Abstract
Conventional chromosome analysis (CCA) and interphase fluorescence in situ hybridization (FISH) was performed in 42 patients with mantle-cell lymphoma (MCL), with BCL1 rearrangement. The t(11;14)(q13;q32) or 11q abnormalities were detected by CCA in 34 cases, 20 of which had additional aberrations. A normal karyotype was observed in 8 cases. Probes detecting the chromosome aberrations that were observed in at least 3 cases by CCA, ie, +12, 13q14 deletion, and 17p deletion, were used for interphase FISH analysis. FISH detected total or partial +12, 13q14 deletion and 17p- in 28.5%, 52.4%, and 26% of the cases, respectively. The presence of these anomalies was not a function of karyotype complexity. Based on the results of CCA/FISH, three groups of increasing karyotype complexity were recognized: group 1, including 11 patients without detectable aberrations in addition to BCL1 rearrangement; group 2, including 14 patients with 1 to 2 additional anomalies; and group 3, including 17 patients with three or more additional anomalies. Clinical parameters associated with shorter survival were male sex (P= .006) and primary lymph-node involvement compared with primary bone marrow involvement (P = .015). Trisomy 12 was the only single cytogenetic parameter predictive of a poor prognosis (P = .006) and the best prognostic indicator was the derived measure of karyotype complexity (P < .0001), which maintained statistical significance in multivariate analysis (P< .0001). We arrived at the following conclusions: 13q14 deletion occurs at a high incidence in MCL; 17p deletion and total/partial +12 are relatively frequent events in MCL, the latter aberration being associated with a shorter survival; and the degree of karyotype complexity has a strong impact on prognosis in this neoplasia.
USING SENSITIVE MOLECULAR cytogenetic techniques, the majority of cases of lymphoma of follicle mantle lineage were shown to carry the 11;14 translocation1-4 and its molecular counterpart, consisting of the juxtaposition of theBCL1 locus and the immunoglobulin heavy-chain locus on the derivative 14q+ chromosome.5-7 This disease, which was recognized as a distinct clinicopathological entity as early as 1982,8 accounts for 3% to 9% of all non-Hodgkin’s lymphomas (NHL) in western countries.9 It more frequently affects middle-aged to elderly males, who usually have advanced disease involving lymph nodes and spleen as well as extranodal sites, especially the gastrointestinal (GI) tract and the bone marrow (BM). Frequently, peripheral blood (PB) involvement also occurs, either during disease evolution10 or at diagnosis, mimicking chronic lymphocytic leukemia (CLL).11
Although a number of hematologic and biologic variables were shown to have correlation with survival in MCL,9,12,13 there is interstudy variability as to the prognostic significance of each of these variables,13-17 partially on account of the limited number of cases studied and of the heterogeneity of patient population.
There is evidence that genetic lesions may help identify prognostically different subgroups of NHL.18-20 Thus, p53 lesions were reproducibly associated with aggressive disease in MCL21,22and in B-cell CLL,23 as was the case with chromosome 17p deletions20,24,25 and aberrations of chromosomes 1, 6, and 1120,26 27 that were found to have prognostic significance in several subtypes of NHL.
In an attempt to better define the incidence and nature of chromosome lesions in MCL and to disclose cytogenetic patterns having prognostic significance, we identified 42 patients with BCL1 involvement and performed conventional cytogenetic analysis (CCA), that identified 13q14 deletion, 17p- and trisomy 12q as the most frequently occurring aberrations in addition to the t(11;14)(q13;q32). These chromosome lesions were subsequently investigated by the more-sensitive interphase fluorescence in situ hybridization (FISH) technique and an analysis was performed of the correlation between these chromosome lesions and salient hematologic parameters.
MATERIALS AND METHODS
Patients and clinical parameters.
Fifty-nine patients with non-Hodgkin’s lymphoma (NHL) of follicle mantle lineage were diagnosed at the Institute of Hematology, University of Ferrara (Ferrara, Italy), over a 10-year period. Histologic diagnosis was performed according to recently summarized criteria9,28 29 on lymph-node specimens and/or bone-biopsy sections.
Of these 59 patients, 42 cases fulfilling the following criteria were included in the present study: (1) karyotype available for review. (2) Presence of the t(11;14) (q13;q32), or the corresponding BCL1involvement as detected by FISH technique on representative lymph-node or PB samples. FISH was shown in previous studies to be a very sensitive method for the detection of rearrangements occurring in the BCL1 locus.2,3 (3) Histologic picture of MCL on lymph-node specimens (28 cases with primary lymph-node involvement); or bone biopsy sections consistent with infiltration by MCL (14 patients with primary involvement of the BM and PB who had no superficial adenopathy available for biopsy). (4) Mantle-cell immunophenotype in cases with leukemic expression, ie, CD5/CD19+, CD22+, CD23−, CD10−, and bright expression of surface immunoglobulins (sIg). Other forms of NHL, CLL, and related disorders (ie, CLL/PL and prolymphocytic leukemia) as well as other chronic (mature) B-cell lymphoproliferative disorders30 were excluded from this analysis, irrespective of the presence of the t(11;14).
Staging procedures included physical examination, a routine laboratory profile, a chest radiograph, and abdomen ultrasonography. When indicated, barium contrast radiography was performed. Computed tomography (CT) scan was performed for staging purposes in 32 cases, bone biopsy was performed in 34 cases (including 14 cases with primary BM involvement). BM aspiration was performed in all cases. PB involvement was studied by light microscopy examination of smears stained by the May-Grunwald-Giemsa method and by immunophenotyping using the following panel of commercially available monoclonal antibodies: anti CD2, CD3, CD5, CD19, CD22, CD23, CD10, and FMC7. Double labeling with anti-CD5/CD19 was performed and the expression of sIg was studied using rabbit antihuman antibodies against the Ig heavy and light chains as previously reported.31
Treatment was not homogeneous. Depending on age, stage, performance status, and on the clinical course the following first-line treatments were used: single-agent therapy (chlorambucil) in 10 cases and multiagent chemotherapy using cyclophosphamide, vincristine, and prednisone, with (26 cases) or without (6 cases) an anthracycline.
Clinical records were surveyed for all cases and the parameters outlined in Table 1 were collected.
Clinical and Biological Parameters at Presentation in 42 Patients With MCL and BCL1 Involvement
| . | Median (Range) . |
|---|---|
| Age yr | 68 (40-88) |
| Hb g/dL | 12.5 (7.8-15.0) |
| WBC × 109/L | 8.5 (1.8-840) |
| Platelet × 109/L | 184 (10-677) |
| LDH U/L | 440 (250-1200) |
| Albumin g/dL | 3.8 (3.1-4.9) |
| No. of cases/total | |
| Splenomegaly | 19/42 (45.2%) |
| Male/female | 29/13 |
| LDH abnormal | 18/41 (43.9%) |
| Advanced stage* | 31/42 (73.8%) |
| B-symptoms | 14/42 (33.3%) |
| BM involvement† | 27/42 (64.3%) |
| Leukemic expression‡ | 31/42 (73.8%) |
| Performance status (ECOG) | |
| 0-1 | 36/42 (85.7%) |
| 2-4 | 6/42 (14.3%) |
| HISTOLOGY | |
| Diffuse/nodular1-153 | 26/2 |
| Classical/blastoid | 39/3 |
| . | Median (Range) . |
|---|---|
| Age yr | 68 (40-88) |
| Hb g/dL | 12.5 (7.8-15.0) |
| WBC × 109/L | 8.5 (1.8-840) |
| Platelet × 109/L | 184 (10-677) |
| LDH U/L | 440 (250-1200) |
| Albumin g/dL | 3.8 (3.1-4.9) |
| No. of cases/total | |
| Splenomegaly | 19/42 (45.2%) |
| Male/female | 29/13 |
| LDH abnormal | 18/41 (43.9%) |
| Advanced stage* | 31/42 (73.8%) |
| B-symptoms | 14/42 (33.3%) |
| BM involvement† | 27/42 (64.3%) |
| Leukemic expression‡ | 31/42 (73.8%) |
| Performance status (ECOG) | |
| 0-1 | 36/42 (85.7%) |
| 2-4 | 6/42 (14.3%) |
| HISTOLOGY | |
| Diffuse/nodular1-153 | 26/2 |
| Classical/blastoid | 39/3 |
Stage III or IV.
Assessed by bone biopsy in 34 cases and by BM aspirate in 8 cases.
Positive cases defined by >10% CD5/CD19-positive cells in the lymphocyte gate.
The histologic architecture of lymphoma growth was assessed on lymph node specimens and not on bone biopsy sections.
CCA.
Cytogenetic investigations were performed on lymph-node and/or PB samples obtained within 3 months of diagnosis in all patients. Single-cell suspensions were prepared, as previously described,32 after collection of a portion of surgically removed lymph node (28 cases, 6 of which were also studied on PB samples), and on PB mononuclear cells obtained by separation over a 1,077 mg/mL density gradient in 14 cases with prominent leukemic involvement. PB and lymph-node cell suspensions containing greater than 90% CD5/CD19+ lymphocytes were cultured for 24 to 72 hours, with and without the following mitogens: phorbol miristate acetate (50 ng/mL), lipopolysaccaride from Escherichia coli (100 mg/mL), and phytohemagglutinin M-form (100 mg/mL). Whenever possible, 20 to 30 metaphases were studied and karyotypes described according to the ISCN.33
Interphase cytogenetics.
Interphase FISH studies were performed using probes detecting BCL1 rearrangements and those chromosome lesions that were found in at least three patients by CCA. These studies were performed on cells taken from the same samples that were used for cytogenetic analysis.
BCL1 involvement was documented as previously reported,34 using the yeast-artificial-chromosome (YAC) probe 214D11, spanning a 390 kb region encompassing the major translocation cluster and the minor translocation clusters of theBCL1 locus at 11q13.35 The presence of three signals in interphase cells (one deriving from the normal allele and two deriving from the split BCL1 allele) was considered indicative of BCL1 involvement. To prevent data misinterpretation due to the presence of trisomy or monosomy 11, a chromosome 11-specific centromeric probe (Oncor, Gaithersburg, MD), or the cosmid cCI11-395, located at 11p15.5, which was obtained from the Japanese Cancer Research Resources Bank,3 were used as control probes. To document the juxtaposition of BCL1/14q sequences, dual-color FISH was performed in those cases without cytogenetic evidence of the t(11;14), using the BCL1 probe and a 14q telomere probe, or the cos a2 IgH constant-region probe (prepared by H. Dohner, Ruprecht-Karls-Universitat, Medizinische Klinik und Poliklinik V, Heidelberg, Germany) because deletions of 13q, 17p, and total or partial trisomy 12 were the most frequent abnormalities in addition to the 11;14 translocation, having been found in at least 3 cases. Probes for the detection of these anomalies in interphase cells were used to increase the sensitivity of our analysis. The 13q14 C21 cosmid, recognizing DNA sequences between the Rb gene and the D13S25 marker,36 was isolated as previously described.34 Simultaneous hybridization with a chromosome 13-telomere probe (Oncor) was performed. The p53.3 cosmid recognizing p53 gene sequences at the 17p13 chromosome band was prepared and distributed by H. Döhner through F. Birg (Institut de Cancérologie e d’Immunologie de Marseille, INSERM 119, Marseille, France) in the context of the Biomed I programme, “E.U. concerted action for cytogenetic diagnosis of hematologic malignancies” (Project leader: A. Hagemeijer, Centre for Human Genetic, K.U.L., Leuven, Belgium). Simultaneous hybridization with a chromosome 17-centromeric probe was performed (Oncor). A commercially available chromosome 12-pericentromeric probe was used for the detection of total/partial trisomy 12 (Oncor).
Hybridization and signal screening.
The hybridization protocol was described in detail in previous studies.3 34 To prevent false-positive results due to inefficient hybridization, signal screening was performed on slides with a high hybridization efficiency, having greater than 80% interphase cells showing two signals with the control probe. The evaluation was performed on a fluorescence microscope (Nikon Italia, Florence, Italy); 200 cells with well-delineated signals were observed and images were captured with a charged-coupled camera device (Genevision, Nikon Italia).
Five control slides obtained from non-neoplastic tissue were tested with each probe. The cut off for the recognition of BCL1involvement and +12 was set at 5% interphase cells with 3 signals, whereas greater than 10% cells with 1 signal were required for the recognition of 13q14 deletion and 17p13 deletion.
Cytogenetic classification.
To analyze the correlation of cytogenetic data and clinicobiologic findings, three cytogenetic groups were identified based on the number of chromosome lesions as detected by CCA and interphase FISH. Group 1, including those BCL1-rearranged patients with a normal karyotype in at least 20 metaphases and those patients with the t(11;14) as the sole change. Group 2, including patients with 1 to 2 aberrations in addition to the t(11;14)/BCL1 rearrangement, and Group 3, defined by those patients with three or more aberrant events in addition to the t(11;14).
Statistical analysis.
Analysis of variance (ANOVA) with Bonferroni correction for multiple comparisons was used in the analysis of continuous variables whereas χ2 test was applied for categorical variables. Patient survival was estimated by the Kaplan-Meier method from the date of diagnosis until death due to any cause or until the last patient follow-up. The survival curves were statistically compared by the log-rank test. Because many statistical tests were undertaken in the evaluation of prognosis, a P value of .02 was used as the criterion for statistical significance. Proportional hazards regression analysis was used to identify the most significant independent prognostic variables on survival. P values less than .05 were considered statistically significant.
RESULTS
Chromosome lesions: CCA and FISH.
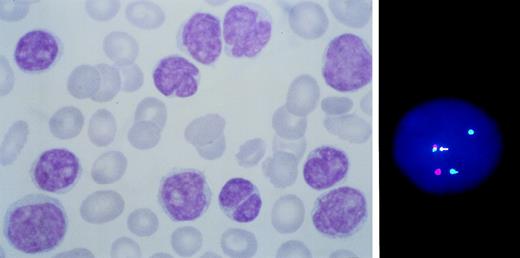
All cases had BCL1 involvement in the area covered by the 390kb YAC probe (Fig 1), resulting in the presence of three signals in 41% to 97% of the interphase cells (median value 73%). The t(11;14)(q13;q32) was documented by chromosome banding in a total of 29 cases, 15 of which had additional aberrations, rearrangements of 11q with complex karyotypes were observed in 5 patients and a normal karyotype was observed at diagnosis in 8 patients. Results are detailed in Table 2.
BCL1 rearranged leukemic MCL with primary BM involvement. Note the heterogeneity of cell size and morphology with irregular nuclear outline (left). A cell showing juxtaposition (arrowed) of BCL1 (green) to IgH (red) sequences is shown on the right.
BCL1 rearranged leukemic MCL with primary BM involvement. Note the heterogeneity of cell size and morphology with irregular nuclear outline (left). A cell showing juxtaposition (arrowed) of BCL1 (green) to IgH (red) sequences is shown on the right.
Karyotypes and Interphase Cytogenetic Findings at Diagnosis in 42 Patients With BCL1 Translocation
| Case No. . | Karyotype [No. of metaphases] . | Sample . | FISH* . | ||
|---|---|---|---|---|---|
| 13q− . | +12 . | 17p− . | |||
| Group 1 | |||||
| 19.30 | 46,XX [20] | PB | — | — | — |
| 8.29 | 46,XX, t(11;14)(q13;q32) [16] | PB | — | — | — |
| 23.31 | 46,XX [20] | PB | — | — | — |
| 28.32 | 46,XY [20] | PB | — | — | — |
| 31.8 | 46,XX,t(11;14)(q13;q32) [18] / 46, XX [2] | LN | — | — | — |
| 35.20 | 46,XY, t(11;14)(q13;q32) [15] | LN | — | — | — |
| 39.25 | 47,XX,t(11;14) (q13;q32) [4] / 46XX [18] | LN | — | — | — |
| 33.16 | 46,XY, t(11;14)(q13;q32) [19] | LN | — | — | — |
| 37.23 | 46,XX,t(11;14)(q13;q32), [20] | LN | — | — | — |
| 41.37 | 46,XY, t(11;14)(q13;q32) [4] / 46,XX [17] | PB | — | — | — |
| 42.40 | 46,XX, t(11;14)(q13;q32) [19] | PB | — | — | — |
| Group 2 | |||||
| 3.10 | 46,XY,t(11;14)(q13;q32) [16] / 46,XY[2] | LN | — | — | 55% |
| 7.19 | 47XY, +15, t(11;14)(q13;q32) [13] | LN | 80% | — | — |
| 9.34 | 46,XX, t(11;14)(q13;q32) [4] / 46, idem, del(13)(q14q21) [2] / 46,XX [12] | PB | 80% | — | — |
| 12.42 | 46,XY [25] | PB | 64% | — | — |
| 15.9 | 46,XY,t(11;14)(q13;q32) [6] / 46,XY [18] | LN | 90% | 58% | — |
| 17.13 | 46,XY,t(11;14)(q13;q32) [4] / 46,XY [16] | LN + PB | 75% | 38% | — |
| 18.22 | 46, XY, t(11;14)(q13;q32) [3] / 46, XY, [19] | LN + PB | 68% | 44% | — |
| 20.38 | 46,XY, t(11;14)(q13;q32) [6]/ 46,XY [18] | PB | 78% | 28% | — |
| 21.26 | 46, XX, der(14)t(11;14)(q13;q32) [2], 46, XX [17] | LN + PB | 70% | — | 75% |
| 22.27 | 46,XX [22] | LN | 66% | — | 62% |
| 24.41 | 46,XX[29] | PB | 90% | — | 45% |
| 30.5† | 46-48,XY, +3,t(11;14)(q13;q32),add(12)(q?) [20] | LN | — | — | — |
| 6.18 | 46,XY [21] | LN | 74% | — | — |
| 34.17 | 46-48XY, t(11;14)(q13;q32), +21 + mar [15] | LN | — | — | — |
| Group 3 | |||||
| 1.3† | 46-49,XY, del(2)(q?), der(3)(q?), +5,+der(11)(q?), +7,+12, del(17)(p1?2) inc [cp18] | LN | — | — | 75% |
| 2.7† | 38-45,X,−X,dup(3)(q21q24),t(6;12)(q23;q12) t(11;X?) (q11;p?), del(11)(q1?3q2?1) [cp8] | LN | — | — | 65% |
| 4.36 | 46-48,XY, add(1)(p?), del (10)(q?) t(11;14)(q13;q32), del(17)(p12) +2 mar [cp15] | PB | 77% | 68% | 59% |
| 5.15 | 45-46XY, −3,del(3)(p21;p23),t(3;11)(q14;q11), t(4;17)(q21;q21), t(8;17)(q12;p13), t(11;14)(q13;q32), −13 [cp19] | LN + PB | 84% | — | — |
| 10.35 | 46,XY, del(1)(p22p34), add(4)(p12), der(13)t(13;?)(q14;?), der(11)t(11;?) (q13;?) inc [10] | PB | 90% | — | — |
| 11.39 | 47,XY, add(1)(p36), add(2)(p22), add(6)(q23), t(11;14)(q13;q32), del(13)(q12q14)+M[cp15] | PB | 74% | — | — |
| 13.1† | 44-47,XY,der(4)t(4;12)(q?;q13), t(11;14) (q13;q32), +der(12)ins(12;13)(q13;q?13q?21), +mar [5] / 47, idem, add(17p) [6] | LN + PB | 80% | 42% | — |
| 14.6† | 47,XY, +der(3)t(3;?)(q?;?),t(11;14)(q13;q32)[6] | LN | 75% | 55% | — |
| 16.11 | 46-75,XY, +3,t(11;14)(q13;q32),+12 [20] | LN | 75% | 38% | — |
| 25.2 | 46,XY [25] | LN + PB | 85% | 40% | 38% |
| 26.4† | 44-46,XY, del(6)(q2?4), t(11;14)(q13;q32), +der(13)t(3;13) (q12;q34), +der(13)t(12;13)(q12;q34), del(17)(p11) [cp8] | LN | 78% | 45% | 75% |
| 27.12 | 43-46,XY, −3,−7, t(11;14)(q13;q32),+12 [12] /46,XY [4] | LN | 64% | 55% | 88% |
| 29.33 | 46,XX, add(10)(q22), t(11;14)(q13;q32) del(13)(q14q21) [9] / 46, idem, add(1)(p36) [6] | PB | 72% | 30% | 66% |
| 32.14 | 46,XY,t(11;14)(q13;q32) [8] / 45, idem, del(1) (p22;p32),inv(6)(p21;q2?3), −15 [4] | LN | — | — | — |
| 38.24 | 46, XY, idic (1)(q10), dup(3)(q25;q27), del(11)(q?) [3] / 47, idem, +14 [14] / 46XY [3] | LN | — | — | — |
| 36.21 | 46,XY, t(11;14)(q13;q32) [19] / 44, idem, −8,−9, del(11)(q13), add(13)(p13), add(17)(p13) [3] | LN | — | — | — |
| 40.28 | 47, XY,add(10)(p15), add(1)(q?),del(2)(p22), add(11)(q?) [15] | LN | — | — | — |
| Case No. . | Karyotype [No. of metaphases] . | Sample . | FISH* . | ||
|---|---|---|---|---|---|
| 13q− . | +12 . | 17p− . | |||
| Group 1 | |||||
| 19.30 | 46,XX [20] | PB | — | — | — |
| 8.29 | 46,XX, t(11;14)(q13;q32) [16] | PB | — | — | — |
| 23.31 | 46,XX [20] | PB | — | — | — |
| 28.32 | 46,XY [20] | PB | — | — | — |
| 31.8 | 46,XX,t(11;14)(q13;q32) [18] / 46, XX [2] | LN | — | — | — |
| 35.20 | 46,XY, t(11;14)(q13;q32) [15] | LN | — | — | — |
| 39.25 | 47,XX,t(11;14) (q13;q32) [4] / 46XX [18] | LN | — | — | — |
| 33.16 | 46,XY, t(11;14)(q13;q32) [19] | LN | — | — | — |
| 37.23 | 46,XX,t(11;14)(q13;q32), [20] | LN | — | — | — |
| 41.37 | 46,XY, t(11;14)(q13;q32) [4] / 46,XX [17] | PB | — | — | — |
| 42.40 | 46,XX, t(11;14)(q13;q32) [19] | PB | — | — | — |
| Group 2 | |||||
| 3.10 | 46,XY,t(11;14)(q13;q32) [16] / 46,XY[2] | LN | — | — | 55% |
| 7.19 | 47XY, +15, t(11;14)(q13;q32) [13] | LN | 80% | — | — |
| 9.34 | 46,XX, t(11;14)(q13;q32) [4] / 46, idem, del(13)(q14q21) [2] / 46,XX [12] | PB | 80% | — | — |
| 12.42 | 46,XY [25] | PB | 64% | — | — |
| 15.9 | 46,XY,t(11;14)(q13;q32) [6] / 46,XY [18] | LN | 90% | 58% | — |
| 17.13 | 46,XY,t(11;14)(q13;q32) [4] / 46,XY [16] | LN + PB | 75% | 38% | — |
| 18.22 | 46, XY, t(11;14)(q13;q32) [3] / 46, XY, [19] | LN + PB | 68% | 44% | — |
| 20.38 | 46,XY, t(11;14)(q13;q32) [6]/ 46,XY [18] | PB | 78% | 28% | — |
| 21.26 | 46, XX, der(14)t(11;14)(q13;q32) [2], 46, XX [17] | LN + PB | 70% | — | 75% |
| 22.27 | 46,XX [22] | LN | 66% | — | 62% |
| 24.41 | 46,XX[29] | PB | 90% | — | 45% |
| 30.5† | 46-48,XY, +3,t(11;14)(q13;q32),add(12)(q?) [20] | LN | — | — | — |
| 6.18 | 46,XY [21] | LN | 74% | — | — |
| 34.17 | 46-48XY, t(11;14)(q13;q32), +21 + mar [15] | LN | — | — | — |
| Group 3 | |||||
| 1.3† | 46-49,XY, del(2)(q?), der(3)(q?), +5,+der(11)(q?), +7,+12, del(17)(p1?2) inc [cp18] | LN | — | — | 75% |
| 2.7† | 38-45,X,−X,dup(3)(q21q24),t(6;12)(q23;q12) t(11;X?) (q11;p?), del(11)(q1?3q2?1) [cp8] | LN | — | — | 65% |
| 4.36 | 46-48,XY, add(1)(p?), del (10)(q?) t(11;14)(q13;q32), del(17)(p12) +2 mar [cp15] | PB | 77% | 68% | 59% |
| 5.15 | 45-46XY, −3,del(3)(p21;p23),t(3;11)(q14;q11), t(4;17)(q21;q21), t(8;17)(q12;p13), t(11;14)(q13;q32), −13 [cp19] | LN + PB | 84% | — | — |
| 10.35 | 46,XY, del(1)(p22p34), add(4)(p12), der(13)t(13;?)(q14;?), der(11)t(11;?) (q13;?) inc [10] | PB | 90% | — | — |
| 11.39 | 47,XY, add(1)(p36), add(2)(p22), add(6)(q23), t(11;14)(q13;q32), del(13)(q12q14)+M[cp15] | PB | 74% | — | — |
| 13.1† | 44-47,XY,der(4)t(4;12)(q?;q13), t(11;14) (q13;q32), +der(12)ins(12;13)(q13;q?13q?21), +mar [5] / 47, idem, add(17p) [6] | LN + PB | 80% | 42% | — |
| 14.6† | 47,XY, +der(3)t(3;?)(q?;?),t(11;14)(q13;q32)[6] | LN | 75% | 55% | — |
| 16.11 | 46-75,XY, +3,t(11;14)(q13;q32),+12 [20] | LN | 75% | 38% | — |
| 25.2 | 46,XY [25] | LN + PB | 85% | 40% | 38% |
| 26.4† | 44-46,XY, del(6)(q2?4), t(11;14)(q13;q32), +der(13)t(3;13) (q12;q34), +der(13)t(12;13)(q12;q34), del(17)(p11) [cp8] | LN | 78% | 45% | 75% |
| 27.12 | 43-46,XY, −3,−7, t(11;14)(q13;q32),+12 [12] /46,XY [4] | LN | 64% | 55% | 88% |
| 29.33 | 46,XX, add(10)(q22), t(11;14)(q13;q32) del(13)(q14q21) [9] / 46, idem, add(1)(p36) [6] | PB | 72% | 30% | 66% |
| 32.14 | 46,XY,t(11;14)(q13;q32) [8] / 45, idem, del(1) (p22;p32),inv(6)(p21;q2?3), −15 [4] | LN | — | — | — |
| 38.24 | 46, XY, idic (1)(q10), dup(3)(q25;q27), del(11)(q?) [3] / 47, idem, +14 [14] / 46XY [3] | LN | — | — | — |
| 36.21 | 46,XY, t(11;14)(q13;q32) [19] / 44, idem, −8,−9, del(11)(q13), add(13)(p13), add(17)(p13) [3] | LN | — | — | — |
| 40.28 | 47, XY,add(10)(p15), add(1)(q?),del(2)(p22), add(11)(q?) [15] | LN | — | — | — |
Abbreviations: BM, bone marrow, PB, peripheral blood; LN, lymph node.
— indicates the absence of the chromosome lesion (% interphase with abnormal hybridization pattern below the cut-off point for positivity); the percentage of interphase nuclei with deletion 13q14 or 17p13 (1 signal) and with +12 (3 signals) is shown in parentheses.
Patients previously published.3
Additional chromosome changes observed in three or more cases were loss of chromosome material at 13q and 17p- and total or partial trisomy 12. Monosomy 13 and structural 13q14 aberrations were detected by CCA in six cases. FISH documented the presence of 13q14 deletion in these cases and disclosed cytogenetically undetected 13q14 deletion in 16 additional cases (percentage of cells with one signal 64% to 90%). Three patients had total/partial trisomy 12 at banding analysis and nine additional patients were shown to carry extra chromosome 12 material in 28% to 68% of the cells by interphase FISH. Three patients had deletion 17p at metaphase banding analysis; cytogenetically undetected 17p13 deletions in 38% to 88% interphase nuclei were observed in eight additional cases. The frequency of these chromosome lesions in patients with complex karyotype (ie, patients with three or more aberrations in addition to the 11;14 translocation) compared with patients with noncomplex karyotype was as follows: 8 of 14 versus 14 of 28 for 13q14 deletion (P = 0.662); 5 of 14 versus 7 of 28 for +12 (P = .469) and 6 of 14 versus 5 of 28 for 17p- (P = .082).
The composition of cytogenetic groups 1, 2, 3 was the following (see Table 2): using data obtained by CCA, 22 patients had the t(11;14) as the sole change or a normal karyotype in more than 20 metaphases (group 1), 6 patients had one to two additional chromosome changes (group 2) and 14 patients had three or more additional aberrant events (group 3); when considering CCA plus interphase cytogenetic findings, 11 patients were found to have no detectable chromosome lesions in addition to the t(11;14)/BCL1 involvement (group 1), 14 patients had one to two additional aberrations (group 2) and 17 patients had three or more additional aberrations (group 3).
Hematologic and clinical features.
The salient hematologic features at presentation are summarized in Table 1. Primary sites of disease involvement at presentation were the lymph-node system in 28 cases, whereas the BM and PB (with or without splenomegaly) were primarily involved in 14 patients. Globally, extranodal involvement of the BM, spleen, gastrointestinal tract and Waldeyer’s ring occurred at presentation in 66% of the cases.
BM involvement usually consisted of interstitial or intertrabecular infiltrates of small- to medium-sized lymphocytes, with round-to-oval nuclei and nuclear indentations. Leukemic expression was found in 31 of 42 cases, having 10% to 98% CD5/C19+ cells in the lymphocyte gate (absolute lymphocyte count ranging between 1 and 800 × 109/L, median 22.4 × 109/L). The morphology of these cells showed heterogeneity of cell size and irregularities of nuclear outline (Fig 1); some small lymphocytes indistinguishable from CLL cells and prolymphocyte-like cells were also present. All cases with leukemic expression were CD5/CD19+, CD22+, FMC7+, CD23−, and CD10− and sIg+ with a bright pattern of expression and surface κ/λ restriction.
Thirteen patients are alive at 12 to 79 months, with a median follow-up of 36 months, whereas 29 died at 2 to 86 months. Median overall survival in this series was 40 and 34 months in the less than 60 and greater than 60 year age groups, respectively.
Survival.
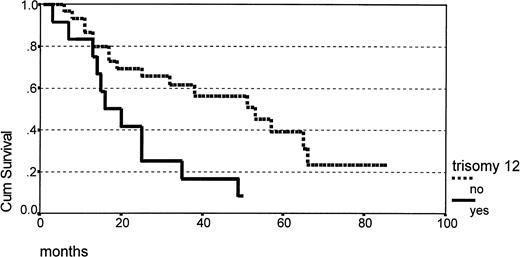
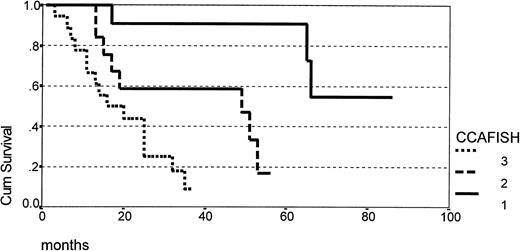
The correlation of clinical outcome and clinicobiologic parameters is summarized in Table 3, showing that male sex and primary lymph-node involvement were predictive of a shorter survival, whereas age, LDH level, advanced stage at presentation, serum albumin level, PB/BM involvement, PS, and splenomegaly were not. Trisomy 12 as detected by FISH was the only single cytogenetic parameter having prognostic significance (Fig 2), however the strongest prognostic indicator of shorter survival in univariate analysis was the degree of karyotype complexity (Table 3 and Fig 3). Except for male sex, which was associated with complex karyotype, the distribution of other salient clinical and hematologic parameters did not show any statistically significant correlation with cytogenetic features, as summarized in Table 4. Few patients presented blastic morphology or a nodular pattern of growth in lymph-node specimens (Table 1), precluding a meaningful analysis of the correlation of these histologic parameters with cytogenetic patterns.
Outcome According to Clinical and Biological Parameters in 42 Patients With t(11;14): Uniparameter Analysis
| Variable . | No. Patients . | Median Survival (Months) . | 95% Confidence Interval . | P . |
|---|---|---|---|---|
| Age yr | ||||
| ≤60 | 13 | 34 | 24-44 | .92 |
| >60 | 29 | 40 | 29-52 | |
| LDH | ||||
| Normal | 23 | 43 | 33-54 | .069 |
| Increased | 18 | 29 | 17-40 | |
| Sex | ||||
| f | 13 | 61 | 49-73 | .006 |
| m | 29 | 31 | 21-42 | |
| Splenomegaly | ||||
| yes | 19 | 33 | 21-45 | .184 |
| no | 23 | 47 | 33-61 | |
| PB involvement | ||||
| no | 11 | 33 | 20-45 | .71 |
| yes | 31 | 47 | 30-52 | |
| BM involvement | ||||
| yes | 27 | 47 | 34-59 | .026 |
| no | 15 | 25 | 17-32 | |
| Primary site of involvement | ||||
| lymph node | 28 | 28 | 21-36 | .015 |
| BM ± spleen | 14 | 55 | 40-71 | |
| Stage | ||||
| advanced | 31 | 34 | 25-43 | .107 |
| initial | 11 | 54 | 33-75 | |
| PS (ECOG) | ||||
| 0-1 | 36 | 42 | 28-56 | .121 |
| 2-4 | 6 | 19 | 7-31 | |
| Serum albumin | ||||
| <4.0 | 29 | 43 | 30-55 | .45 |
| ≥4.0 | 13 | 34 | 21-47 | |
| +12 by FISH | ||||
| yes | 12 | 22 | 13-30 | .006 |
| no | 30 | 47 | 36-59 | |
| 17p by FISH | ||||
| yes | 11 | 28 | 18-39 | .073 |
| no | 31 | 45 | 33-57 | |
| 13q by FISH | ||||
| yes | 22 | 30 | 22-38 | .053 |
| no | 20 | 48 | 33-64 | |
| CCA | ||||
| 1 | 21 | 64 | 50-78 | |
| 2 | 7 | 24 | 18-30 | .0001 |
| 3 | 14 | 12 | 8-16 | |
| CCA + FISH | ||||
| 1 | 11 | 86* | Not reached at 86 months | |
| 2 | 14 | 48 | 36-60 | <.0001 |
| 3 | 17 | 15 | 9-21 |
| Variable . | No. Patients . | Median Survival (Months) . | 95% Confidence Interval . | P . |
|---|---|---|---|---|
| Age yr | ||||
| ≤60 | 13 | 34 | 24-44 | .92 |
| >60 | 29 | 40 | 29-52 | |
| LDH | ||||
| Normal | 23 | 43 | 33-54 | .069 |
| Increased | 18 | 29 | 17-40 | |
| Sex | ||||
| f | 13 | 61 | 49-73 | .006 |
| m | 29 | 31 | 21-42 | |
| Splenomegaly | ||||
| yes | 19 | 33 | 21-45 | .184 |
| no | 23 | 47 | 33-61 | |
| PB involvement | ||||
| no | 11 | 33 | 20-45 | .71 |
| yes | 31 | 47 | 30-52 | |
| BM involvement | ||||
| yes | 27 | 47 | 34-59 | .026 |
| no | 15 | 25 | 17-32 | |
| Primary site of involvement | ||||
| lymph node | 28 | 28 | 21-36 | .015 |
| BM ± spleen | 14 | 55 | 40-71 | |
| Stage | ||||
| advanced | 31 | 34 | 25-43 | .107 |
| initial | 11 | 54 | 33-75 | |
| PS (ECOG) | ||||
| 0-1 | 36 | 42 | 28-56 | .121 |
| 2-4 | 6 | 19 | 7-31 | |
| Serum albumin | ||||
| <4.0 | 29 | 43 | 30-55 | .45 |
| ≥4.0 | 13 | 34 | 21-47 | |
| +12 by FISH | ||||
| yes | 12 | 22 | 13-30 | .006 |
| no | 30 | 47 | 36-59 | |
| 17p by FISH | ||||
| yes | 11 | 28 | 18-39 | .073 |
| no | 31 | 45 | 33-57 | |
| 13q by FISH | ||||
| yes | 22 | 30 | 22-38 | .053 |
| no | 20 | 48 | 33-64 | |
| CCA | ||||
| 1 | 21 | 64 | 50-78 | |
| 2 | 7 | 24 | 18-30 | .0001 |
| 3 | 14 | 12 | 8-16 | |
| CCA + FISH | ||||
| 1 | 11 | 86* | Not reached at 86 months | |
| 2 | 14 | 48 | 36-60 | <.0001 |
| 3 | 17 | 15 | 9-21 |
Abbreviations: CCA, patients subdivided in group 1, 2, 3 based solely on conventional cytogenetics; CCA + FISH, group 1, 2, and 3 compiled using metaphase banding analysis and interphase cytogenetic data.
Survival according to the presence (n = 12) or absence (n = 30) of total/partial trisomy 12 (P .006).
Survival according to the presence (n = 12) or absence (n = 30) of total/partial trisomy 12 (P .006).
Survival according to the degree of karyotype complexity as defined by CCA + FISH analysis (group 1 = 11 patients; group 2 = 14 patients; group 3 = 17 patients) (P < .0001).
Survival according to the degree of karyotype complexity as defined by CCA + FISH analysis (group 1 = 11 patients; group 2 = 14 patients; group 3 = 17 patients) (P < .0001).
Clinicopathologic Findings in Correlation With Cytogenetics in 42 MCL
| . | +12 . | 13q− . | 17p− . | Cytogenetic Groups . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Yes n = 12 . | No n = 30 . | Yes n = 22 . | No n = 20 . | Yes n = 11 . | No n = 31 . | 1 n = 11 . | 2 n = 14 . | 3 n = 17 . | |
| Male | 11 | 18 | 17 | 12 | 6 | 23 | 4 | 10 | 15 |
| Female | 1 | 12 | 5 | 8 | 5 | 8 | 7 | 4 | 2 |
| P = .045 | P = .227 | P = .226 | P = .015 | ||||||
| Stage | |||||||||
| Advanced | 10 | 21 | 17 | 14 | 9 | 22 | 6 | 11 | 14 |
| Initial | 2 | 9 | 5 | 6 | 2 | 9 | 5 | 3 | 3 |
| P = .375 | P = .592 | P = .482 | P = .232 | ||||||
| PS 0-1 | 11 | 25 | 19 | 17 | 9 | 27 | 11 | 12 | 13 |
| PS 2-4 | 1 | 5 | 3 | 3 | 2 | 4 | 0 | 2 | 4 |
| P = .486 | P = .90 | P = .667 | P = .221 | ||||||
| Splenomegaly | |||||||||
| Yes | 6 | 13 | 13 | 6 | 6 | 13 | 3 | 8 | 8 |
| No | 6 | 17 | 9 | 14 | 5 | 18 | 8 | 6 | 9 |
| P = .695 | P = .059 | P = .470 | P = .324 | ||||||
| Albumin | |||||||||
| <4.0 gr/dl | 6 | 23 | 13 | 16 | 6 | 23 | 7 | 11 | 11 |
| >4.0 gr/dl | 5 | 7 | 9 | 4 | 5 | 8 | 4 | 3 | 6 |
| P = .091 | P = .143 | P = .226 | P = .639 | ||||||
| BM involved | 6 | 21 | 16 | 11 | 5 | 22 | 8 | 11 | 8 |
| BM not involved | 6 | 9 | 6 | 9 | 6 | 9 | 3 | 3 | 9 |
| P = .222 | P = .231 | P = .129 | P = .151 | ||||||
| PB involved | 9 | 22 | 17 | 14 | 7 | 24 | 8 | 9 | 14 |
| PB not involved | 3 | 8 | 5 | 6 | 4 | 7 | 3 | 5 | 3 |
| P = .912 | P = .592 | P = .372 | P = .521 | ||||||
| Age <60 | 3 | 10 | 8 | 5 | 4 | 9 | 2 | 4 | 7 |
| Age >60 | 9 | 20 | 14 | 15 | 7 | 22 | 9 | 10 | 10 |
| P = .598 | P = .426 | P = .426 | P = .426 | ||||||
| LDH normal | 6 | 17 | 11 | 12 | 4 | 19 | 8 | 8 | 7 |
| LDH elevated | 6 | 12 | 11 | 7 | 7 | 11 | 2 | 6 | 10 |
| P = .613 | P = .397 | P = .123 | P = .145 | ||||||
| Extranodal disease | |||||||||
| No | 3 | 11 | 8 | 6 | 3 | 11 | 6 | 4 | 4 |
| Yes | 9 | 19 | 14 | 14 | 8 | 20 | 5 | 10 | 13 |
| P = .469 | P = .662 | P = .142 | P = .212 | ||||||
| . | +12 . | 13q− . | 17p− . | Cytogenetic Groups . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Yes n = 12 . | No n = 30 . | Yes n = 22 . | No n = 20 . | Yes n = 11 . | No n = 31 . | 1 n = 11 . | 2 n = 14 . | 3 n = 17 . | |
| Male | 11 | 18 | 17 | 12 | 6 | 23 | 4 | 10 | 15 |
| Female | 1 | 12 | 5 | 8 | 5 | 8 | 7 | 4 | 2 |
| P = .045 | P = .227 | P = .226 | P = .015 | ||||||
| Stage | |||||||||
| Advanced | 10 | 21 | 17 | 14 | 9 | 22 | 6 | 11 | 14 |
| Initial | 2 | 9 | 5 | 6 | 2 | 9 | 5 | 3 | 3 |
| P = .375 | P = .592 | P = .482 | P = .232 | ||||||
| PS 0-1 | 11 | 25 | 19 | 17 | 9 | 27 | 11 | 12 | 13 |
| PS 2-4 | 1 | 5 | 3 | 3 | 2 | 4 | 0 | 2 | 4 |
| P = .486 | P = .90 | P = .667 | P = .221 | ||||||
| Splenomegaly | |||||||||
| Yes | 6 | 13 | 13 | 6 | 6 | 13 | 3 | 8 | 8 |
| No | 6 | 17 | 9 | 14 | 5 | 18 | 8 | 6 | 9 |
| P = .695 | P = .059 | P = .470 | P = .324 | ||||||
| Albumin | |||||||||
| <4.0 gr/dl | 6 | 23 | 13 | 16 | 6 | 23 | 7 | 11 | 11 |
| >4.0 gr/dl | 5 | 7 | 9 | 4 | 5 | 8 | 4 | 3 | 6 |
| P = .091 | P = .143 | P = .226 | P = .639 | ||||||
| BM involved | 6 | 21 | 16 | 11 | 5 | 22 | 8 | 11 | 8 |
| BM not involved | 6 | 9 | 6 | 9 | 6 | 9 | 3 | 3 | 9 |
| P = .222 | P = .231 | P = .129 | P = .151 | ||||||
| PB involved | 9 | 22 | 17 | 14 | 7 | 24 | 8 | 9 | 14 |
| PB not involved | 3 | 8 | 5 | 6 | 4 | 7 | 3 | 5 | 3 |
| P = .912 | P = .592 | P = .372 | P = .521 | ||||||
| Age <60 | 3 | 10 | 8 | 5 | 4 | 9 | 2 | 4 | 7 |
| Age >60 | 9 | 20 | 14 | 15 | 7 | 22 | 9 | 10 | 10 |
| P = .598 | P = .426 | P = .426 | P = .426 | ||||||
| LDH normal | 6 | 17 | 11 | 12 | 4 | 19 | 8 | 8 | 7 |
| LDH elevated | 6 | 12 | 11 | 7 | 7 | 11 | 2 | 6 | 10 |
| P = .613 | P = .397 | P = .123 | P = .145 | ||||||
| Extranodal disease | |||||||||
| No | 3 | 11 | 8 | 6 | 3 | 11 | 6 | 4 | 4 |
| Yes | 9 | 19 | 14 | 14 | 8 | 20 | 5 | 10 | 13 |
| P = .469 | P = .662 | P = .142 | P = .212 | ||||||
The Cox regression model for survival showed that among the above-reported variables having prognostic significance in univariate analysis, only the measure of karyotype complexity as assessed by FISH and CCA maintained prognostic significance, with a P < .0001 (4,7805 hazard ratio; 2.2884 to 9.9874, 95% confidence limits).
DISCUSSION
A preliminary methodological problem in this study was represented by the definition of the inclusion criteria, given the heterogeneity of clinicopathological manifestations of MCL.9,12,21,37 Because lymphoma of follicle mantle lineage have in common a specific genetic marker and the immunophenotypic profile,9 we included patients with the t(11;14)/BCL1 rearrangement and with cytoimmunologic features characteristic of MCL. These biological markers of MCL were particularly important in distinguishing those cases presenting with primary BM and PB involvement from other leukemic NHL and chronic B-cell disorders, which may present similar morphologic features. Inclusion criteria in this series accounted on the one hand for the absence of cases with primary extranodal disease, all biopsy material from extranodal sites having been sent to the pathologist for histologic diagnosis and, on the other hand, for the relatively high number of cases with primary BM involvement and leukemic expression. Other hematologic features in our patients (Tables 1 and 3) did not differ significantly as compared with those reported in recent studies,12,13,15,16 with frequent presentation in advanced stage (74% of the cases, compared with 77% to 91%), male sex preponderance (2:1 compared with 1.6:1 up to 3:1), relatively old age (68 years, compared with 62 to 65 years) and short survival (34 and 40 months in the greater than 60 and less than 60 year age groups, compared with 43 to 56 months median overall survival). The histologic features, with a majority of cases presenting diffuse infiltration pattern and few cases displaying a predominantly blastic morphology, are in line with previous observations.12
The overall cytogenetic picture and the chromosome lesions that were found in this series in addition to BCL1involvement/t(11;14)(q13;q32) improve our knowledge on the clinicobiologic significance of cytogenetics in MCL, showing that total/partial trisomy 12 and a derived measure of karyotype complexity may have a correlation with survival. 13q14 deletion, total or partial trisomy 12, and 17p13 deletion occurred at a relatively high frequency in this series and were not simply a function of karyotype complexity, suggesting that these anomalies are acquired early during the cytogenetic evolution of MCL.
We found a relatively high incidence of normal karyotype (19%) and isolated t(11;14) (33%) in newly diagnosed BCL1+MCL. These findings add to the data by Pittaluga et al29who described a 55% and 8% incidence of normal karyotype and t(11;14) as the sole change, respectively, in 38 cases and with the data collected by Johansson et al,38 who presented a 19.8% incidence of isolated t(11;14) in 91 cases published in the literature. FISH studies,39 however, showed that cytogenetic findings in MCL are partially influenced by suboptimal culture conditions and inadequate banding resolution, having three consequences: (1) detection of normal karyotypes due to divisions occurring in residual normal lymphocytes; (2) detection of the primary chromosome change defining the stemline with failure to show additional anomalies present in the sidelines; and (3) impossibility to recognize subtle rearrangements, especially small deletions, occurring in the context of abnormal karyotypes. Point 1 is illustrated in our study by eight BCL1rearranged cases with apparently normal karyotype. Though representing a laboratory artifact, the finding of a normal karyotype in low-grade lymphomas is clinically important, in that it reflects low mitotic activity of the neoplastic clone, a feature that was associated with a favorable prognosis in B-CLL40 and in follicle center cell lymphomas in which the percentage of normal metaphases in lymph-node samples was associated with a more favorable outcome.20 24Our finding that patients with normal cytogenetics or with t(11;14)/BCL1 rearrangement as the only detectable change (group 1) fared better than patients with additional chromosome changes represents, to the best of our knowledge, the first demonstration that MCL may benefit of cytogenetic investigations for a more accurate assessment of prognosis.
Point 2 is better illustrated in our series by those patients having a t(11;14) as the sole aberration by CCA, who were shown to carry extra chromosome 12 material in 26% to 58% of the interphase cells (cases 15.9; 17.13; 18.22; 20.38; 29.33). The possibility that in some patients with complex karyotype (cases 13.1; 14.6; 26.4), partial trisomy 12 was not recognized at karyotyping due to the presence of marker chromosomes, should be considered. It is noteworthy that in B-CLL, CCA was reported to underestimate the incidence of +12 compared with interphase cytogenetics, both in cases with normal karyotypes41 and with complex rearrangements.42,43 The acquisition of extra chromosome 12 material was detected much less frequently in follicle center cell lymphoma and in marginal zone B-cell lymphoma in three studies using comparative genomic hybridization,44-46 suggesting that this chromosome imbalance may be preferentially associated with CD5+ B-cell lymphoproliferative disorders. These considerations have practical implications, because total/partial trisomy 12 was the only single cytogenetic parameter that was predictive of an adverse outcome in this series, and point at the importance of FISH for the refinement of cytogenetic diagnosis in MCL.
Point 3 is illustrated by those cases having cytogenetically undetected 13q14 and 17p13 deletions. Our findings are reassuring with respect to the specificity of metaphase banding analysis and show that the sensitivity of CCA in detecting loss of chromosome material is unsatisfactory.
The high incidence of 13q14 deletions (52.3%) that was found in this series is a relatively new finding, this anomaly having been associated with 40% of B-CLL studied by molecular cytogenetics.36,43,47 Interestingly, 13q14 deletion was found to occur by FISH at a relatively high incidence in atypical CLL carrying the 11;14 translocation34 and in a recent study on MCL.48 Because a 8.8% incidence of 13q14 deletion was found at our Institution in 91 samples of B-NHL excluding MCL and small lymphocytic lymphoma (data not shown), we suggest that loss of genetic material involving the 13q14 region may represent an important step in the transformation of CD5+ B-cell neoplasias.
Whereas the 13q-anomaly carries a favorable prognostic significance in B-CLL, no statistically significant survival difference was noted in our patients with and without 13q14 deletion. A trend towards a shorter survival was observed instead in 13q-patients, partially accounted for by the presence of additional aberrations in half of the cases. It is noteworthy that 13q14 deletions are found, albeit at a lower frequency, in a spectrum of lymphoid neoplasias,49 including multiple myeloma, in which they have been associated with an inferior prognosis.50 51
The 26.2% incidence of 17p13 deletion in this series is comparable with the 18% incidence that was reported by Clodi et al,52who analyzed by interphase FISH 79 unselected NHL using a 17p13/p53 probe. No correlation was found in this study and in the analysis by Clodi et al52 between deletion involving this region and survival; hence it is reasonable to assume that p53 gene deletion per se does not have a major impact on prognosis in NHL. As for the case with 13q14 deletions, the possibility should be considered that more cases need to be studied to allow a single cytogenetic parameter to reach statistical significance on survival analysis, even more so that a number of chromosome changes was frequently found in association with each of these anomalies.
This consideration prompted us to define a measure of karyotype complexity to compare survival among different patient groups. Derived measures of karyotype complexity, ie, modal chromosome number, number of translocation breakpoints, and number of marker chromosomes were shown to have prognostic significance in low-grade NHL and in follicle center cell lymphomas.20,26 The difference in survival according to the number of chromosome lesions that was observed in our study was evident using data obtained by CCA and the observed difference had an optimal statistical significance using CCA plus FISH data. This classification clearly depends on the probes that we chose to test in interphase cells, based on the identification of the most frequent additional changes by CCA. Other chromosome regions that may potentially have prognostic significance20,26 were shown to be deleted or rearranged in a fraction of MCL,38 the most frequent being 6q15-21, 6q25, 1p21-32, and 1q21-23. Attention was devoted in a recent study on a cycline-dependent kinase (CDK) inhibitor, referred to as p16INK4a and located on chromosome 9p21, the deletion of which was shown to be associated with a high proliferation index.53 Interestingly, the gene encoding for a closely related CDK inhibitor, p18INK4c, was mapped to 1p31-35,54 in a chromosome segment that was deleted in two of our cases. In general, it is reasonable to predict that, as more cases will be studied, new recurring chromosome breaks will be recognized that will allow for the definition of a wider panel of FISH probes, possibly resulting in the identification of other clinically important genomic imbalances.
Supported by BMH-1 EU CA: CT 94-1703, and by C.N.R., ACRO project and M.U.R.S.T, fondi 40% and 60%.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal