the control of the MMCP-4 promoter into IC-2 cells. Luciferase activity was enhanced when we used the reporter plasmids containing the MMCP-4 promoter starting from both nt-1556 and -791 (Fig 8B). In contrast, the luciferase activity did not increase when we used the reporter plasmid containing the MMCP-4 promoter starting from nt-313. Transcription activity was abolished when the CATGTG motif (nt-383 to -378) was mutated, but not when the CAGATG motif (nt-366 to -361) was mutated. When we compared the reporter gene activity between IC-2 and NIH/3T3 cells, no significant difference was detectable in spite of the expression of +-MITF in IC-2 cells.
(A) Expression of MMCP-4 mRNA in IC-2 cells. The blot was hybridized with 32P-labeled cDNA probe of MMCP-4 or of β-actin. (B) Luciferase reporter gene promoter assay in IC-2 cells that expressed +-MITF. The luciferase gene under control of the normal, deleted, or mutated MMCP-4 promoter was introduced into the IC-2 mast cell line with electroporation. Bars indicate the standard error of three assays.
(A) Expression of MMCP-4 mRNA in IC-2 cells. The blot was hybridized with 32P-labeled cDNA probe of MMCP-4 or of β-actin. (B) Luciferase reporter gene promoter assay in IC-2 cells that expressed +-MITF. The luciferase gene under control of the normal, deleted, or mutated MMCP-4 promoter was introduced into the IC-2 mast cell line with electroporation. Bars indicate the standard error of three assays.
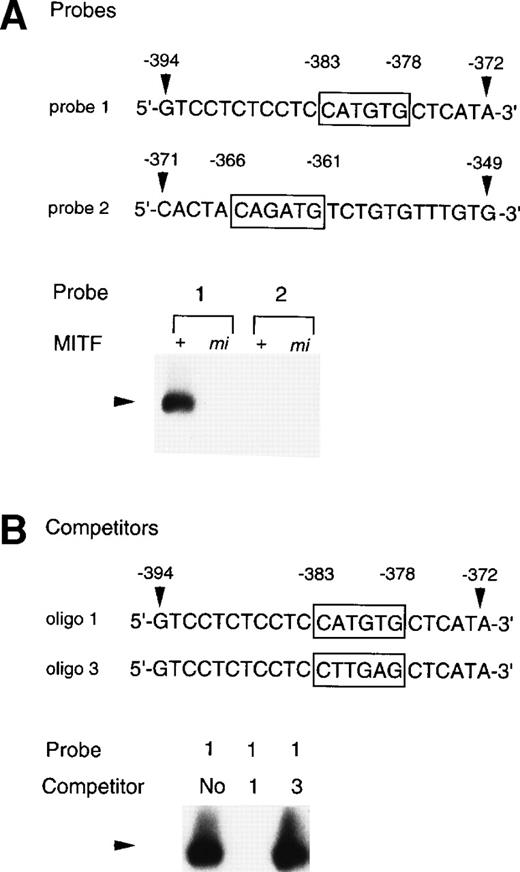
To examine whether the +-MITF protein practically bound the CATGTG motif (nt-383 to -378) and/or the CAGATG motif (nt-366 to -361), EGMSA was performed with each oligonucleotide probe containing the CATGTG or CAGATG motif. The 5′-GTCCTCTCCTCCATGTGCTCATA-3′ oligonucleotide (probe 1: the hexametric motif is shown by the underline) and the 5′-CACTACAGATGTCTGTGTTTGTG-3′ oligonucleotide (probe 2) were synthesized and labeled. EGMSA was performed using GST-+-MITF or GST-mi-MITF fusion protein. No band was detected in the sample that contained no proteins (data not shown). A retarded band was observed in the sample containing the probe 1 and GST-+-MITF but not in the sample containing the probe 1 and GST-mi-MITF (Fig 9A). In contrast to the probe 1, the probe 2 was bound by neither GST-+-MITF nor GST-mi-MITF (Fig9A). To examine the specificity of the binding of the GST-+-MITF to probe 1, we added unlabeled oligonucleotide that had the identical sequence with probe 1 (oligo 1, Fig 9B) to the reaction mixture containing the GST-+-MITF and probe 1. The binding of the GST-+-MITF to probe 1 was completely inhibited (Fig 9B). We then performed competition binding with the oligonucleotide 3, in which the hexametric motif of the probe 1 was mutated from CATGTG to CTTGAG (oligo 3, Fig 9B). The addition of the unlabeled oligo 3 did not affect the binding of GST-+-MITF to the probe 1 (Fig 9B).
Binding of +-MITF to the CATGTG motif in 5′ flanking region of the MMCP-4 gene. (A) EGMSA using GST-+-MITF and GST-mi-MITF fusion proteins. The 5′-GTCCTCTCCTCCATGTGCTCATA oligonucleotide containing a CATGTG motif (probe 1, nt -394 to -372; numbers refer to the sequence shown in Fig 5) and the 5′-CACTACAGATGTCTGTGTTTGTG-3′ oligonucleotide containing a CAGATG motif (probe 2, nt -371 to -349) were used (CANNTG motifs are boxed). The DNA-protein complex is indicated by an arrowhead. (B) Competitive DNA binding assay with GST-+-MITF. Two competitors were synthesized. The oligo 1 was identical to the probe 1; oligo 3 had the mutation at the CATGTG motif (to CTTGAG). DNA-protein complexes are indicated by an arrowhead.
Binding of +-MITF to the CATGTG motif in 5′ flanking region of the MMCP-4 gene. (A) EGMSA using GST-+-MITF and GST-mi-MITF fusion proteins. The 5′-GTCCTCTCCTCCATGTGCTCATA oligonucleotide containing a CATGTG motif (probe 1, nt -394 to -372; numbers refer to the sequence shown in Fig 5) and the 5′-CACTACAGATGTCTGTGTTTGTG-3′ oligonucleotide containing a CAGATG motif (probe 2, nt -371 to -349) were used (CANNTG motifs are boxed). The DNA-protein complex is indicated by an arrowhead. (B) Competitive DNA binding assay with GST-+-MITF. Two competitors were synthesized. The oligo 1 was identical to the probe 1; oligo 3 had the mutation at the CATGTG motif (to CTTGAG). DNA-protein complexes are indicated by an arrowhead.
DISCUSSION
The amount of MMCP-4 mRNA was significantly greater in +/+ CMC than inmi/mi and tg/tg CMC. In situ hybridization also suggested the higher content of MMCP-4 mRNA in the skin mast cells of +/+ mice when compared with that in the skin mast cells ofmi/mi and tg/tg mice. Although the poor expression of the MMCP-7 gene in C57BL/6 strain is due to a splice-site mutation causing instability of its transcript,50 the nucleotide sequence of MMCP-4 cDNA obtained from CMC of C57BL/6-mi/mi mice was identical to that of CMC of C57BL/6-+/+ mice. Thus, the poor content of MMCP-4 mRNA in mi/mi CMC was not attributable to the abnormal structure of the MMCP-4 transcript. Because the nuclear run-on assay showed the lack of the significant amount of the MMCP-4 transcript in mi/mi CMC, the primary reason why the MMCP-4 gene was not expressed in mi/mi mast cells was a decreased rate of the transcription. Moreover, the overexpression of +-MITF but not ofmi-MITF in mi/mi CMC normalized the transcription of the MMCP-4 gene. These results showed the involvement of +-MITF in the transcription of the MMCP-4 gene.
We have reported the poor expression of MMCP-517 and granzyme B55 in mi/mi CMC. In the present study, we also showed the poor expression of MMCP-4 and cathepsin G in mi/miCMC. The expression of genes encoding all MMCP-5, granzyme B, MMCP-4, and cathepsin G appeared to be affected by MITF. Genes encoding granzyme B, cathepsin G, and mast cell chymases are located on chromosome 14 (from 5′ to 3′).28 These results suggest that MITF may affect some global regulatory element responsible for organizing the domains in which these genes lie, or some LCR element that may interact with multiple local regulatory sequences. In fact, Pham et al54 suggested that there is probably a LCR for the granzymes and another LCR for cathepsin G and the downstream chymases. MITF might be involved in the regulatory mechanism of the single LCR or LCRs that may regulate the expression of these proteases.
Although the single LCR or LCRs appeared to regulate the expression of genes that encode mast cell–specific or hematopoietic cell-specific proteases, we could not exclude the other possibility that MITF could activate all of these genes in their different and appropriate compartments. GST-+-MITF fusion protein but not GST-mi-MITF fusion protein bound the CATGTG motif in the 5′-upstream region of the MMCP-4 gene. When the luciferase construct containing the MMCP-4 promoter with the CATGTG motif was transfected into NIH/3T3 fibroblasts, the coexpression of +-MITF but not of mi-MITF significantly increased the luciferase activity, suggesting that the +-MITF transactivated the luciferase gene through the direct binding with the CATGTG motif. We have also reported the direct and functional binding of MITF with the proximal transcriptional element of granzyme B.55 The direct binding of MITF with the 5′-upstream region of MMCP-5 gene was not detectable,17 and the interaction of MITF with the 5′-upstream region of cathepsin G gene has not been examined. We are now planning to investigate the interaction of MITF with 5′ flanking regions of all mast cell–specific chymases located on chromosome 14.
We previously reported that the expression of the MMCP-6 gene was also deficient in both skin mast cells and CMCs of mi/mimice.10,11,14 Overexpression of +-MITF in mi/mi CMC normalized the transcription of the MMCP-6 gene.14The +-MITF protein bound the CANNTG motifs in the 5′-upstream region of the MMCP-6 gene appeared to transactivate it. Although MMCP-4 is a chymase and the MMCP-6 is a tryptase, their transactivation mechanism by +-MITF appeared to be similar. Both MMCP-6 and MMCP-7 are tryptase, and genes encoding MMCP-6 and MMCP-7 are located on chromosome 17. If the expression of MMCP-7 is also deficient in mi/mi CMC, there is a possibility that a LCR in chromosome 17 might regulate the expression of both MMCP-6 and MMCP-7.
The expression of MMCP-5 was also deficient in mi/miCMC.17 In contrast with MMCP-4 and MMCP-6, the expression of MMCP-5 was not deficient in the skin mast cells of mi/mimice. This was attributable to SCF-dependent expression of MMCP-5 mRNA in mi/mi mice. In the culture condition without SCF, the overexpression of the +-MITF but not of mi-MITF normalized the poor expression of the MMCP-5 gene in mi/mi CMC, indicating the involvement of +-MITF in transactivation of the MMCP-5 gene.17 Although +-MITF directly bound CANNTG motifs in the promoter regions of the MMCP-4 and MMCP-6 genes, +-MITF did not bind to the CAGTTG motif in the promoter region of the MMCP-5 gene. The +-MITF appeared to transactivate the MMCP-5 gene indirectly. The expression of MMCP-5 gene might be regulated only by the LCR(s) on chromosome 14, whereas the expression of MMCP-4 might be regulated by both the LCR(s) and the 5′-upstream region of the MMCP-4 gene. Although both MMCP-4 and MMCP-5 are chymases, their transactivation mechanisms by +-MITF appeared to be different.
ACKNOWLEDGMENT
The authors thank Dr H. Arnheiter of NIH for VGA9-tg/tg mice, Dr S. Nagata of Osaka University for pEF-Bos, Dr K. Nakajima of Osaka University for pSPLuc, Kirin Brewery Company Ltd for rmSCF, Dr H. Hamada of Osaka University for technical advice on the nuclear run-on assay, and Dr M. Yamamoto of Tsukuba University for valuable discussion.
Supported by Grants from the Ministry of Education, Science and Culture, the Ministry of Health and Welfare, the Ryoichi Naito Foundation for Medical Research, the Joint Research Project under the Japan-Korea Basic Scientific Promotion Program, and the Organization for Pharmaceutical Safety and Research. Y-M.L. is a postdoctoral fellow supported by the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Yukihiko Kitamura, MD, Department of Pathology, Osaka University Medical School, Yamada-oka 2-2, Suita 565-0871, Japan.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal