Abstract
The t(11;14)(q13;q32) chromosomal translocation, which is the hallmark of mantle cell lymphoma (MCL), is found in approximately 30% of multiple myeloma (MM) tumors with a 14q32 translocation. Although the overexpression of cyclin D1 has been found to be correlated with MM cell lines carrying the t(11;14), rearrangements of theBCL-1/cyclin D1 regions frequently involved in MCL rarely occur in MM cell lines or primary tumors. To test whether specific 11q13 breakpoint clusters may occur in MM, we investigated a representative panel of primary tumors by means of Southern blot analysis using probes derived from MM-associated 11q13 breakpoints. To this end, we first cloned the breakpoints and respective germ-line regions from a primary tumor and the U266 cell line, as well as the germ-line region from the KMS-12 cell line. DNA from 50 primary tumors was tested using a large panel of probes, but a rearrangement was detected in only one case using the KMS-12 breakpoint probe. Our results confirm previous findings that the 11q13 breakpoints in MM are scattered throughout the 11q13 region encompassing the cyclinD1 gene, thus suggesting the absence of 11q13 breakpoint clusters in MM.
CHROMOSOMAL translocations affecting the immunoglobulin heavy chain (IGH) locus on 14q32 represent the mechanism of activation of a number of proto-oncogenes in B-cell lymphoid neoplasms.1 The t(11;14)(q13;q32) chromosomal translocation is associated with approximately 70% to 90% of mantle cell lymphomas (MCL)2-5 and leads to the overexpression of the cyclin D1 gene.6-9 The breakpoints on chromosome 11q13 were initially found clustered in a 1-kb region, named the major translocation cluster (MTC), of the BCL-1 locus.2 Further investigations have shown that breakpoints may occur telomeric of the MTC in a region of about 120 kb between the MTC and the cyclin D1 loci.3 Molecular analyses, including fluorescence in situ hybridization (FISH), have also demonstrated that breakpoints may occur either centromeric or telomeric of this 120 kb region in some cases.4,5 9
Multiple myeloma (MM) is a malignant proliferation of bone marrow plasma cells that is characterized by a wide spectrum of clinical entities and whose molecular pathogenesis is still largely unknown.10,11 Cytogenetic analyses in MM are limited and difficult mainly because of the low proliferation rate of malignant plasma cells. However, in about 20% to 40% of tumors with an abnormal karyotype, a 14q+ marker has been reported.12,13 This marker is generally the consequence of translocation events involving the IGH locus on chromosome 14q32. Interestingly, in almost 30% of cases with cytogenetically detectable 14q+, the marker is the result of a t(11;14)(q13;q32) chromosomal translocation. However, rearrangements of the BCL-1/cyclin D1 regions frequently involved in MCL rarely occur in MM.9,14-16 Molecular characterization of breakpoints in a limited number of MM-derived cell lines carrying the t(11;14)(q13;q32) chromosomal translocation suggested that they are scattered over a relatively large area encompassing theBCL-1/cyclin D1 region.9,16 However, overexpression of cyclin D1 is generally associated with the t(11;14) in MM cell lines, thus suggesting that it represents the target gene of the translocation.7,9 16 Therefore, the currently available probes specific for the 11q13 breakpoints may not be useful for detecting BCL-1/cyclin D1 rearrangements in MM.
In an attempt to investigate this point further, we cloned the genomic regions from the 11q13 breakpoints of three different MM cases (two cell lines and a primary tumor). The breakpoints were scattered along the 11q13 region at various distances from the cyclin D1 locus, as observed by FISH analysis. Specific 11q13 probes from these regions were used in Southern blot analyses to search for genomic rearrangements in a representative panel of primary MM tumors that had been previously found to be negative for BCL-1/cyclinD1 rearrangements. With these new probes, we were able to detect rearrangements in only one of the 50 MM tumors investigated. Our results confirm previous findings that 11q13 breakpoint in MM are scattered over the 11q13 region encompassing the cyclin D1 gene and suggest the absence of 11q13 breakpoint clusters in MM.
MATERIALS AND METHODS
Pathological samples.
Bone marrow or peripheral blood samples from 50 MM patients investigated by Southern blot analysis were collected during the course of standard diagnostic procedures. The diagnosis and clinical staging of MM was made according to the criteria described by Durie and Salmon.17 These samples came from a larger series of 88 previously investigated primary MM tumors.18 Forty patients were at first diagnosis: six in stage I (indolent phase), 22 in stage II, and 12 in stage III; five patients were evaluated at clinical relapse and five were affected by plasma cell leukemia (four at diagnosis and one at relapse). Twenty-one of the patients were male and 29 female; their median age was 61 years (range, 42 to 80). Monoclonal component was as follows: IgG (35 patients), IgA (11), κ/λ chain 30/16; κ chain (3) λ chain (1). No conventional cytogenetic analyses (G-banding) of these samples were available.
The previously reported tumor LB41118 was derived from a 69-year old male patient affected by IgGk-type plasma cell leukemia with 2 months survival; no karyotype was available in this case. Case AC97 was a 74-year-old female with a κ-type MM in clinical stage III that had the following karyotype: 46,XX, del(1)(p13p22), inv(9)(p12;q13), t(11;14)(q13;q32), del(13)(q22q31). The patient showed a partial clinical response to conventional chemotherapy and is still alive 2 years after diagnosis.
The MM-derived cell lines U266 and KMS-12 have been previously reported; a cytogenetically detectable t(11;14)(q13;q32) chromosomal translocation has been found in the KMS-12, but not the U266 cell line.19,20 No evidence of any rearrangement of theBCL-1 locus or cyclin D1 gene has been observed in either cell line, whereas cyclin D1 overexpression has been detected in both7 16 (and present study). The U266 cell line was obtained from the American Type Culture Collection (Rockville, MD); the KMS-12 cell line was kindly provided by Dr T. Otsuki (Okayama, Japan).
DNA preparation and Southern blot analysis.
Mononuclear cell suspensions with more than 95% viability were prepared from the pathological samples by means of Ficoll-Hypaque gradient centrifugation; the percentage of malignant plasma cells identified by immunocytomorphologic analyses was between 22% and 98%. DNA from pathological samples and cell lines was purified by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation.21 A total of 10 μg of genomic DNA was digested with BamHI, EcoRI, or HindIII restriction enzymes, electrophorized in a 0.7% agarose gel, and then denaturated, neutralized, and transferred to nylon filters (Amersham International, Amersham, UK). The filters were hybridized to32P-labeled probes according to the manufacturer’s specifications, washed in 0.5 × SSC (NaCl/Na citrate)/1% sodium dodecyl sulfate (SDS) for 1 hour at 60°C and then autoradiographed using an intensifying screen at −80°C.21,IGH gene rearrangement was analyzed using previously described probes22-24; the probes used for the rearrangement analysis of the BCL-1/cyclinD1 locus were MTC, p94, and cyclin D1 cDNA.2,3 6
Molecular cloning.
The identification of a t(11;14)(q13;q32) chromosomal translocation in one primary case of MM (LB411) and the U266 cell line was made possible following a Southern blot approach recently reported by us,18 which allows the identification of putative switch-translocated IGH alleles on the basis of the absence of any linkage between different IGH regions. A quite similar approach has been previously reported by others.25 We reasoned that, as a result of immunoglobulin gene recombination during maturation of B cells, the rearranged joining-switch-constantIGH regions are generally contained on a novel BamHI restriction fragment, and that a translocation event involving the switch region should therefore generate a rearranged BamHI fragment containing the 3′ constant region, but not the 5′ joining sequences of the IGH gene. In our Southern blot assay, the DNA was digested with BamHI restriction enzyme, and the filters subsequently hybridized with the JH, Cμ, Cα1, and Cγ1 probes. The identification of rearranged IGH alleles as potential candidates for switch-mediated chromosomal translocations was based on the absence of comigration between the DNA fragments containing constant IGH regions and those positive for the JH probe. Recombinant phage clones containing translocated Cα-rearranged IGH alleles from case LB411 and the U266 cell line were obtained by complete digestion of genomic DNA withBamHI, the subsequent ligation of gel-purified fractions into λEMBL3 phage vectors (Stratagene, La Jolla, CA) and screening with the Cα1 probe. The germ-line regions of chromosome 11 were isolated by screening a genomic library of human placenta DNA (Clontech, San Diego, CA) using probes derived from recombinant clones. The normal 11q13 region encompassing the breakpoint in KMS-12 was cloned by screening a phage genomic library with a 193-bp fragment specific for the KMS-12 breakpoint region, which was obtained by polymerase chain reaction (PCR) amplification of genomic DNA using a pair of previously reported primers.16 The library screening and plaque isolation were performed according to established procedures.21 The inserts were analyzed using restriction enzyme mapping and then subcloned into plasmid vector pGEM3 (Promega, Madison, WI).
DNA sequencing.
DNA sequence analysis was performed on restriction fragments cloned into pGEM3 plasmid (Promega) by “dideoxy” chain-termination analysis using the Sequenase sequencing kit (USB, Cleveland, OH).
cDNA amplification.
The synthesis of the first strand cDNA was performed as previously described.18 PCR amplifications were made by diluting 5 μL of first-strand cDNA from each individual case into a 50-μL PCR mixture. A 196-bp fragment encompassing exons 3 and 4 of cyclinD1 gene6 was amplified using the following primers: sense 5′-AACAGATCATCCGCAAACAC-3′; antisense 5′-TCACACTTGATCACTCTGGA-3′. Thirty cycles of amplification were performed at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds.
FISH.
The chromosome preparations were hybridized in situ with probes labeled with biotin or directly with the fluorochrome, Cy3 (Amersham, Little Chalfont, UK) by nick translation, as described21 with minor modifications.26,27 Briefly, 200 ng of labeled probe were used for each experiment, and hybridization was performed at 37°C in 2 × SSC, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulphate, 5 μg Cot1 DNA (Boehringer, Mannheim, Germany), and 3 μg of sonicated salmon sperm DNA in a volume of 10 μL. Posthybridization washing was at 60°C in 0.1 × SSC (three times). Biotin-labeled DNA was detected using fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories, Burlingame, CA). The chromosomes were identified by simultaneous 4′,6′-diaminido-2-phenylindole dihydrochloride (DAPI) staining, which produces a Q-banding pattern. Chromosome 11 and 14 painting probes were obtained by Alu-PCR amplification of the somatic cell hybrids retaining only the human chromosome 11 or 14.28 Digital images were obtained using a Leica DMR epifluorescence microscope equipped with a CCD camera (Cohu, Inc, San Diego, CA). FITC-avidin, Cy3, and DAPI fluorescence signals were detected using specific filters and recorded separately as gray scale images. Pseudocoloring and image merging were performed using Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Probes for FISH analysis.
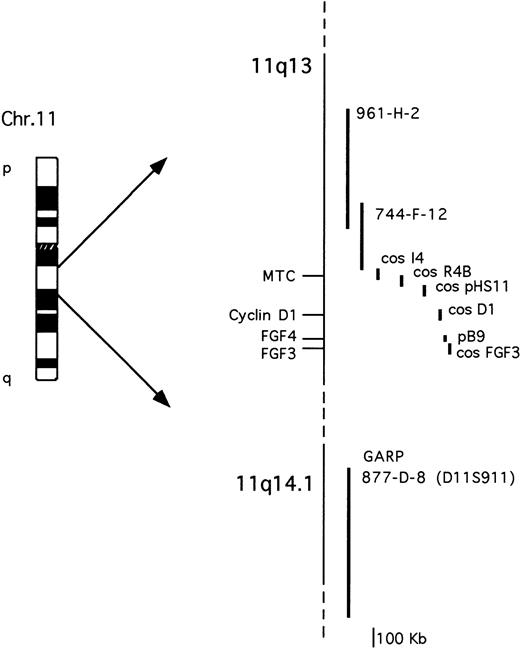
The probes used are described in Fig 1. The cosmid clones representative of the cyclin D1 and FGF3loci were isolated by screening a cosmid library of human placenta (Clontech) with probes specific for cyclin D16 orFGF3/int-2 cDNA.29 The FGF4/hst-1 locus was investigated using an 8-kb genomic fragment containing the 5′ of the gene.30 The cosmid clones I4, R4B, and pHS-11 encompassing the MTC region have been previously described.9 All of the YACs were obtained from the human CEPH2 library (YAC Screening Center, DIBIT, Milan, Italy): YAC clones 961-H-2 (800 kb) and 744-F-12 (330 Kb) have been previously reported31 and are both negative for the MTC region; YAC clone 877-D-8 (1080 kb) was identified by the presence of the D11S911 marker, telomeric of the GARP locus on 11q14.9,32The IGH locus was analyzed using a phage clone containing the 18-kb Cα1 BamHI germ-line fragment.23
Schematic representation of the chromosome 11q13-11q14.1 probes used for the molecular and FISH analyses. See Materials and Methods for further details.
Schematic representation of the chromosome 11q13-11q14.1 probes used for the molecular and FISH analyses. See Materials and Methods for further details.
RESULTS
Cloning of a t(11;14)(q13;q32) in a MM primary tumor.
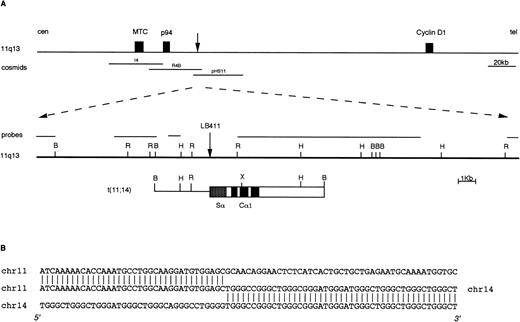
We have recently reported the identification of IGH rearranged alleles as potential candidates for switch-mediated chromosomal translocations in 21 of 88 cases of primary MM tumors investigated.18 To confirm the presence of chromosomal breakpoints in these alleles, we cloned the rearranged fragments from three cases (two Cα and one Cμ rearrangement). In two of these tumors, molecular cloning and FISH analyses showed the presence of a novel t(4;14)(p16.3;q32) chromosomal translocation.18 In the third MM tumor (case LB411), a t(11;14)(q13;q32) chromosomal translocation was identified by cloning a 10-kb BamHI Cα1 rearranged fragment (Fig 2) that was apparently negative for JH sequences.18 This fragment was isolated as described in Materials and Methods and used as a probe in the FISH analysis of metaphase spreads from mitogen-stimulated normal peripheral blood lymphocytes. This clone hybridized to both chromosome 14q32 and chromosome 11q13, thus indicating that the rearrangement was a result of a t(11;14)(q13;q32) chromosomal translocation (data not shown). About 25 kb of the normal genomic region surrounding the breakpoint on chromosome 11q13 were cloned by isolating two overlapping phages from a normal genomic library (Fig 2). None of this genomic region hybridized with probes specific for the BCL-1 locus (MTC, p94, and full-length cyclin D1 cDNA), thus confirming the absence of rearrangements in these regions observed in Southern blot analysis of LB411 DNA (data not shown). Subsequently, this region was hybridized with a set of 11q13 genomic clones (see Fig 1); cosmids R4B and pHS-11 were both positive, thus indicating that the breakpoint in case LB411 occurred approximately 40 to 45 kb telomeric of the MTC region and consequently 65 to 70 kb centromeric of the cyclinD1 gene. Interestingly, reverse transcriptase (RT)-PCR analysis showed that this case was overexpressing cyclin D1 (data not shown).
Molecular cloning of the chromosomal breakpoint from case LB411. (A) Schematic representation of the breakpoint and the respective 11q13 germ-line region. From the top: a diagram of theBCL-1/cyclin D1 locus where the cosmid clones I4, R4b, pHS11 are located is shown; the 11q13 germ-line region and the probes used for the Southern and Northern blot analyses are shown as solid lines. The vertical arrow indicates the breakpoint position. In the t(11;14) breakpoint clone, chromosome 14 is indicated by open boxes with black or stippled boxes representing different IGH regions and chromosome 11 is shown as a solid line. Restriction enzyme symbols: B, BamHI; R, EcoRI; H, HindIII; X,XhoI. (B) The nucleotide sequence of the breakpoint region and its alignment with 11q13 and 14q32 germ-line sequences are shown.
Molecular cloning of the chromosomal breakpoint from case LB411. (A) Schematic representation of the breakpoint and the respective 11q13 germ-line region. From the top: a diagram of theBCL-1/cyclin D1 locus where the cosmid clones I4, R4b, pHS11 are located is shown; the 11q13 germ-line region and the probes used for the Southern and Northern blot analyses are shown as solid lines. The vertical arrow indicates the breakpoint position. In the t(11;14) breakpoint clone, chromosome 14 is indicated by open boxes with black or stippled boxes representing different IGH regions and chromosome 11 is shown as a solid line. Restriction enzyme symbols: B, BamHI; R, EcoRI; H, HindIII; X,XhoI. (B) The nucleotide sequence of the breakpoint region and its alignment with 11q13 and 14q32 germ-line sequences are shown.
Cloning and mapping of the 11q13 breakpoint in MM cell line U266.
We investigated cyclin D1 expression in a panel of MM-derived cell lines (U266, KMM1, JJN3, OPM2, Karpas, IM9, Sultan, LP-1) for which karyotypic analyses have been reported to be negative for the t(11;14)(q13;q32) chromosomal translocation. Interestingly, Northern blot analysis demonstrated a high level of cyclin D1 expression in U266 cell line (Fig 3A). On the basis of this evidence, we looked for the presence of an illegitimateIGH recombination in U266 DNA by Southern blot, as for case LB411. As shown in Fig 3B, hybridization of BamHI-digested DNA with the Cα1 probe detected a rearranged fragment that did not comigrate with the rearranged JH allele. A phage library was constructed using BamHI-digested DNA from the U266 cell line and screened with the Cα1 probe. The rearranged Cα1 fragment was isolated and tested on normal metaphases by FISH analysis, which showed hybridization with chromosomes 14q32 and 11q13 (data not shown) that was consistent with the presence of a t(11;14)(q13;q32) chromosomal translocation. We next cloned about 20 kb of the normal genomic region surrounding the breakpoint by isolating a recombinant phage from a normal genomic library (Fig 3C). This genomic region did not hybridize with MTC, p94, or full-length cyclin D1 cDNA and with any of the available 11q13 clones (see Fig 1).
Molecular analysis of the U266 MM cell line. (A) Northern blot analysis of human MM cell lines showing cyclin D1 overexpression in U266. The length of the cyclin D1 transcripts is shown in kb. β-actin hybridization is shown for loading quantification. (B) Southern blot analysis of the IGH locus in the U266 cell line. The DNA was digested with BamHI restriction enzyme, and the nylon filter was subsequently hybridized with the probes specified below. The germ-line bands are indicated by dashes. The arrow indicates the cloned “illegitimate” IGHrearranged fragment. (C) Molecular cloning of the chromosomal breakpoint in the U266 cell line. The breakpoint and the respective 11q13 germ-line region are shown; the probes used for Southern and Northern blot analyses are shown in the diagram. The vertical arrow indicates the breakpoint position. Chromosome 14 is represented by open boxes with black or stippled boxes showing different IGHregions and chromosome 11 is represented as a solid line. Restriction enzyme symbols: B, BamHI; R, EcoRI; H,HindIII; X, XhoI. (D) The nucleotide sequence of the breakpoint region and its alignment with 14q32 and 11q13 germ-line sequences are shown.
Molecular analysis of the U266 MM cell line. (A) Northern blot analysis of human MM cell lines showing cyclin D1 overexpression in U266. The length of the cyclin D1 transcripts is shown in kb. β-actin hybridization is shown for loading quantification. (B) Southern blot analysis of the IGH locus in the U266 cell line. The DNA was digested with BamHI restriction enzyme, and the nylon filter was subsequently hybridized with the probes specified below. The germ-line bands are indicated by dashes. The arrow indicates the cloned “illegitimate” IGHrearranged fragment. (C) Molecular cloning of the chromosomal breakpoint in the U266 cell line. The breakpoint and the respective 11q13 germ-line region are shown; the probes used for Southern and Northern blot analyses are shown in the diagram. The vertical arrow indicates the breakpoint position. Chromosome 14 is represented by open boxes with black or stippled boxes showing different IGHregions and chromosome 11 is represented as a solid line. Restriction enzyme symbols: B, BamHI; R, EcoRI; H,HindIII; X, XhoI. (D) The nucleotide sequence of the breakpoint region and its alignment with 14q32 and 11q13 germ-line sequences are shown.
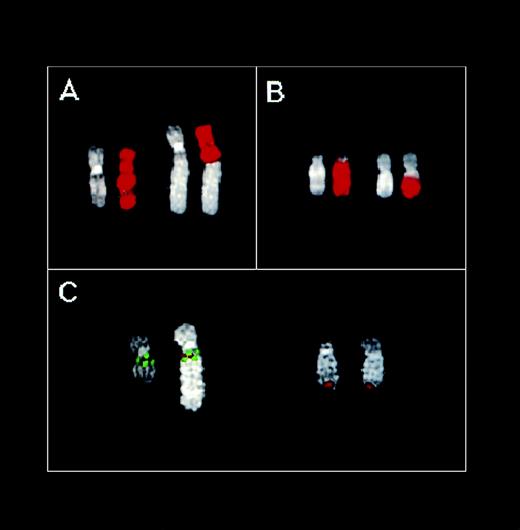
To define the location of the U266 11q13 breakpoint more precisely, we performed FISH analyses on metaphases spreads. As shown in Fig 4A, the chromosome 11 painting probe hybridized with two chromosomes on U266 metaphases: an apparently normal chromosome 11 and a structurally altered chromosome consisting of the 11pter→11q13-14 region and extra material of unknown origin. Hybridizations with probes specific for the MTC region, and thecyclin D1, FGF4, and FGF3 loci, indicated that all of these regions are contained in the putative 11q13 region of this abnormal chromosome (data not shown). Hybridization with the chromosome 14 painting probe clearly detected two different chromosomes: an apparently normal chromosome 14 and an abnormal chromosome with a large amount of chromosome 14 material on its long arm. (Fig 4B). Thus, no detectable exchange of material between chromosomes 11 and 14 was observed with painting probes hybridization: a finding consistent with the absence of a cytogenetically detectable t(11;14)(q13;q32) in U266 cell line. Interestingly, the Cα1 clone was clearly detected at 11q13-14 on the abnormal chromosome 11 (colocalized with each of the 11q13 probes tested above; Fig 4C and data not shown), as well as in the telomeric region of both chromosomes recognized by the 14 painting probe (Fig 4C). These findings indicated the presence of a t(11;14) and suggested that the breakpoint on 11q13 may be located telomeric of theFGF3 locus. Interestingly, the YAC clone 877-D-8 specific for a region telomeric of the GARP locus on 11q14 hybridized to the abnormal chromosome 11 in U266 metaphases, thus suggesting that this locus is still retained. Triple-hybridization experiments using MTC (green), Cα1 (red), and 877-D-8 (green) probes showed that the Cα region is apparently located between the MTC and GARP loci (Fig4C). These findings indicate that the t(11;14) translocation in U266 is the result of a complex chromosome rearrangement, as is also suggested by the presence of extra material on the abnormal chromosome 11. The FISH experiments performed in an attempt to elucidate the origin of the extra material on this chromosome gave very complex results; however, chromosomes 3 and 4 are apparently involved (data not shown).
FISH analysis of the U266 MM cell line. (A) Partial U266 metaphase hybridized with the chromosome 11 painting probe. (B) Partial U266 metaphase hybridized with the chromosome 14 painting probe. DAPI counterstaining is shown for each chromosome. (C) Partial U266 metaphase cohybridized with cosmid I4 (green), YAC 877-D-8 (green), and C1 (red) probes. For details concerning these probes, see text and Fig 1.
FISH analysis of the U266 MM cell line. (A) Partial U266 metaphase hybridized with the chromosome 11 painting probe. (B) Partial U266 metaphase hybridized with the chromosome 14 painting probe. DAPI counterstaining is shown for each chromosome. (C) Partial U266 metaphase cohybridized with cosmid I4 (green), YAC 877-D-8 (green), and C1 (red) probes. For details concerning these probes, see text and Fig 1.
Mapping of 11q13 breakpoints in MM tumors with a cytogenetically detectable t(11;14)(q13;q32) chromosomal translocation.
We used FISH analysis to investigate the approximate location of the 11q13 breakpoints in the cell line KMS-12 and a primary MM tumor (AC97), both carrying a t(11;14)(q13;q32) chromosomal translocation.
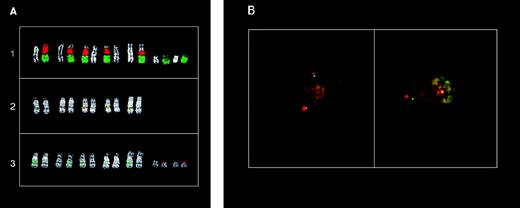
While this work was in progress, Chesi et al16 reported the cloning of a t(11;14) breakpoint in the KMS-12; more recently, Vaandrager et al33 have reported that this breakpoint is located approximately 215 kb centromeric of the MTC region and is juxtaposed to the Cγ2 of the IGH locus. Therefore, only part of our results will be presented. In particular, double-color FISH on KMS-12 metaphase spreads with the painting probes specific for chromosomes 11 and 14 showed the presence of five putative 14 (der) chromosomes containing material from chromosome 11 (Fig 5A1). FISH analyses further demonstrated that the MTC, cyclin D1, FGF4, andFGF3 loci were all contained in these chromosomes and apparently juxtaposed to sequences recognized by the Cα1 probe (Fig 5A2 and data not shown). The YAC clones 961-H-2 and 744-F-12 located centromeric of the MTC region (see Fig 1) did not hybridize with the 14 (der) chromosomes (Fig 5A3 and data not shown), but with two small and structurally-altered chromosomes recognized by the painting and centromeric-specific probes of chromosome 11 (Fig 5A1, 3, and data not shown). These results suggest that the breakpoint is centromeric of the MTC region and are consistent with previously reported data.33
FISH analysis of the 11q13 breakpoints in MM tumors with a cytogenetically detectable t(11;14)(q13;q32) chromosome translocation. (A) FISH analysis of the KMS12 cell line: (1) partial metaphase cohybridized with chromosome 11 (green) and chromosome 14 (red) painting probes; (2) partial metaphase cohybridized with cosmid I4 (MTC-green) and C1 (red) probes; (3) partial metaphase cohybridized with YAC 961-H-2 (red) and C1 (green) probes. DAPI counterstaining is shown for each chromosome. (B) Double-color interphase FISH analysis of MM tumor AC97. Left: colocalized signals in cohybridization experiments with YAC 744-F-12 (green) and pHS11 (red) probes; right: dissociated signals in cohybridization experiments with pHS11 (red) and cyclin D1 (green) probes. See scheme in Fig 1for the probes used.
FISH analysis of the 11q13 breakpoints in MM tumors with a cytogenetically detectable t(11;14)(q13;q32) chromosome translocation. (A) FISH analysis of the KMS12 cell line: (1) partial metaphase cohybridized with chromosome 11 (green) and chromosome 14 (red) painting probes; (2) partial metaphase cohybridized with cosmid I4 (MTC-green) and C1 (red) probes; (3) partial metaphase cohybridized with YAC 961-H-2 (red) and C1 (green) probes. DAPI counterstaining is shown for each chromosome. (B) Double-color interphase FISH analysis of MM tumor AC97. Left: colocalized signals in cohybridization experiments with YAC 744-F-12 (green) and pHS11 (red) probes; right: dissociated signals in cohybridization experiments with pHS11 (red) and cyclin D1 (green) probes. See scheme in Fig 1for the probes used.
Case AC97 carried a t(11;14)(q13;q32) chromosomal translocation (see Materials and Methods; data not shown) without any evidence of the involvement of the BCL-1/cyclin D1 regions at Southern blot analysis (data not shown). No data concerning cyclin D1 expression were available in this case. Two-color FISH analysis in interphase nuclei with probes of the 11q13 region showed that the breakpoint in this case is apparently located between MTC and thecyclin D1 locus. In particular, colocalization of signals (red and green) were detected on interphase nuclei when YAC 744-F-12 was used together with the pHS11 clone (Fig 5B) and when the cyclinD1 cosmid was cohybridized with the FGF3-specific clone (data not shown), but dissociation of signals was observed when the pHS11 andcyclin D1 cosmids were used (Fig 5B).
Lack of clusters of 11q13 breakpoints in MM.
Because the t(11;14)(q13;q32) translocation is recurrently found in MM, we investigated by Southern blot whether any of the cloned 11q13 breakpoints were involved in the translocation in other MM cases using the probes described in Fig 6, which are specific for case LB411 and cell lines U266 and KMS-12. We tested DNA from 50 MM primary tumors for which karyotype data were not available. Furthermore, all of these cases have been found to be negative for rearrangements of the MTC and p94 regions and cyclin D1 locus (data not shown). Southern blot analysis was performed on BamHI DNA digests and, when possible, on EcoRI and HindIII digests. Rearrangements were detected in only one tumor (patient LB104) and involved the 11q13 region where the breakpoint of the KMS-12 cell line is located (Fig 6). Case LB104 was a 72-year-old female patient affected by an IgAλ-type MM in clinical stage IIA at diagnosis and with 32 months survival. Cyclin D1 expression was evaluated by RT-PCR in only 11 of the 50 MM patients (including case LB104). Specific amplified fragments were detected in case LB104 and in an IgGλ-type MM tumor (case LB413) from a 75-year-old female patient in clinical stage IIIB at diagnosis with 13 months survival (data not shown).
Southern blot analysis of MM tumors using 11q13 probes. (A) DNA was digested with HindIII restriction enzyme and hybridized with the 193 bp probe (see Materials and Methods) located in proximity of the KMS-12 breakpoint16; a rearrangement was observed in one tumor (case LB104; see text). (B) DNA was digested withBamHI restriction enzyme and hybridized with the A/H 0.6 kb probe located in proximity of the LB411 breakpoint. (C) DNA was digested with HindIII restriction enzyme and hybridized with the B/R 0.4 kb probe located in proximity of U266 breakpoint. The rearrangement patterns of KMS12, LB411, and U266 with the specified 11q13 probes are respectively shown in (A, B, and C). Germ-line fragments are indicated by dashes. The diagrams represent the 11q13 germ-line regions; probes used in the Southern blot analyses are shown. Restriction enzyme symbols: H, HindIII; B, BamHI; R,EcoRI; A, AvaI.
Southern blot analysis of MM tumors using 11q13 probes. (A) DNA was digested with HindIII restriction enzyme and hybridized with the 193 bp probe (see Materials and Methods) located in proximity of the KMS-12 breakpoint16; a rearrangement was observed in one tumor (case LB104; see text). (B) DNA was digested withBamHI restriction enzyme and hybridized with the A/H 0.6 kb probe located in proximity of the LB411 breakpoint. (C) DNA was digested with HindIII restriction enzyme and hybridized with the B/R 0.4 kb probe located in proximity of U266 breakpoint. The rearrangement patterns of KMS12, LB411, and U266 with the specified 11q13 probes are respectively shown in (A, B, and C). Germ-line fragments are indicated by dashes. The diagrams represent the 11q13 germ-line regions; probes used in the Southern blot analyses are shown. Restriction enzyme symbols: H, HindIII; B, BamHI; R,EcoRI; A, AvaI.
DISCUSSION
Conventional cytogenetic analyses of MM are generally a difficult task because of the low proliferation rate of malignant plasma cells. Abnormal karyotypes have been reported only in about 40% of MM and at a higher frequency in plasma cell leukemia; interestingly, in about 20% to 40% of MM patients with an abnormal karyotype a 14q+ marker is observed.12,13 In about 30% of the cases, the 14q+ marker originates through a t(11;14)(q13;q32) chromosomal translocation, which is mainly associated with MCL and involves the cyclin D1 gene on 11q13.12 However, rearrangements of theBCL-1/cyclin D1 regions involved in MCL have been rarely demonstrated in cases of MM carrying such a translocation (mainly cell lines) or in unselected tumors.9,14-16 The overexpression of cyclin D1 has been observed in the majority of MM-derived cell lines carrying the t(11;14), thus suggesting that this translocation may lead to the disregulation of this gene in MM, as it does in MCL.7,9 16 The main aim of the present study was to investigate whether specific regions on 11q13 are associated with 11q13 breakpoints in MM. To this end, we cloned and mapped different 11q13 breakpoints, isolated the corresponding germ-line regions, and evaluated in Southern blot a representative panel of MM primary tumors for genomic rearrangements.
In case LB411, the breakpoint was located between MTC andcyclin D1, approximately 40 kb telomeric of the MTC. In case AC97 carrying a t(11;14), FISH analysis allowed us to map the breakpoint in this region, in which the breakpoints from the MM cell lines XG1 and SK-MM2 have also been found to be located.9,16 In the U266 cell line, the breakpoint is telomeric of the cyclin D1 locus between the FGF3 andGARP loci; however this rearrangement is complex because the Cα1 region is located to the abnormal chromosome 11, and apparently the GARP locus is still retained on this chromosome. As far as we know, this is the second MM tumor in which the 11q13 breakpoint has been mapped telomeric of cyclin D1. Raynaud et al9reported a similar finding in the XG2 cell line; however, thecyclin D1 gene was not found to be expressed in this cell line. Given the large distance between FGF3 and GARP, it can be suggested that the breakpoint in U266 may be closer to theFGF3 gene, at a distance that can affect cyclin D1 expression. In the KMS-12 cell line, the breakpoint is centromeric of the MTC. During the course of this study, Vaandrager et al33 reported that the breakpoint in this cell line is located 215 kb centromeric of the MTC region; Southern blot analysis showed that the breakpoint in our case LB104 appears to be located in the same region. A breakpoint centromeric of the MTC locus has been previously reported in the XG5 MM cell line.9 Taken together, these data confirm the notion that 11q13 breakpoints in MM are scattered along the 11q13 (see scheme in Fig 7) and suggest that they may occur either centromeric of the MTC, between the MTC and cyclin D1 loci, or telomeric of the cyclin D1. Interestingly, we found by RT-PCR that the presence of 11q13 breakpoints in tumors LB411 and LB104 correlates with cyclin D1 overexpression (data not shown).
Schematic representation of the MM breakpoints location in the 11q13 region. For details, see Raynaud et al9(XG1,XG2,XG5 cell lines), Meeus et al15 (two MM cases), Chesi et al16 (SK-MM2 cell line), and Vaandrager et al33 (KMS-12 cell line). The arrows indicate cases for which the breakpoint has been cloned; the dashes indicate cases for which the breakpoint has been approximately mapped by FISH or Southern blot analyses.
Schematic representation of the MM breakpoints location in the 11q13 region. For details, see Raynaud et al9(XG1,XG2,XG5 cell lines), Meeus et al15 (two MM cases), Chesi et al16 (SK-MM2 cell line), and Vaandrager et al33 (KMS-12 cell line). The arrows indicate cases for which the breakpoint has been cloned; the dashes indicate cases for which the breakpoint has been approximately mapped by FISH or Southern blot analyses.
The availability of 11q13 probes specific for breakpoints in MM would make it possible to investigate by Southern blot analysis whether distinct breakpoint clusters may occur in MM. As far as we know, this is the first study regarding the screening of MM tumors using such an approach; however, our analysis with probes from three distinct 11q13 regions representative of MM breakpoints detected a rearrangement in only one case, further confirming the notion that 11q13 rearrangements associated with MM are scattered over a large region encompassing thecyclin D1 gene. These findings suggest that other technical approaches, such as FISH on interphase nuclei,4 are needed to assess the frequency and involvement of 11q13 breakpoints in MM.
AKNOWLEDGMENT
We are grateful to Dr S. Raynaud and P. Delli Bovi for providing us with cosmid clones I4, R4B, pHS-11, and FGF4 clone, respectively, and to Dr T. Otsuki for providing us with the KMS-12 cell line. We thank G. Ciceri for her expert technical assistance.
Supported by grants from the Associazione Italiana Ricerca sul Cancro (AIRC) (to A.N. and M.R.) and the Ministero della Sanità to Ospedale Maggiore IRCCS (Ricerca Corrente 1994).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Antonino Neri, MD, Servizio Ematologia Istituto di Scienze Mediche, Università di Milano, Ospedale Maggiore di Milano, IRCCS Via Francesco Sforza 35, 20122 Milano, Italy; e-mail: filobus@imiucca.csi.unimi.it.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal