Abstract
In patients with an atypical stem-cell myeloproliferative disorder with lymphoma (B or T cell), myeloid hyperplasia, and eosinophilia, the chromosome 8p11-12 region is the site of a recurrent breakpoint that can be associated with three different partners, 6q27, 9q32-34, and 13q12. Rearrangements are supposed to affect a pluripotent stem cell capable of myeloid and lymphoid differentiation and to involve the same 8p11-12 gene. The t(8;13) translocation has recently been shown to result in a fusion between the FGFR1 gene that encodes a tyrosine kinase receptor for fibroblast growth factors and a novel gene, FIM (also called RAMP or ZNF198), belonging to a novel family of zinc finger genes. In the present study, we have cloned the t(6;8)(q27;p11) translocation in two patients and found a fusion between FGFR1 and a novel gene, FOP(FGFR1Oncogene Partner), located on chromosome band 6q27. This gene is alternatively spliced and ubiquitously expressed. It encodes a protein containing two regions of putative leucine-rich repeats putatively folding in -helices and separated by a hydrophobic spacer. The two reciprocal fusion transcripts were evidenced by reverse transcription-polymerase chain reaction in the tumoral cells of the patients. The predicted chimeric FOP-FGFR1 protein contains the FOP N-terminus leucine-rich region fused to the catalytic domain of FGFR1. It may promote hematopoietic stem cell proliferation and leukemogenesis through a constitutive phosphorylation and activation of the downstream pathway of FGFR1.
SPECIFIC RECURRENT chromosomal translocations have been described for many malignant tumors, including leukemias, lymphomas, and sarcomas.1 Characterization of translocation breakpoints has shown fusion of genes frequently encoding potential transcription factors and nucleic acid-binding proteins or, in some instances, tyrosine kinases,2 which lead to the generation of chimeric products and/or alteration of gene expression, thereby providing a putative oncogenic stimulus.3-5
A recently described distinct myeloproliferative disorder (MPD) is associated with recurrent translocations in which the 8p11-12 chromosomal region is rearranged with three different partners, 6q27, 9q32-34, and 13q12.6 This hematological malignancy is characterized by myeloid hyperplasia, eosinophilia, and T- or B-cell lymphoblastic lymphoma and generally progresses to acute myeloid leukemia (AML). The biphenotypic feature of this MPD strongly suggests that the genetic abnormality occurs in the hematopoietic stem cell. The 8p11-12 breakpoint has been recently characterized.7,8 It is distinct from the breakpoints in other translocations evidenced in AML, ie, t(8;16)(p11;p13)9 and its t(8;14)(p11;q11.1),10 and t(8;19)(p11;q13)11variants and t(8;22)(p11;q13).12,13 The MOZ gene is involved in the t(8;16) translocation14 and in AML with inv(8)(p11;q13).15 We have demonstrated that the three different translocations associated with MPD have the same localization of the 8p11-12 breakpoint and that cosmids specific of the fibroblast growth factor receptor 1 (FGFR1) gene spanned the breakpoint.16FGFR1 encodes a receptor for members of the fibroblast growth factor family.17 Four groups at least have cloned the t(8;13) breakpoint. All studies showed that this translocation results in the fusion of the FGFR1 gene to a previously unidentified 13q12 gene named FIM,18RAMP,19 or ZNF198.20,21 The putative oncogenic fusion protein contains zinc finger motifs, contributed by the product of the 13q12 gene, and the catalytic domain of FGFR1.18-22 The chimeric protein is relocalized to the cytoplasm20 and has a constitutive tyrosine kinase activity.18 These findings strongly suggest that the genesis of the 8p11-12 MPD syndrome is due to altered FGFR1 signal transduction pathways.
Among the rearrangements specific of the 8p11-12 MPD, the t(6;8)(q27;p11) is a rare recurrent translocation and, to date, only four cases have been described.16,23,24 In the present study, we report that FGFR1 is fused to a novel gene we namedFOP (FGFR1OncogenePartner) in two patients. In both cases, the translocation results in the fusion of a putative leucine-rich domain of FOP to the protein kinase domain of FGFR1. The two reciprocal fusion transcripts are expressed. The t(6;8) breakpoint occurs within FGFR1 intron 8, as is the case for the t(8;13) translocation.18
MATERIALS AND METHODS
Patients.
The two patients with a t(6;8)(q27;p11) studied have been partially described in a previous work.16 The clinical phenotype in each case was consistent with the 8p11-12 MPD.6
Patient no. 1 was a 27-year-old man with a white blood cell count (WBC) of 62 × 109/L with 48% myeloid cells. The bone marrow aspirate was extremely hypercellular, with, respectively, 10% and 28% of eosinophilic and lymphoid cell population. A 46,XY,t(6;8)(q27;p11) karyotype was found in the 49 bone marrow-derived metaphases studied. A 7-month treatment with hydroxyurea reduced the WBC. Two years later, a polycythemia vera was diagnosed. Five years later, the disease progressed to acute myeloid leukemia, and the patient died 6 months later after incomplete remission.
Patient no. 2 was a 19-year-old man. When the patient was 12 years of age, a polycythemia vera associated with a cutaneo-mucous lichen was diagnosed. Complete hematological remission was obtained after treatment with hydroxyurea, but the skin disorder was unresponsive to the immunosuppressive treatment. When the patient was 19 years of age, the number of polymorphonuclear neutrophils increased to 20 × 109/L. Bone marrow examination evidenced hyperplasia in the three myeloid lineages without fibrosis or eosinophilia. At that time, chest x-rays showed enlargement of thymus and histological examination after exeresis showed lymphoid hyperplasia. One year later, cervical and axillary lymphadenopathies appeared and histological examination displayed infiltration by myeloid blast cells. Acute transformation into AML4 with 40% blasts in bone marrow was diagnosed. Bone marrow karyotype showed a t(6;8)(q27;p11) in the 32 mitoses analyzed. After high-dose chemotherapy, a return to chronic phase was obtained. Nine months later, a Lyell syndrome was diagnosed and the patient died from septic shock.
Human cell lines.
The following cell lines were cultured as recommended by the supplier: IE8, pre-B/B stage acute lymphoblastic leukemia (ALL); JY, B-ALL; U937, histiocytic lymphoma; KG1, myeloblastic/promyelocytic AML; SU-DHL-1, Ki-1 lymphoma cells bearing a t(2;5)(p23;q35); Daudi, Burkitt’s lymphoma; HSB-2, MO, and MOLT-4, T-ALL; HEL, erythroleukemia AML; HL60, promyelocytic AML treated or not with phorbol esters; MIA PaCa-2, pancreas carcinoma; IMR-90, lung fibroblast; A549, lung carcinoma.
All cell lines were purchased from the American Type Culture Collection (ATCC; Rockville, MD), except for IE8 (gift from T. LeBien, Medical School, University of Minnesota, Minneapolis, MN) and SU-DHL-1 (gift from R. Rimokh, Hôpital E. Herriot, Lyon, France).
Fractionation of peripheral blood cells.
Peripheral blood cells were isolated from blood samples obtained from healthy donors (Centre Régional de Transfusion Sanguine, Marseille, France) and fractionated in the four major hematopoietic nucleated populations (B and T lymphocytes, granulocytes, and monocytes) as previously described.25 The percentage of purified normal cells was more than 90%, as assessed by immunophenotype analysis.
cDNA cloning and sequencing.
The fusion transcript was analyzed with a Marathon cDNA amplification kit (Clontech, Palo Alto, CA). First-strand cDNA was synthesized from 2 μg of t(6;8) total RNA (case no. 1) by using anFGFR1-specific antisense primer (F5R, 5′-ATGGACAGGTCCAGGTACTCC-3′). Second-strand cDNA was synthesized and adapter ligated according to the manufacturer’s protocol. The fusion transcript was amplified with anFGFR1-specific antisense primer (F-4R, 5′-ACTCTGGTGGGTGTAGATCCG-3′) and adapter primer AP1 from the Marathon kit. Polymerase chain reaction (PCR) conditions were as follows: initial denaturation at 95°C for 5 minutes, then 95°C for 1 minute, 60°C for 1 minute, and 68°C for 4 minutes for 30 cycles, with a final elongation step at 68°C for 10 minutes. One tenth of the first-round PCR product was reamplified with a nested-specific FGFR1-specific antisense primer (F-3R, 5′-CTTGGAGGCATACTCCACGAT-3′) and adapter primer AP2. PCR conditions were the same as for the first-round amplification. The second-round PCR product was precipitated in ethanol and subcloned in the pUC18 vector by using the Sure Clone Ligation kit (Pharmacia, Uppsala, Sweden). Nucleotide sequences were obtained using the T7Sequencing kit (Pharmacia) and the forward and reverse sequencing primers for pUC18. Products were analyzed on 5% polyacrylamide/urea sequencing gels.
The full-length FOP cDNA was obtained by screening the SCID t(8;13) library we previously constructed.18 Approximately 106 plaque-forming units were screened with a 1.5-kbNot I-EcoRI fragment from cDNA clone zs55g02. Clone sequencing was performed as described above or at Génome Express (Grenoble, France) on an automated sequencer (Applied Biosystems 373; Applied Biosystems, Foster City, CA). Comparisons with GenBank and dbEST entries were made using BLASTN and TBLASTN.26 Comparisons with protein databases (EMBL; Swissprot, Geneva, Switzerland) used BLASTX, TBLASTX.27 Protein alignment was performed using multiple sequence alignment software28(http://www.toulouse.inra.fr/multalin.html).
Northern blot analyses.
The multiple tissue Northern blots (Clontech; no. 7759-1, 7760-1, and 7767-1) were hybridized according to the manufacturer’s instructions by using, as a probe, the 1.5-kb Not I-EcoRI fragment from cDNA clone zs55g02 labeled with [32P]dCTP in random priming reaction.
Determination of the genomic structure of FOP.
The genomic structure of the gene was deduced based on the cDNA sequences of FOP, ESTs retrieved from dbEST database (http://www.ncbi.nlm.nih.gov/irx/dbST/dbest_query.html), and the genomic sequence from Pl artificial chromosome (PAC) 167A14 (accession no. Z94721). Sequence comparisons were performed using TBLASTN.26 Intron position was deduced from the determination of both 5′ and 3′ limits of the exons.29
For alternative splicing analysis, reverse transcription-PCR (RT-PCR) experiments were performed. Three primer sets from the 5′ end (sense, 5′-GGAGCGTACCCTGCTGCG-3′; antisense, 5′-CCTTCGAGACCTTGCAGC-3′), middle part (sense, 5′-TTAGTGGCTAGTCTTGTTGC-3′; antisense, 5′-TAAAGAAGGGGCTCCCGC-3′), and 3′ end (sense, 5′-GAAGCAAGCAGGAAGTCTGG-3′; antisense, 5′-GCAGTGTTTTTGAAATGCAAGG-3′) of FOP sequence were used to amplify FOP transcripts. PCR reactions were performed in a Perkin Elmer-Cetus (Norwalk, CT) apparatus using an equivalent of 500 ng of reversed transcribed RNA and 50 pmol of each oligonucleotide as primers. The first cycle was run as follows: denaturation at 95°C for 5 minutes, annealing at 60°C for 30 seconds, and synthesis at 72°C for 1 minute. The next 30 cycles were run using the same conditions except that the denaturation step was only 30 seconds. For the last cycle, the synthesis time was 7 minutes. PCR products were gel-purified in agarose 1%, subcloned and sequenced as described above.
Analysis of gene expression by RT-PCR.
RT-PCR reactions were performed from 2 μg of total RNAs as previously described.18 Fusion transcripts were analyzed for the two t(6;8) patients. FOP-FGFR1 fusion transcript was amplified with the FOP-specific sense (FOP-1F, 5′-CATTCTCCACCAAAGTCACCA-3′) and FGFR1-specific antisense (F-9.2, 5′-CATACTCAGAGACCCCTGCTAGC-3′) primers, giving a 162-bp product. FGFR1-FOP fusion transcript was amplified with FGFR1-specific sense (FA, 5′-ATCATCTATTGCACAGGGGCC-3′) and FOP-specific antisense (FOP-1R, 5′-CCCGCTTGTCTTCTTCTTACC-3′) primers that generated a 197-bp product. PCR conditions were as described above.
The wild-type FOP and FGFR1 transcripts were amplified using FOP-1F/FOP-1R (PCR product of 101 bp) and FA/F-9.2 (PCR product of 258 bp) primer pairs, respectively. PCR conditions were as described above.
The human β2 microglobulin primer pair (sense, 5′-CCAGCAGAGAATGGAAAGTC-3′; antisense, 5′-GATGCTGCTTACATGTCTCG-3′; PCR product of 268 bp) was used as a control for RT and PCR efficiency.
RESULTS
Cloning of the MPD t(6;8) breakpoint.
To clone the putative fusion gene, we did PCR amplification of cDNA 5′ ends (5′-RACE) from t(6;8) malignant cell total RNA (case 1) by using FGFR1-specific oligonucleotides. The resulting PCR products were cloned. Two chimeric clones of 400 and 800 bp were sequenced, showing that a sequence not previously characterized was fused in a continuous reading frame to FGFR1 at nucleotide 1272 from ATG (accession no. M34185). Database searches with the novel sequence showed identity with human DNA sequences from one PAC, PAC 167A14 (EMBL Z94721) localized on chromosome 6q27, but did not show significant similarity or identity to any known gene or protein. We named this novel gene FOP. ESTs were shown to correspond to PAC 167A14 and we studied some of these cDNA clones. Sequencing of cDNA clone zs55g02 from which two ESTs (AA286845 and AA286846) are available in dbEST showed that it contained the sequences surrounding the breakpoint identified in PCR products from patient RNA.
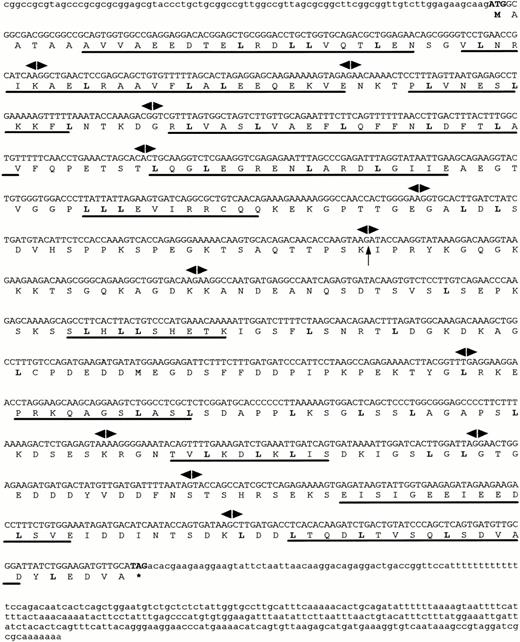
Cloning of FOP gene.
A cDNA library we previously constructed18 was screened with the insert from cDNA clone zs55g02 as a probe. Two of the largest clones isolated were sequenced. They all shared identical sequences with ESTs AA286845 and AA286846, one of which had a 5′ extended end of 150 bp and was approximately 1.7 kb in length. The cDNA and deduced aminoacid sequences of FOP are shown in Fig 1. Within the full-length cDNA of 1,630 bp (EMBL: Y18046), a single open reading frame of 1,197 nucleotides starts with the putative ATG at nt 85. The sequence surrounding this codon matches the consensus sequence for optimal translation initiation, as determined by NetStart 1.0, a program for translation start prediction (http://www.cbs.dtu.dk/services/NetStart/). A stop codon at nt 1,282 is followed by a 338-bp long 3′ untranslated sequence, including a polyadenylation signal upstream to a poly(A) tail. It encodes a putative protein of 399 amino acid residues (aa).
Sequences of the human FOP gene. A full-lengthFOP cDNA clone is shown with the predicted amino acid sequence of an open reading frame. Leucine residues that may correspond to a novel type of leucine-rich motifs are in bold. Putative -helices are underlined. Arrowheads indicate exon/intron boundaries. The arrow corresponds to the t(6;8) breakpoint. The asterisk indicates the stop codon.
Sequences of the human FOP gene. A full-lengthFOP cDNA clone is shown with the predicted amino acid sequence of an open reading frame. Leucine residues that may correspond to a novel type of leucine-rich motifs are in bold. Putative -helices are underlined. Arrowheads indicate exon/intron boundaries. The arrow corresponds to the t(6;8) breakpoint. The asterisk indicates the stop codon.
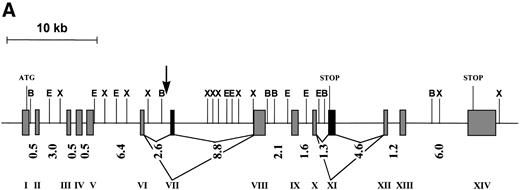
The genomic structure of FOP was deduced based on the cDNA sequences, ESTs retrieved from dbEST, and the genomic sequence from PAC 167A14. It is composed of 13 exons spanning more than 40 kb of genomic DNA. The size of the exons is highly homogeneous, whereas the size of introns is heterogeneous, ranging from 0.5 to 8.8 kb (Fig 2A). All of the intron/exon boundaries agree with the canonic acceptor and donor splice sites.29
(A) Schematic representation of the genomic structure of the FOP gene. All exons are represented in the 5′ to 3′ order of genomic DNA: shaded and solid boxes represent constant and alternatively spliced exons, respectively. Arabic and roman numbers indicate the size in kilobases of introns and the position of exons, respectively. The arrow indicates the breakpoint. Restriction enzyme sites are BamHI (B), EcoRI (E), andXba I (X). (B) Similarities of the deduced human FOP protein (Hs) with EST protein translation from Mus musculus (Mm),Danio rerio (Dr), Leishmania major (Lm), andOryza sativa (Os). Shading indicates common (boxed) and similar amino acid residues. Numbers at the top of each alignment indicate amino acid positions in FOP. Asterisks show conserved leucine residues.
(A) Schematic representation of the genomic structure of the FOP gene. All exons are represented in the 5′ to 3′ order of genomic DNA: shaded and solid boxes represent constant and alternatively spliced exons, respectively. Arabic and roman numbers indicate the size in kilobases of introns and the position of exons, respectively. The arrow indicates the breakpoint. Restriction enzyme sites are BamHI (B), EcoRI (E), andXba I (X). (B) Similarities of the deduced human FOP protein (Hs) with EST protein translation from Mus musculus (Mm),Danio rerio (Dr), Leishmania major (Lm), andOryza sativa (Os). Shading indicates common (boxed) and similar amino acid residues. Numbers at the top of each alignment indicate amino acid positions in FOP. Asterisks show conserved leucine residues.
Multiple FOP cDNA variants were identified as the result of alternative splicing. In some cDNA clones an additional exon, exon 11 (Fig 2A), was identified. Variant transcripts resulting from splices of either exon 7 or exon 11 were cloned by RT-PCR and also found in several human and murine ESTs (including AA919880) and a human EST (N46560), respectively. It must be pointed out that, when exon 11 is not spliced out, a shorter protein of 331 aa can be deduced as the result of the presence of an in-frame stop codon. Moreover, an alternative splice is suspected in the FOP 3′ noncoding part.
FOP sequence motifs.
Analysis of the transcript containing 1,197-bp FOP coding sequence suggests that FOP encodes a protein of 399 amino acid residues with a predicted molecular mass of 44.3 kD. FOP is a protein largely hydrophilic and contains, in its N- and C-termini, several regions folding in α-helices, as determined using GOR secondary structure prediction program.30 These two regions are interrupted by a hydrophobic spacer region (Fig 1). The α-helix regions contain repeats with the consensus sequence L-X2-L-X3-5-L-X3-5-L, in one-third of which the leucine is substituted by either a valine or an isoleucine.
Evidence for a novel gene family.
The EST database (NCBI, Bethesda, MD) was screened for sequences similar to the FOP gene. Several human ESTs, whose deduced amino acid sequence (after translation in the 6 reading frames) has similarities to portions of the FOP protein, were found. These results are evidence for a multigenic family. EST similarities were found not only in vertebrates including human, mouse (AA103209,AA919880), and zebrafish (AA497344), but also in more distant phyla such as monocellular eucaryotes (leishmania, AI034687) and plants (rice, RICS1091A). The alignment of the deduced N-terminus of the FOP protein with translated nonhuman ESTs displaying sequence similarities and encoding putative orthologs is shown in Fig 2B.
Gene expression.
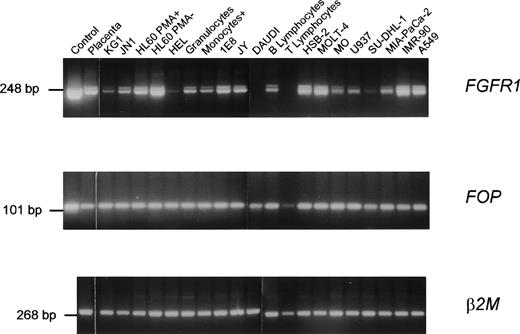
Expression of the wild-type FOP gene was investigated by Northern blot analysis of poly(A)+ RNAs of various human tissues. Two major transcripts of approximately 1.2 and 1.7 kb were detected in all samples (Fig 3). A high level of expression and an additional transcript of approximately 3 kb were detected in the heart. Using RT-PCR analysis, FOP expression was found in normal hematopoietic cells as well as in different malignant cell lines (Fig 4). Similarly, FGFR1expression was found in normal and tumoral cells (Fig 4), in agreement with previous studies.18,31 32FOP andFGFR1 were expressed in the two t(6;8) patients’ samples (Fig 5, rows 1 and 4 in the middle and right panels, respectively).
FOP expression. Clontech Northern blots with the indicated poly(A)+ RNAs were hybridized with anFOP probe derived from cDNA insert of clone zs55g02. β-Actin was used as a control. The marker sizes (in kilobases) are indicated on the left.
FOP expression. Clontech Northern blots with the indicated poly(A)+ RNAs were hybridized with anFOP probe derived from cDNA insert of clone zs55g02. β-Actin was used as a control. The marker sizes (in kilobases) are indicated on the left.
Expression of FGFR1 and FOP genes. RT-PCR products were obtained from a variety of tissues and normal and tumoral hematopoietic cells using specific primer pairs of each gene. Each panel is a photograph of the ethidium bromide-stained agarose gel in which PCR products were electrophoresed. Labels at the top of rows indicate the source of the material. The first row corresponds to cDNA controls: a 2.5-kb FGFR1 cDNA (pFLG-16) containing the complete coding sequence (upper panel)49 and the FOP cDNA clone zs55g02 (middle panel), respectively. The other rows correspond to RNAs from a variety of lympho-hematopoietic cell lines (listed in Materials and Methods) and mature peripheral (T lymphocytes, B lymphocytes, and monocytes) blood cells purified as indicated in Materials and Methods. β2 microglobulin (β2M) amplification was used as a control.
Expression of FGFR1 and FOP genes. RT-PCR products were obtained from a variety of tissues and normal and tumoral hematopoietic cells using specific primer pairs of each gene. Each panel is a photograph of the ethidium bromide-stained agarose gel in which PCR products were electrophoresed. Labels at the top of rows indicate the source of the material. The first row corresponds to cDNA controls: a 2.5-kb FGFR1 cDNA (pFLG-16) containing the complete coding sequence (upper panel)49 and the FOP cDNA clone zs55g02 (middle panel), respectively. The other rows correspond to RNAs from a variety of lympho-hematopoietic cell lines (listed in Materials and Methods) and mature peripheral (T lymphocytes, B lymphocytes, and monocytes) blood cells purified as indicated in Materials and Methods. β2 microglobulin (β2M) amplification was used as a control.
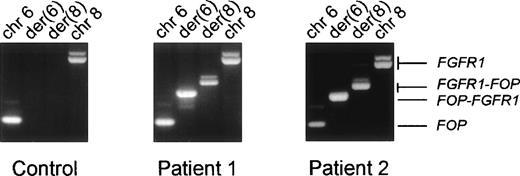
Expression of the fusion transcripts. RT-PCR were performed using RNAs from the erythroleukemia HEL cell line used as a control (left panel) and from the two t(6;8) patients’ malignant cell samples and specific primers located near the translocation breakpoint. The respective chromosomal positions and the transcripts identified in each reaction are indicated at the top and on the right, respectively.
Expression of the fusion transcripts. RT-PCR were performed using RNAs from the erythroleukemia HEL cell line used as a control (left panel) and from the two t(6;8) patients’ malignant cell samples and specific primers located near the translocation breakpoint. The respective chromosomal positions and the transcripts identified in each reaction are indicated at the top and on the right, respectively.
Predicting the transcription of chimeric messages over the breakpoint, we designed primers from FOP and FGFR1 to do RT-PCR on RNA from the two t(6;8) patients. Both FGFR1-FOP andFOP-FGFR1 reciprocal transcripts were detected in patients (Fig 5, rows 2 and 3 in the middle and right panels, respectively) but not in normal and other tumoral samples (Fig 5, left panel, rows 2 and 3, respectively). They proved to encode in-frame fusion junctions on sequence analysis (Fig6).
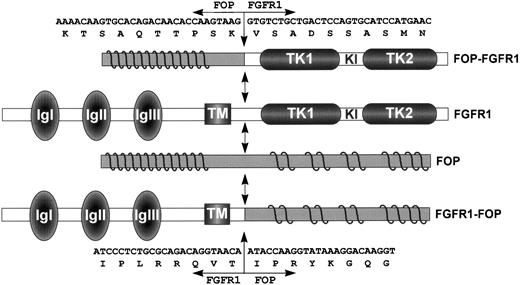
Schematic representation of FGFR1, FOP, and both FOP-FGFR1 and FGFR1-FOP fusion proteins. FGFR1 domains are indicated as follows: IgI, IgII, and IgIII, the three Ig-like domains; TM, the transmembrane domain; and TK1 and TK2, the tyrosine kinase 1 and 2 subdomains interrupted by a kinase insert (KI). Waved lines in the N- and C-terminal parts of FOP represent the leucine-rich repeats. FGFR1-FOP retains the extracellular and transmembrane domains of FGFR1, whereas FOP-FGFR1 fusion protein contains the FGFR1 catalytic domain. Double arrows indicate the t(6;8) breakpoint. Nucleotide and amino acid breakpoint sequences are indicated for both resultant chimeric products.
Schematic representation of FGFR1, FOP, and both FOP-FGFR1 and FGFR1-FOP fusion proteins. FGFR1 domains are indicated as follows: IgI, IgII, and IgIII, the three Ig-like domains; TM, the transmembrane domain; and TK1 and TK2, the tyrosine kinase 1 and 2 subdomains interrupted by a kinase insert (KI). Waved lines in the N- and C-terminal parts of FOP represent the leucine-rich repeats. FGFR1-FOP retains the extracellular and transmembrane domains of FGFR1, whereas FOP-FGFR1 fusion protein contains the FGFR1 catalytic domain. Double arrows indicate the t(6;8) breakpoint. Nucleotide and amino acid breakpoint sequences are indicated for both resultant chimeric products.
Characterization of the fusion junctions.
We next determined the nucleotide sequence spanning the breakpoints on both parental and translocated chromosomes. On chromosome 8, the break occurred in FGFR1 intron 8, 139 bp upstream of exon 9, which encodes the intracellular juxtamembrane domain. Two variants of theFGFR1-FOP fusion transcribed from chromosome der(8) were obtained. They differ by the use of alternativeFGFR1 5′ splice sites located six nucleotides apart at the 3′ end of exon 8. They lead to inclusion or not of two amino acids, VT (Fig 3), in the juxtamembrane region, a feature described for wild-type FGFR1,33,34 and in the fusion product resulting from the t(8;13) translocation.18 On chromosome 6, the break occurred in FOP intron 6, 424 bp downstream of exon 6.FOP intron 6 is 2.6-kb long and contains two AT rich regions, a CA(n) repeat region and a subtype of long terminal repeat (LTR/MaLR), located downstream of the breakpoint. The chimeric sequence 5′-ATGGTCTTT-3′ of the rearranged intron from theFOP-FGFR1 fusion gene results from the sequences 5′-ATGGTTTTT-3′ and 5′-ATGGTCTTT-3′ ofFOP intron 6 and FGFR1 intron 8, respectively. It is noteworthy that the breakpoint occurs in an identical stretch of nucleotides (but for one) in both FOP and FGFR1wild-type introns. The characteristics of the wild-type FGFR1 and FOP proteins, the fusion proteins FOP-FGFR1 and FGFR1-FOP, are depicted in Fig 6.
DISCUSSION
In this study, we describe the fusion of the FGFR1 gene with a previously uncharacterized gene, FOP, in the t(6;8) translocation involved in the myeloproliferative syndrome linked to the 8p11-12 chromosomal region. Two novel leukemia-specific fusion genes,FGFR1-FOP and FOP-FGFR1, were identified as the result of the translocation. In contrast to many chromosome translocations, both chimeric gene derivatives are expressed in patient cells. This observation is consistent with two works in which the expression of both FIM-FGFR1 andFGFR1-FIM RNAs in cells with t(8;13) associated with the same syndrome.18 22 In both t(8;13) and t(6;8) translocations, the breakpoints in the FGFR1 gene lie in intron 8.
To date, two of the three chromosomal translocations consistently associated with myeloproliferative disorder linked to the 8p11-12 chromosomal region have been cloned. The FGFR1 partners are unrelated novel genes. Our preliminary results from the molecular study of the t(8;9) translocation demonstrate that the 9q32-34 gene is also a novel gene that is different from the two others (Popovici et al, manuscript in preparation). In each fusion, the N-terminal region of the novel proteins, postulated to trigger protein dimerization, is fused to the catalytic domain of FGFR1. In the t(8;13), the 13q12 gene encodes a member of a novel family of zinc finger proteins.18-21 In the present work, we show that the t(6;8) translocation is associated with a leukemic aberrant protein in which a putative leucine-rich N-terminal region encoded by the widely expressed chromosome 6 FOP gene. Indeed, FOP encodes a protein that has, in its N- and C-terminal parts, some structural features involving putative α-helices. The comparison of aligned internal FOP sequences show considerable conservation of leucines that may correspond to a novel type of leucine-rich repeats. Because these motifs are different from the consensus leucine-rich repeats present in a large family of proteins involved in various functions and found in a great variety of species,35 36 we postulate that FOP may define a novel family of leucine-rich proteins. In addition, we retrieved from databases several ESTs with sequence similarities from humans and from distant phyla. Altogether, these results suggest that FOP belongs to a multigenic family and is highly conserved during evolution.
To date, proteins containing leucine-rich repeats are thought to be involved in protein-protein interactions, and at least half of them take part in signal transduction.35 We previously showed that the putative oncogenic fusion gene involved in the t(8;13) translocation has a constitutive tyrosine kinase activity.18 Thus, the oncogenic role of theFOP-FGFR1 fusion gene product may be mediated by constitutive phosphorylation of the FGFR1 kinase domain triggered by dimerization of FOP leucine-rich motif.
Distinct species of human FOP cDNAs derived from differential splicing were identified. This raises the possibility that, although the FOP gene is expressed ubiquitously in various tissues and organs, the expression of its alternative splicing products may be regulated in a tissue-specific fashion.
The FGFR1 gene encodes a tyrosine kinase receptor for members of the fibroblast growth factor family.17 FGFR1 is one of the four FGF receptors responsible for mediating cellular responses to fibroblast growth factor signaling, which include mitogenesis, mesoderm induction, angiogenesis, chemotaxis, and neuronal survival.37,38 Signals from FGFRs appear to control differentiation as well as proliferation. FGFR1 is specifically involved in a hereditary craniosynostosis syndrome39 and may contribute to tumorigenesis in human cancers. FGFR2 constitutive activation after chromosomal rearrangement was shown in rat osteosarcoma cells.40 Activating mutations associated with translocation involving FGFR3 and the IgH locus were described in some cases of multiple myeloma.41FGFR1 may be the key gene for driving the 8p11 amplification process in breast cancers.42-45 Its likely oncogenic role in the 8p11-12 MPD has been documented in previous works16 18-21 and is strengthened by this study. These data therefore provide a new example of an oncogene implicated in both solid tumor formation and leukemogenesis.
Our present data suggest not only that alteration of transcriptional cascades mediated by FGFR1 signaling may be a mechanism of leukemic transformation, but also that FGFR1 may play a role in normal hematopoietic development. The finding of acquired chromosomal translocations that are consistently associated with specific tumor types supports the hypothesis of lineage-specific mechanisms of tumorigenesis.46 In the case of 8p11-12 MPD, the crucial event for transformation is likely to take place in the hematopoietic stem cell, as previously postulated,6 leading to the disruption of normal hematopoiesis of both lymphoid and myeloid lineages. Increased expression of FGF2 and its receptors has been recently shown in CD34+ hematopoietic progenitors from patients suffering from an MPD with myelofibrosis and myeloid metaplasia.47 Molecular reagents derived from t(6;8) or t(8;13) may be used to facilitate the expansion of stem cells by stimulating their growth and/or survival and by overcoming negative regulatory signals.
In MPD, proliferative activity in the bone marrow is seldom limited to a single lineage, but often involves the erythroid, granulocytic, and/or megakaryocytic series.48 It may be of interest to note that, whereas overlapping clinical features are found in all patients with MPD linked to 8p11-12,6 a polycythemia vera was diagnosed in both patients with a t(6;8) translocation. This suggests that the chromosome 6 gene, FOP, may be an important gene for normal proliferation and differentiation of the erythroid lineage. A better understanding of the molecular processes affected by the fusion of the genes involved in such an MPD may shed light on the role of FGFR1 and its multiple fusion partners in leukemogenesis and on the biology of the normal hematopoietic stem cell.
ACKNOWLEDGMENT
The authors thank Drs C. Mawas and D. Maraninchi for encouragement and comments. We are grateful to Drs F. Birg, V. Ollendorff, and P. Pontarotti for helpful discussions. Thanks are also due to J. Adélaı̈de for occasional technical help and J. Simonetti for patient no. 2 karyotype.
Supported by INSERM, Institut Paoli-Calmettes, and grants from the Ligue Nationale contre le Cancer, Comité du Var de la Ligue Nationale contre le Cancer, and FEGEFLUC. C.P. and B.Z. were recipients of a fellowship from the Société Française d’Hématologie and People’s Republic of China, respectively.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Marie-Josèphe Pébusque, PhD, Institut de Cancérologie et d’Immunologie de Marseille, Laboratoire d’Oncologie Moléculaire, INSERM U119, 27 Bd Leı̈ Roure, 13009 Marseille, France; e-mail:pebusque@marseille.inserm.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal