To the Editor:
Infection of host cells by human immunodeficiency virus type 1 (HIV-1) is determined by binding of the viral envelope to the CD4 molecule as well as to a coreceptor such as CCR5 or CXCR4.1 Expression of CCR5 is apparently restricted to certain cell types (ie, lymphocytes, monocyte/macrophages, or CD34+ progenitor cells),2-4 which are susceptible to macrophage (M)-tropic (or R5) strains of HIV-1. Because most primary isolates of HIV-1 use CCR5 to enter cells and levels of CCR5 expression correlate well with the infectability with M-tropic HIV-1 in vitro,2 it is important to delineate the cellular and molecular mechanisms whereby expression of CCR5 is regulated. The present study demonstrates that the transcription factor GATA-1 can upregulate CCR5 promoter activity.
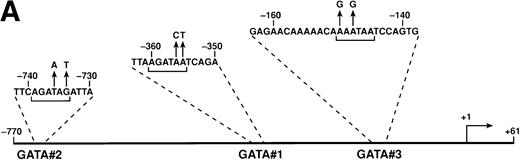
The CCR5 promoter region contains at least three GATA-binding elements (Fig 1A). We have previously identified one of those elements as a GATA-1 binding site4 and have designated it as GATA#1. Characterization of the further upstream region demonstrated another GATA binding site (GATA#2) between −734 and −739 relative to the transcription start site (TSS4; data not shown). Further characterization of the promoter showed that one of the DNase-protected areas (FP-5)4 contains a nonconsensus GATA element (Fig 1B), designated as GATA#3. Therefore, we tested whether transcription factors of GATA family can modulate CCR5 promoter activity in transient expression assays. Plasmid pGL-CCR5(WT) contains the CCR5 promoter region between −770 and +61 relative to the TSS, followed by the luciferase gene, and plasmids pGL-CCR5ΔGATA#1, pGL-CCR5ΔGATA#2, pGL-CCR5ΔGATA#3, and pGL-CCR5ΔGATA#1+2+3 have mutations on these GATA-binding sites individually or in combination (see Fig 1A for mutated nucleotides). The indicated CCR5 promoter-luciferase reporter construct was cotransfected with either a GATA-1 expression vector (pMT-GATA1; kindly provided by S.H. Orkin, Harvard University, Boston, MA5) or its parent plasmid (pMT2T) into PM1 cells (CCR5-positive,2,4 CD4+T-lymphoid cell line) or THP-1 cells (CCR5-positive,4monocytoid cell line). Expression of GATA1, but not GATA-2 or GATA-3 (data not shown), upregulated CCR5 promoter activity up to 15-fold; however, mutation on GATA#1 markedly reduced GATA-1–mediated transactivation of the CCR5 promoter (Fig2). Whereas mutation on GATA#2 or GATA#3 alone had only modest effects on the promoter activity, mutation on all three GATA elements further diminished GATA-1 responsiveness compared with mutation on GATA#1 alone (Fig 2). These results indicate that GATA-1 transcription factor upregulates CCR5 promoter activity through interaction with several GATA elements on the promoter.
Identification of GATA elements on the CCR5 promoter region. (A) Map and sequence of the CCR5 promoter region. Location and sequences of three GATA elements (GATA#1, GATA#2, and GATA#3) are shown. The sequence shown for GATA#3 was also used as an oligonucleotide probe (F5) for gel-mobility shift assays (see B). Nuleotides shown in upward arrows indicate mutated nucleotides in plasmids pGL-CCR5▵GATA#1, pGL-CCR5▵GATA#2, pGL-CCR5▵GATA#3, and pGL-CCR5▵GATA#1+2+3. Brackets indicate GATA elements. (B) Identification of GATA#3 as a binding site for GATA-1 by gel mobility shift assays. A radiolabeled oligonucleotide F5 corresponding to FP-5 (a DNase-protected area in footprinting assays4) was incubated with nuclear extracts from PM1 cells. Lanes 1 and 9 represent probe alone. Where indicated, a 50- or 500-fold molar excess of oligonucleotide indicated above the figure was added as a competitor. F5-m has mutation on the GATA element as in plasmid pGL-CCR5▵GATA#3. For GATA or AP1 consensus oligonucleotide, see Moriuchi et al.4 Gel shift disruption experiments (lanes 9 through 14) were performed by adding either control Ab or monoclonal antibodies to either GATA-1, GATA-2, or GATA-3 (Santa Cruz Biotechnology, Inc, Santa Cruz, CA), as indicated above the figure. The arrow indicates the GATA-1 complex. NS, nonspecific bands; FP, free probe. The experiments were repeated twice with similar results.
Identification of GATA elements on the CCR5 promoter region. (A) Map and sequence of the CCR5 promoter region. Location and sequences of three GATA elements (GATA#1, GATA#2, and GATA#3) are shown. The sequence shown for GATA#3 was also used as an oligonucleotide probe (F5) for gel-mobility shift assays (see B). Nuleotides shown in upward arrows indicate mutated nucleotides in plasmids pGL-CCR5▵GATA#1, pGL-CCR5▵GATA#2, pGL-CCR5▵GATA#3, and pGL-CCR5▵GATA#1+2+3. Brackets indicate GATA elements. (B) Identification of GATA#3 as a binding site for GATA-1 by gel mobility shift assays. A radiolabeled oligonucleotide F5 corresponding to FP-5 (a DNase-protected area in footprinting assays4) was incubated with nuclear extracts from PM1 cells. Lanes 1 and 9 represent probe alone. Where indicated, a 50- or 500-fold molar excess of oligonucleotide indicated above the figure was added as a competitor. F5-m has mutation on the GATA element as in plasmid pGL-CCR5▵GATA#3. For GATA or AP1 consensus oligonucleotide, see Moriuchi et al.4 Gel shift disruption experiments (lanes 9 through 14) were performed by adding either control Ab or monoclonal antibodies to either GATA-1, GATA-2, or GATA-3 (Santa Cruz Biotechnology, Inc, Santa Cruz, CA), as indicated above the figure. The arrow indicates the GATA-1 complex. NS, nonspecific bands; FP, free probe. The experiments were repeated twice with similar results.
GATA-1 upregulates CCR5 promoter activity through interaction with GATA elements on the promoter. Twenty million PM1 cells were transfected with 10 μg of the indicated CCR5 promoter-luciferase construct along with 10 μg of either pMT2T or pMT-GATA-1, and luciferase assays were performed 40 hours after transfection, as described previously.4 Results are reported as means ± SD from four independent experiments. Similar results were obtained for THP-1 cells (data not shown).
GATA-1 upregulates CCR5 promoter activity through interaction with GATA elements on the promoter. Twenty million PM1 cells were transfected with 10 μg of the indicated CCR5 promoter-luciferase construct along with 10 μg of either pMT2T or pMT-GATA-1, and luciferase assays were performed 40 hours after transfection, as described previously.4 Results are reported as means ± SD from four independent experiments. Similar results were obtained for THP-1 cells (data not shown).
GATA-1 is critical for gene expression in hematopoietic progenitor cells5 or eosinophils,6,7 which have been shown to be infectable with M-tropic HIV-13 8; therefore, GATA-1 may play a role in the regulation of CCR5 expression and thereby in the susceptibility of these cells to infection with M-tropic HIV-1.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal