Abstract
Interleukin-1 receptor antagonist (IL-1Ra) is produced by hepatocytes with characteristics of an acute-phase protein. To examine the role of IL-4 and IL-13 in production of IL-1Ra, human primary hepatocytes and HepG2 human hepatoma cells were cultured in the presence of IL-4 or IL-13 in combination with IL-1β and/or IL-6. The results indicated that both IL-4 and IL-13 amplified the stimulatory effect of IL-1β on production of IL-1Ra protein and messenger RNA (mRNA) by both human primary hepatocytes and HepG2 cells. IL-1Ra refers to three different peptides, one secreted (sIL-1Ra) and two intracellular (icIL-1RaI and icIL-1RaII), derived from the same gene. sIL-1Ra and icIL-1RaI are the products of two different mRNA, whereas icIL-1RaII is synthesized by alternative translation initiation mainly from sIL-1Ra mRNA. Our results show that both sIL-1Ra and icIL-1RaII, but not icIL-1RaI, are produced by HepG2 cells and human hepatocytes. Transient transfection experiments as well as mRNA stability studies indicated that IL-4 stimulated sIL-1Ra production primarly at the level of transcription. Gel retardation assays showed that IL-4 induced the formation of a STAT6-DNA complex with a STAT6 binding element within the sIL-1Ra promoter, but had no effect on IL-1–induced NF-κB binding activity. In contrast to IL-1Ra, production of C-reactive protein by human primary hepatocytes was stimulated by IL-6 and decreased by the addition of IL-4.
THE ACUTE-PHASE PROTEINS (APP) include several plasma proteins, the levels of which are modified during inflammatory conditions. These proteins have different biologic functions and they contribute to both initiation and modulation of inflammatory responses.1 Recently, we showed that interleukin-1 (IL-1) receptor antagonist (IL-1Ra) is produced by hepatocytes and regulated as an APP.2 IL-1Ra has been shown to have antiinflammatory properties in several experimental animal models of diseases, further emphasizing the role of APP as modulators of the inflammatory response.
Although IL-6 is considered the major inducer of APP, other cytokines such as IL-1, tumor necrosis factor (TNF)-α, transforming growth factor (TGF)-β, interferon (IFN)γ, and IL-8 can also stimulate the production of APP. In addition, changes in APP production by hepatocytes are not always mediated by only one cytokine but are influenced by a combination of cytokines, cytokine receptors, and hormones.1
IL-4 is a cytokine that is produced primarily by CD4+ T lymphocytes and mast cells.3,4 IL-4 inhibits the production of IL-1, TNF-α, IL-8, and prostaglandin E2,5-7 whereas it increases the production of IL-1Ra by monocytes stimulated by lipopolysaccharide (LPS).5 These observations suggest that IL-4 could have antiinflammatory properties. IL-13, another T-cell–derived cytokine, shares several biologic activities with IL-4. However, IL-4 can induce unique responses in certain cells, such as T lymphocytes, which do not respond to IL-13.8 IL-4 has been reported to decrease the production of APP such as haptoglobin, C-reactive protein (CRP), and albumin by human primary hepatocytes, whereas it had no effect on other APP.9 In addition, IL-4 was shown to decrease the effects of IL-1β, TNF-α, and IL-6 on lipogenesis in mouse hepatocytes in vivo.10 The effects of IL-13 on hepatocytes have not been reported.
The studies included herein show that both IL-4 and IL-13 amplified the stimulatory effects of IL-1β on production of IL-1Ra by hepatocytes. In contrast, IL-4 downregulated the IL-6–induced production of CRP by hepatocytes.
MATERIALS AND METHODS
Reagents.
IL-1β, IL-6, and IL-4 were purchased from R&D Systems, Inc (Minneapolis, MN); the LPS contamination in each cytokine preparation was less than 0.1 ng/mg protein according to the manufacturer. IL-13 was generously donated by DNAX (Palo Alto, CA). Rabbit antihuman CRP antibodies were purchased from Dako (Glostrup, Denmark).
Cell cultures.
Human primary hepatocytes were isolated from livers obtained from 11 organ donors through a collaboration with the International Institute for the Advancement of Medicine (Scranton, PA), a nonprofit tissue and organ bank. Human livers used for hepatocyte isolation were obtained under the United Network for Organ Sharing specifications and guidelines but were determined unsuitable for orthotopic liver transplantation due to physical trauma to a portion of the organ, anoxia, or a high interstitial fat content. All livers had been serologically screened and found negative for human immunodeficiency virus (HIV), human T-cell lymphotrophic virus (HTLV), hepatitis B, and hepatitis C before acceptance. Human hepatocytes were isolated as previously described.11 Viable hepatocytes (200 × 103 cells/mL), as assessed by Trypan blue exclusion, were seeded on 24-well culture Primaria plates (Becton Dickinson, Franklin Lakes, NJ) in RPMI 1640 medium (Mediatech, Inc, Herndon, VA) supplemented with penicillin, streptomycin, and 10% fetal calf serum (FCS) (Summit Biotechnology, Greeley, CO). After 12-hour culture, the cells were placed in medium supplemented with 5% FCS. After 24-hour culture, the cells were placed in medium supplemented with 0.2% bovine serum albumin (BSA). The cells were stimulated after 48 hours of culture. Supernatants were collected at 18 hours after stimulation and kept frozen at −20°C until assayed for IL-1Ra and CRP content.
HepG2 cells were purchased from the American Type Culture Collection (Rockville, MD) and cultured in 24-well plates (Becton Dickinson) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with penicillin, streptomycin, and 10% FCS. After the cells reached confluence, the medium was renewed, and stimulants added. The supernatants were collected at 18 hours and kept frozen until assayed for IL-1Ra content.
IL-1Ra enzyme-linked immunosorbent assay (ELISA).
IL-1Ra concentrations were measured in culture supernatants from human primary hepatocytes and from HepG2 cells using a sandwich ELISA, as previously described.2 The sensitivity of the assay was 78 pg/mL.
CRP ELISA.
CRP concentrations were measured in culture supernatants from human primary hepatocytes using a sandwich ELISA, as previously described.12 The sensitivity of the assay was 0.4 ng/mL.
Western blot analysis.
Protein extracts were obtained from HepG2 cells, human primary hepatocytes, and the human keratinocyte cell line A431 using Trizol (Life Technologies, Gaithersburg, MD) as recently described.13 Twenty μg total protein were heated in reducing buffer and electrophoresed on 17.5% polyacrylamide gels followed by electrophoretic transfer to polyvinylidene difluoride (PVDF) membrane (DuPont-NEN, Boston, MA). Western blot analysis was then performed as described.2
IL-1Ra messenger RNA (mRNA) analysis by ribonuclease protection assay.
Total RNA was prepared from HepG2 cells and human primary hepatocytes cultured in 100-mm plates. After 16 hours of stimulation, if not otherwise mentioned, the cells were lysed in Trizol (Life Technologies) and RNA was prepared according to the manufacturer’s instructions.
A specific probe for human IL-1Ra isoforms mRNA was generated by polymerase chain reaction (PCR) using the following primers (5′ to 3′): 5′ icIL-1Ra, GCA TGA ATT CCA GGT ACT GCC CGG GTG CTA CTT TAT; and 3′ common IL-1Ra, GCA TAA GCT TTG CCT CCA GCT GGA GTC TGG TCT C. The PCR products were cleaved with EcoRI andHindIII and then cloned into Bluescript SK+ plasmid (Stratagene, La Jolla, CA). Absence of mutation was verified by sequencing. Generation of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was previously described.11 The RNase protection assays were performed as recently described.2Briefly, 10 μg total RNA was hybridized with the32P-labeled riboprobe and RNase-treated. The RNA protected fragments were resolved on denaturing 6% polyacrylamide-8 mol/L urea gels. Autoradiography was immediately performed and quantitation of IL-1Ra and GAPDH mRNA fragments was performed by PhosphorImager (ImageQuant; Molecular Dynamics Inc, Sunnydale, CA).
HepG2 cell transfection studies.
The psRA-294Luc refers to the 294-bp of 5′-flanking DNA with respect to the first exon of sIL-1Ra. Transfections were performed using a modification of the calcium phosphate method as described.2 Briefly, 0.5 × 106 HepG2 cells were seeded in 60-mm dishes in complete medium. After 24 hours of culture, 0.5 mL of calcium precipitate containing 7.5 μg plasmid DNA was added to each plate. The cells then were incubated at 37°C for 4 hours and subjected to a glycerol shock for 2 minutes. After 24 hours the cytokines were added. The cells were harvested after 24 hours and luciferase activity was assessed as described previously.2Luciferase activity was normalized to total protein concentration of the cell lysate (protein assay kit; BioRad Laboratories, Richmond, CA).
Nuclear extracts.
Nuclear extracts were prepared from untreated HepG2 cells or cells stimulated by IL-1β, IL-4, or a combination of both cytokines for 30 minutes as previously described.2 Protease and phosphatase inhibitors were added to the buffers, including 1 μg/mL leupeptin, 1 μg/mL aprotonin, 1 μg/mL pepstatin, 10 mmol/L β-glycerophosphate, and 1 mmol/L sodium vanadate. Protein concentrations were measured using a Bio-Rad protein assay kit. Nuclear proteins were kept at −80°C until used for electrophoretic mobility shift assay (EMSA).
EMSA.
For NF-κB, 5 μg of nuclear extracts were incubated in 20 mL of EMSA-binding reaction consisting of 10 mmol/L HEPES (pH 7.9), 80 mmol/L KCl, 1 mmol/L EDTA (pH 8.0), 1 mmol/L ethyleneglycoltetracetic acid (EGTA) (pH 8.0), 6% glycerol, and 4 μg poly (dI:dC) at 15°C for 15 minutes before adding 20,000 cpm of32P-labeled oligonucleotide probe for an additional 15 minutes at 15°C. For STAT6, the binding reaction was performed with 10 μg of nuclear extracts in 20 mmol/L HEPES (pH 7.9), 50 mmol/L KCl, 0.1 mmol/L EDTA (pH 8.0), 1 mmol/L dithiothreitol (DDT), 5% glycerol, 200 μg/mL BSA, and 3 μg poly (dI:dC) at 4°C for 15 minutes before adding 50,000 cpm of 32P-labeled oligonucleotide probe for an additional 15 minutes at room temperature (RT). The reaction was analyzed on a 6% nondenaturing polyacrylamide gel in 0.5× (0.25× for STAT6) Tris-borate/EDTA buffer. After electrophoresis, the gel was fixed in 10% acetic acid, dried under vacuum, and autoradiography was performed immediately.
For DNA competition studies, the binding reaction was conducted in the presence of nonspecific competitor together with a 100-fold molar excess of unlabeled probe. For antibody supershift assays, the extracts were preincubated either with antibodies against the p65 (RelA) subunit of NF-κB, p50 subunit of NF-κB, or STAT6. Rabbit IgG was used as control. Both antibodies and consensus sequence oligonucleotides were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Oligonucleotides used in EMSA:
NF-κB wild type (WT): ATG CGA GGA GGG TAT TTC CGC TTC T;
NF-κB Mutant: ATG CGA GGA Gct cga g TC CGC TTC T;
Consensus NF-κB: ATG TGA GGG GAC TTT CCC AGG;
SBE1 WT: GC TCT TCT TCC CAG GAA CTC AAT;
SBE1 Mutant: GC TCT TCT g CC CAG Gc A CTC AAT;
Consensus STAT6: GTA TTT CCC AGA AAA GGA AC; and
SBE2 WT: AT CTT AAT TTT GGG GAA ATT GCA C
(SBE = STAT6 binding element).
Double-stranded WT oligonucleotides were labeled with 32P dNTPs by fill-in reaction of 5′ overhangs using theKlenow enzyme (Promega, Madison, WI) and isolated by two passages over Sephadex G-50 spin columns (Pharmacia Biotech, Uppsala, Sweden).
Statistical analysis.
Student’s t-test (unpaired, two tailed) was used for comparisons between specified different conditions.
RESULTS
Production of IL-1Ra by HepG2 cells stimulated in the presence of IL-4 and IL-13.
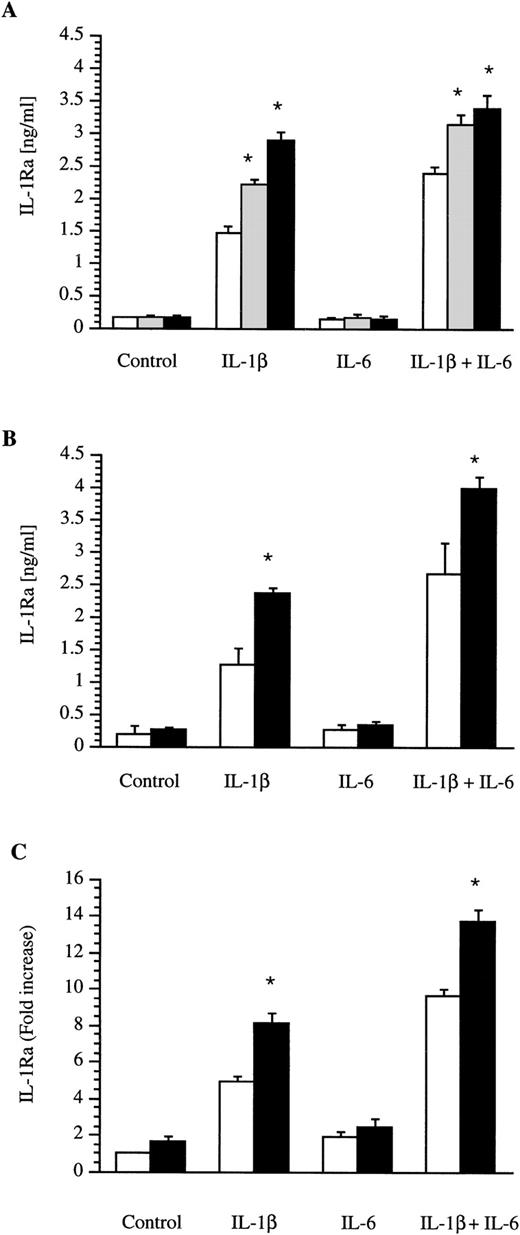
Confluent human hepatoma HepG2 cells were cultured in the presence of IL-1 β, IL-6, IL-4, and IL-13 alone or in combination. Culture supernatants were collected after 18 hours and the IL-1Ra concentrations were determined by ELISA. As recently described, IL-1β alone or in combination with IL-6 was able to upregulate the production of IL-1Ra (Fig 1). Concentrations of IL-1β and IL-6 used in these experiments were previously shown to maximally induce IL-1Ra.2 IL-4 alone or in the presence of IL-6 did not stimulate the production of IL-1Ra (Fig 1A). In contrast, IL-4 at concentrations of 1 ng/mL or 10 ng/mL significantly enhanced the effect of IL-1β or the combination of IL-1β and IL-6 on IL-1Ra production. Dose-response studies with IL-4 concentrations ranging from 0.1 to 100 ng/mL showed that a concentration of 10 ng/mL maximally induced IL-1Ra production in the presence of IL-1 β (data not shown). The combination of IL-1β and IL-4 was almost as potent as the combination of IL-1β and IL-6. In addition, IL-13 at a concentration of 10 ng/mL also amplified the effect of IL-1β or the combination of IL-1β and IL-6 on IL-1Ra production (Fig 1B).
Production of IL-1Ra by the HepG2 human hepatoma cell line and by human primary hepatocytes. HepG2 cells were cultured in the absence (control) or presence of IL-1β, IL-6, and a combination of IL-1β and IL-6 without (□) or with the addition of 1 ng/mL (▩) or 10 ng/mL (▩) IL-4 (A). Similar experiments were performed with the addition (▩) of 10 ng/mL IL-13 in HepG2 cells (B) and of 10 ng/mL IL-4 in human primary hepatocytes (C). After 18 hours of stimulation, levels of IL-1Ra were measured by ELISA in culture supernatants. Values represent the mean ± SEM of three experiments. *P < 0.01 in comparison with cells cultured in the absence of IL-4 or IL-13.
Production of IL-1Ra by the HepG2 human hepatoma cell line and by human primary hepatocytes. HepG2 cells were cultured in the absence (control) or presence of IL-1β, IL-6, and a combination of IL-1β and IL-6 without (□) or with the addition of 1 ng/mL (▩) or 10 ng/mL (▩) IL-4 (A). Similar experiments were performed with the addition (▩) of 10 ng/mL IL-13 in HepG2 cells (B) and of 10 ng/mL IL-4 in human primary hepatocytes (C). After 18 hours of stimulation, levels of IL-1Ra were measured by ELISA in culture supernatants. Values represent the mean ± SEM of three experiments. *P < 0.01 in comparison with cells cultured in the absence of IL-4 or IL-13.
Production of IL-1Ra by human primary hepatocytes stimulated with IL-4.
Human primary hepatocytes obtained from livers unsuitable for transplantation were cultured in the presence of IL-1β, IL-6, and IL-4 alone or in combination. Culture supernatants were collected after 18-hour stimulation and the IL-1Ra concentrations were determined by ELISA. A similar pattern of IL-1Ra production was found in hepatocytes from three liver donors. However, due to large variations in basal and induced IL-1Ra values between donors, the results are expressed as fold-increase. As shown in Fig 1C, IL-4 significantly enhanced the stimulatory effect of IL-1β on production of IL-1Ra by human primary hepatocytes. IL-4 alone or in combination with IL-6 had a slight but not significant enhancing effect on IL-1Ra production. The effect of IL-13 on human primary hepatocytes was examined in one donor and the results were similar to those observed with IL-4 (data not shown).
Western blot analysis.
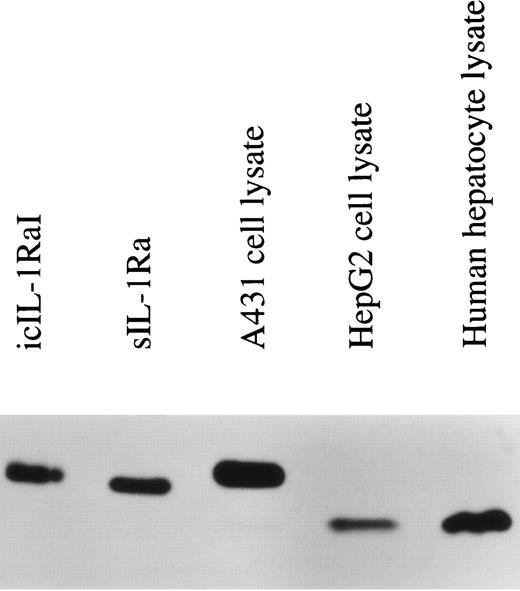
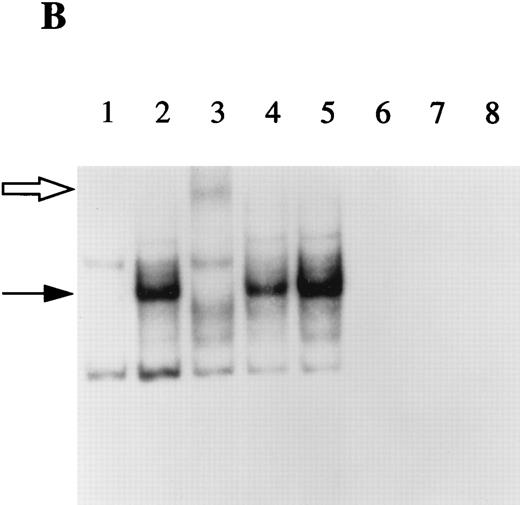
The term IL-1Ra refers to three different proteins that are encoded by the same gene, but are generated by alternate RNA splicing of two different first exons. One form is secreted (17 kD sIL-1Ra) and one form remains intracellular (now termed 18 kD icIL-1RaI). In addition, our laboratory has recently described another intracellular smaller molecular mass variant of IL-1Ra (16 kD icIL-1RaII).14 This latter isoform is produced by alternative translation initiation mainly from sIL-1Ra mRNA. In previous studies, we observed that HepG2 cells expressed only the sIL-1Ra mRNA. In addition, by Western blot analysis, we detected the presence of sIL-1Ra protein in culture supernatants and of 16 kD icIL-1RaII in HepG2 lysates.2 To examine whether the expression of this smaller molecular weight IL-1Ra isoform was also present in normal human hepatocytes, protein extracts were prepared from cell lysates and examined by Western blot analysis. As shown in Fig 2, 16 kD icIL-1RaII, but not 18 kD icIL-1RaI, was present in both HepG2 cells (lane 4) and human primary hepatocytes (lane 5). In contrast, 18 kD icIL-1RaI was present in the lysates of the human keratinocyte cell line A431 (lane 3).
IL-1Ra size isoforms in the lysates of HepG2 cells, human primary hepatocytes, and the human keratinocyte cell line A431. Twenty micrograms of lysate proteins from HepG2 cells and human primary hepatocytes stimulated by IL-1β and IL-4 and from unstimulated A431 cells were electrophoresed on a 17.5% polyacrylamide gel, followed by electroblotting onto a PVDF membrane. Western blot analysis was performed using a mouse monoclonal antibody against human IL-1Ra that recognized all the described isoforms. Lane 1, recombinant icIL-1RaI; lane 2, recombinant sIL-1Ra; lane 3, A431 lysate; lane 4, HepG2 lysate; lane 5, human primary hepatocyte lysate.
IL-1Ra size isoforms in the lysates of HepG2 cells, human primary hepatocytes, and the human keratinocyte cell line A431. Twenty micrograms of lysate proteins from HepG2 cells and human primary hepatocytes stimulated by IL-1β and IL-4 and from unstimulated A431 cells were electrophoresed on a 17.5% polyacrylamide gel, followed by electroblotting onto a PVDF membrane. Western blot analysis was performed using a mouse monoclonal antibody against human IL-1Ra that recognized all the described isoforms. Lane 1, recombinant icIL-1RaI; lane 2, recombinant sIL-1Ra; lane 3, A431 lysate; lane 4, HepG2 lysate; lane 5, human primary hepatocyte lysate.
mRNA of IL-1Ra isoforms determined by RNase protection assay.
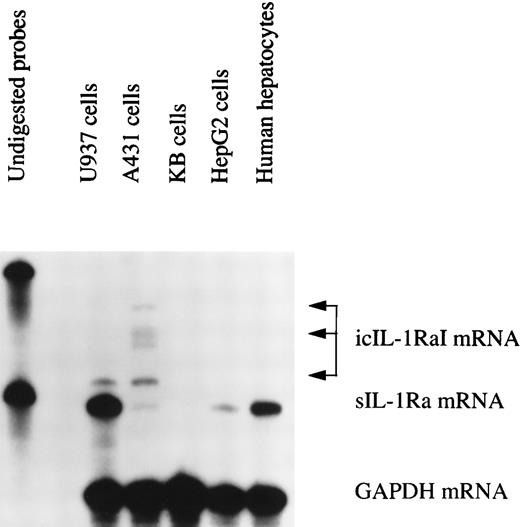
To further characterize the expression of the different IL-1Ra isoform mRNAs in human primary hepatocytes, we created a riboprobe that specifically recognized both IL-1Ra transcripts as two different protected size fragments after RNase digestion. The 5′ end of the riboprobe included the icIL-1RaI mRNA transcription start site and its 3′ end finished 250 bases after the splice site, into the IL-1Ra common region. As shown in Fig 3, both HepG2 cells and human primary hepatocytes produced only sIL-1Ra mRNA (lane 5 and 6). U937 cells were able to express both transcripts (lane 2), although sIL-1Ra mRNA was predominently produced. Interestingly, A431 cells also produced both IL-1Ra transcripts (lane 3) but, in contrast to U937 cells, they predominently produced icIL-1RaI mRNA. In addition, we also detected the presence of several icIL-1RaI mRNA size products. The monocytic cell line U937 produced predominately a smaller size transcript, whereas A431 cells produced mainly longer transcripts. These differences are probably related to the use of different transcription start sites.13 15 In KB cells, included as negative controls, we did not detect any IL-1Ra isoform mRNA (lane 4).
IL-1Ra mRNA isoforms. Total RNA from HepG2 cells and human primary hepatocytes stimulated with IL-1β and IL-4, from human keratinocyte cell lines A431 and KB, and from the U937 human monocytic cell line cultured with PMA and LPS were examined by RNase protection assay using a riboprobe that recognized the different IL-1Ra mRNA isoforms. Ten micrograms of total RNA were hybridized simultaneously with 32P-labeled riboprobes complementary to IL-1Ra and GAPDH mRNA, then digested with RNase A and RNase T1 (see Materials and Methods). The protected fragments were analyzed in a 6% denaturing polyacrylamide gel. Lane 1, undigested probes; lane 2, U937 cells; lane 3, A431 cells; lane 4, KB cells; lane 5, HepG2 cells; lane 6, human primary hepatocytes.
IL-1Ra mRNA isoforms. Total RNA from HepG2 cells and human primary hepatocytes stimulated with IL-1β and IL-4, from human keratinocyte cell lines A431 and KB, and from the U937 human monocytic cell line cultured with PMA and LPS were examined by RNase protection assay using a riboprobe that recognized the different IL-1Ra mRNA isoforms. Ten micrograms of total RNA were hybridized simultaneously with 32P-labeled riboprobes complementary to IL-1Ra and GAPDH mRNA, then digested with RNase A and RNase T1 (see Materials and Methods). The protected fragments were analyzed in a 6% denaturing polyacrylamide gel. Lane 1, undigested probes; lane 2, U937 cells; lane 3, A431 cells; lane 4, KB cells; lane 5, HepG2 cells; lane 6, human primary hepatocytes.
Quantitation of sIL-1Ra mRNA production by HepG2 cells.
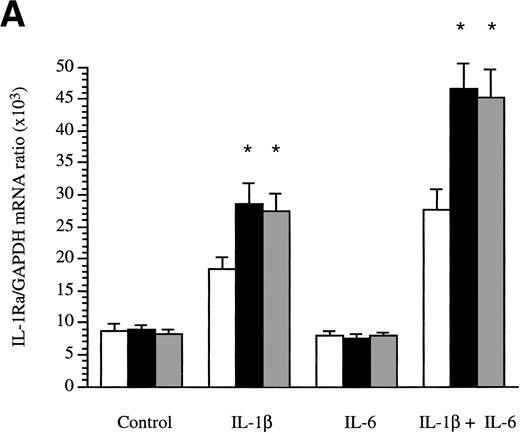
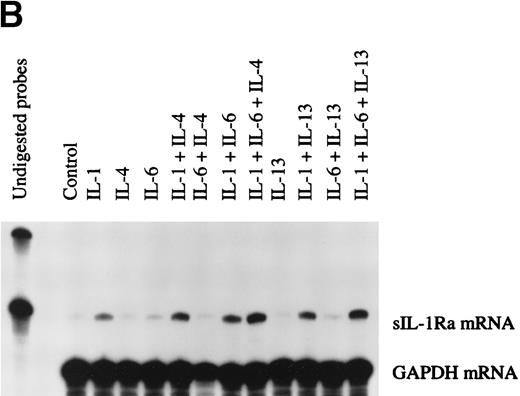
The production of sIL-1Ra mRNA by HepG2 cells cultured in the absence or presence of cytokines was examined by RNase protection assay. Time-course studies showed that sIL-1Ra mRNA production was increased 2 hours after stimulation by IL-1β and that elevated sIL-1Ra mRNA levels were maintained for up to 48 hours (data not shown). In addition, consistent with the results obtained in culture supernatants, both IL-4 and IL-13 enhanced the effect of IL-1β on production of sIL-1Ra mRNA, but neither cytokine exhibited any effect alone or in combination with IL-6 (Fig 4A).
Determination of IL-1Ra mRNA levels in HepG2 cells and in human primary hepatocytes by RNase protection assay. HepG2 cells were cultured in the absence (control) or presence of IL-1β , IL-6, and a combination of IL-1β and IL-6 both without (□) and with the addition of 10 ng/mL IL-4 (▩) or 10 ng/mL IL-13 (▩) for 16 hours (A). Ten micrograms of total RNA from the HepG2 cells were hybridized simultaneously with 32P-labeled riboprobes complementary to IL-1Ra and GAPDH mRNA, then digested with RNase A and RNase T1. The protected fragments were analyzed in a 6% denaturing polyacrylamide gel. The intensity of the protected RNA fragments was analyzed by phosphorImager. The results represent IL-1Ra/GAPDH ratio × 103. Values represent the mean ± SEM of three experiments. *P < .05 in comparison with cells cultured in the absence of IL-4 or IL-13. Human primary hepatocytes were cultured in the absence (control) or presence of IL-1β, IL-6, and a combination of IL-1β and IL-6, both without and with the addition of 10 ng/mL IL-4 or 10 ng/mL IL-13 for 16 hours (B). Ten micrograms of total RNA from the hepatocytes were examined by RNase protection assay as described above. These results are representative of three different experiments.
Determination of IL-1Ra mRNA levels in HepG2 cells and in human primary hepatocytes by RNase protection assay. HepG2 cells were cultured in the absence (control) or presence of IL-1β , IL-6, and a combination of IL-1β and IL-6 both without (□) and with the addition of 10 ng/mL IL-4 (▩) or 10 ng/mL IL-13 (▩) for 16 hours (A). Ten micrograms of total RNA from the HepG2 cells were hybridized simultaneously with 32P-labeled riboprobes complementary to IL-1Ra and GAPDH mRNA, then digested with RNase A and RNase T1. The protected fragments were analyzed in a 6% denaturing polyacrylamide gel. The intensity of the protected RNA fragments was analyzed by phosphorImager. The results represent IL-1Ra/GAPDH ratio × 103. Values represent the mean ± SEM of three experiments. *P < .05 in comparison with cells cultured in the absence of IL-4 or IL-13. Human primary hepatocytes were cultured in the absence (control) or presence of IL-1β, IL-6, and a combination of IL-1β and IL-6, both without and with the addition of 10 ng/mL IL-4 or 10 ng/mL IL-13 for 16 hours (B). Ten micrograms of total RNA from the hepatocytes were examined by RNase protection assay as described above. These results are representative of three different experiments.
sIL-1Ra mRNA production by human primary hepatocytes.
To examine the effect of IL-4 and IL-13 on production of sIL-1Ra mRNA by human hepatocytes, the cells were cultured 16 hours in the absence or presence of cytokines. Consistent with the results obtained in culture supernatants, both IL-4 and IL-13 increased the effect of IL-1β on sIL-1Ra mRNA levels but had no effect alone or when used in combination with IL-6 (Fig 4B).
IL-4 enhanced the effect of IL-1β on sIL-1Ra promoter activity.
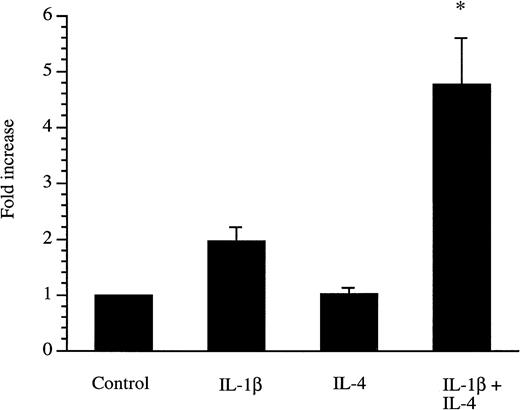
We recently showed that the sIL-1Ra promoter was active in HepG2 cells and that this promoter contained response elements for transcription factors NF-κB and C/EBP that play important role in the response to IL-β and IL-6.2 To examine whether IL-4 is able to stimulate the activity of the sIL-1Ra promoter, HepG2 cells were transfected with a plasmid containing the 294-bp sIL-1Ra promoter fragment coupled with the luciferase reporter gene and cultured in the presence or absence of IL-4 and/or IL-1β. The results showed that luciferase activity was increased by IL-1β alone and that, consistent with results observed at the endogenous gene level, IL-4 significantly enhanced the stimulatory effect of IL-1β (Fig 5).
Secreted IL-1Ra promoter transfections. HepG2 cells were transfected with a construct containing the 294-bp sIL-1Ra promoter fragment coupled with the luciferase reporter gene. The cells were then cultured in the absence (control) or presence of IL-1β and/or IL-4. After 24 hours of stimulation, the cell lysates were assayed for luciferase activity (see Materials and Methods). The data are expressed as fold increase over unstimulated cells. The results represent the mean ± SEM of three experiments. *P < .05 in comparison with cells cultured in the presence of IL-1β alone.
Secreted IL-1Ra promoter transfections. HepG2 cells were transfected with a construct containing the 294-bp sIL-1Ra promoter fragment coupled with the luciferase reporter gene. The cells were then cultured in the absence (control) or presence of IL-1β and/or IL-4. After 24 hours of stimulation, the cell lysates were assayed for luciferase activity (see Materials and Methods). The data are expressed as fold increase over unstimulated cells. The results represent the mean ± SEM of three experiments. *P < .05 in comparison with cells cultured in the presence of IL-1β alone.
To examine whether the effect of IL-4 on sIL-1Ra production was also mediated at a posttranscriptional level, HepG2 cells and human primary hepatocytes were cultured in the presence of IL-1β alone or with a combination of IL-1β and IL-4. After 4 hours of stimulation, the medium was changed and α-amanitin was added to block transcription. The results of these experiments indicated that IL-4 did not increase sIL-1Ra mRNA half-life (data not shown).
IL-4 induces STAT6 binding activity.
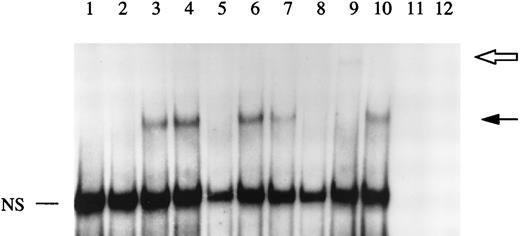
The data described above indicated that IL-4 regulated IL-1Ra gene expression at the level of transcription. The effects of IL-4 and IL-13 on transcriptional regulation of several genes are mediated through STAT6.16-18 In addition, some investigators recently described the presence of two potential STAT6 binding sites within the sIL-1Ra promoter, STAT6 binding element 1 and 2 (SBE1 and SBE2), located between bases −247 and −238 and bases −208 and −200, respectively.19 They showed that SBE1, but not SBE2, was involved in the regulation of IL-1Ra production in response to IL-4 in macrophages.19 To examine the effect of IL-4 on STAT6 binding activity, gel retardation assays were performed using nuclear extracts from HepG2 cells, cultured in the presence or absence of IL-4 and/or IL-1β, and a 32P-labeled oligonucleotide containing SBE1. As shown in Fig 6, treatment with IL-4 markedly enhanced the STAT6 binding activity (lane 3). Stimulation with both IL-4 and IL-1β produced a pattern similar to that observed with IL-4 alone (lane 4). In contrast, we did not detect any STAT6 binding activity in nuclear extracts from both unstimulated and IL-1β–stimulated HepG2 cells (lanes 1 and 2). To further characterize the protein binding to our oligonucleotide, competition studies were performed using nuclear extracts from IL-4–stimulated HepG2 cells. The formation of the DNA-binding complex was completely competed away by unlabeled wild-type SBE1 and by an oligonucleotide containing the STAT6 consensus region (lanes 5 and 8), but not by a mutated oligonucleotide (lane 6).
EMSA of STAT6 binding activity. The DNA binding activity of nuclear extracts from HepG2 cells cultured in the presence or absence of IL-1β and/or IL-4 for 30 minutes were examined using a 32P-labeled oligonucleotide from the sIL-1Ra promoter region containing the recently characterized STAT6 binding element (SBE1). Competition studies were performed with nuclear extracts from HepG2 cells stimulated with IL-4 (see Materials and Methods). Lane 1, unstimulated; lane 2, IL-1β; lane 3, IL-4; lane 4, IL-1β and IL-4; lane 5, competition with the cold probe; lane 6, competition with the cold probe mutated in the potential STAT6 binding site; lane 7, competition with the cold oligonucleotide containing another potential STAT6 binding site within the sIL-1Ra promoter (SBE2); lane 8, competition with the cold oligonucleotide containing the STAT6 consensus element; lane 9, preincubation of nuclear extracts with antibodies against STAT6 before the addition of the32P-labeled SBE1 probe; lane 10, preincubation of nuclear extracts with control IgG. Lanes 11 and 12 contain the SBE1 probe and the antibodies against STAT6 or control IgG, respectively, without nuclear extracts. The dark arrow shows the STAT6-DNA complex. The open arrow represents the supershifted complex. NS = nonspecific protein-DNA binding. These results are representative of three different experiments.
EMSA of STAT6 binding activity. The DNA binding activity of nuclear extracts from HepG2 cells cultured in the presence or absence of IL-1β and/or IL-4 for 30 minutes were examined using a 32P-labeled oligonucleotide from the sIL-1Ra promoter region containing the recently characterized STAT6 binding element (SBE1). Competition studies were performed with nuclear extracts from HepG2 cells stimulated with IL-4 (see Materials and Methods). Lane 1, unstimulated; lane 2, IL-1β; lane 3, IL-4; lane 4, IL-1β and IL-4; lane 5, competition with the cold probe; lane 6, competition with the cold probe mutated in the potential STAT6 binding site; lane 7, competition with the cold oligonucleotide containing another potential STAT6 binding site within the sIL-1Ra promoter (SBE2); lane 8, competition with the cold oligonucleotide containing the STAT6 consensus element; lane 9, preincubation of nuclear extracts with antibodies against STAT6 before the addition of the32P-labeled SBE1 probe; lane 10, preincubation of nuclear extracts with control IgG. Lanes 11 and 12 contain the SBE1 probe and the antibodies against STAT6 or control IgG, respectively, without nuclear extracts. The dark arrow shows the STAT6-DNA complex. The open arrow represents the supershifted complex. NS = nonspecific protein-DNA binding. These results are representative of three different experiments.
As previously reported,19 SBE2 only partially competed for the STAT6-DNA complex (lane 7), indicating that SBE2 bound STAT6 with a lower affinity than SBE1. In addition, when gel retardation assays were performed using SBE2 as a 32P-labeled probe, we did not detect the presence of the STAT6-DNA complex (data not shown). To examine whether the binding activity induced by IL-4 was due to STAT6, immunological reactivity of the complexed protein was tested using antibodies raised against STAT6. The results showed that the STAT6-DNA complex was largely competed away with the presence of a higher molecular weight band (supershift) when nuclear extracts were pretreated in the presence of anti-STAT6 antibodies (lane 9). In contrast, control IgG had no effect (lane 10).
Taken together these findings indicate that the IL-4–induced binding activity in HepG2 cell nuclear extracts was due to the presence of STAT6 and confirmed the presence of a STAT6 binding site (SBE1) within the sIL-1Ra promoter.
IL-4 does not alter the IL-1–induced NF-κB binding activity in HepG2 cells.
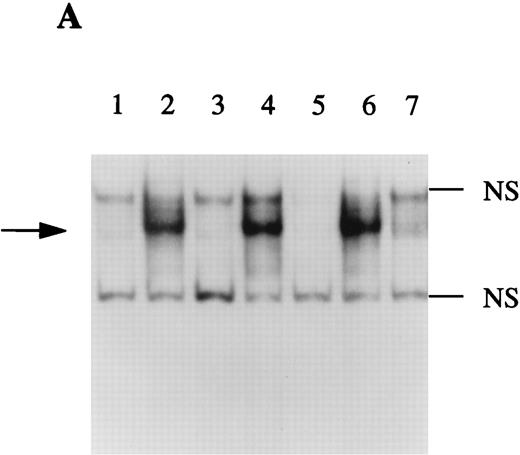
IL-4 has been shown to decrease the NF-κB binding activity in macrophages.20,21 In contrast, nothing is known regarding the effect of this cytokine on NF-κB activation in hepatocytes. We recently showed that NF-κB was involved in the regulation of production sIL-1Ra by HepG2 cells after IL-1β stimulation and the results presented herein showed that IL-4 enhanced the stimulatory effect of IL-1β on IL-1Ra production. To examine the effect of IL-4 on NF-κB activity in HepG2 cells, gel retardation assays were performed using nuclear extracts from HepG2 cells as described above and a 32P-labeled oligonucleotide containing the previously described NF-κB binding site within the sIL-1Ra promoter.2 As shown in Fig 7A, IL-1 β induced the formation of the NF-κB-DNA complex (lane 2) and addition of IL-4 did not modify the NF-κB activity (lane 4). In contrast, we did not detect any NF-κB binding activity in the nuclear extracts from unstimulated or IL-4–stimulated HepG2 cells (lanes 1 and 3). To ensure that the binding activity was due to NF-κB, competition studies were performed with unlabeled oligonucleotides. The presence of the complex was competed away by the wild-type nucleotide (lane 5) and by an oligonucleotide containing the consensus NF-κB binding site (lane 7), but not by a mutated oligonucleotide (lane 6). In addition, as shown in Fig 7B, the complex was totally supershifted when nuclear extracts were incubated with antibodies against the p65 (RelA) subunit of NF-κB (lane 3), whereas control immunoglobulins had no effect (lane 5). Interestingly, antibodies against the p50 subunit of NF-κB had also little effect (lane 4), indicating that p65 was the most abundant protein in this complex.
EMSA of NF-κB binding activity. The DNA binding activity of nuclear extracts from HepG2 cells cultured in the presence or absence of IL-1β and/or IL-4 for 30 minutes were examined using a 32P-labeled oligonucleotide from the sIL-1Ra promoter containing the NF-κB binding site (A). Competition studies were performed with nuclear extracts from HepG2 cells stimulated with IL-1β (see Materials and Methods). Lane 1, unstimulated; lane 2, IL-1β; lane 3, IL-4; lane 4, IL-1β and IL-4; lane 5, competition with the cold probe; lane 6, competition with the cold probe mutated in the NF-κB binding site; lane 7, competition with the cold oligonucleotide containing the NF-κB consensus region. Characterization of the protein present in the NF-κB–DNA complex was performed using specific antibodies against p65(RelA) and p50 (B). Lane 1, unstimulated; lane 2, IL-1β; lane 3, preincubation of nuclear extracts of IL-1β–stimulated HepG2 cells with antibodies against NF-κB p65 (RelA) before the addition of the 32P-labeled NF-κB probe; lane 4, preincubation with antibodies against p50; lane 5, preincubation with control IgG; lanes 6 to 8 contain the NF-κB probe and the antibodies against the p65 subunit, p50 subunit, and control IgG, without nuclear extracts. The dark arrow shows the NF-κB–DNA complex. The open arrow represents the supershifted complex. NS = nonspecific protein-DNA binding. These results are representative of three different experiments.
EMSA of NF-κB binding activity. The DNA binding activity of nuclear extracts from HepG2 cells cultured in the presence or absence of IL-1β and/or IL-4 for 30 minutes were examined using a 32P-labeled oligonucleotide from the sIL-1Ra promoter containing the NF-κB binding site (A). Competition studies were performed with nuclear extracts from HepG2 cells stimulated with IL-1β (see Materials and Methods). Lane 1, unstimulated; lane 2, IL-1β; lane 3, IL-4; lane 4, IL-1β and IL-4; lane 5, competition with the cold probe; lane 6, competition with the cold probe mutated in the NF-κB binding site; lane 7, competition with the cold oligonucleotide containing the NF-κB consensus region. Characterization of the protein present in the NF-κB–DNA complex was performed using specific antibodies against p65(RelA) and p50 (B). Lane 1, unstimulated; lane 2, IL-1β; lane 3, preincubation of nuclear extracts of IL-1β–stimulated HepG2 cells with antibodies against NF-κB p65 (RelA) before the addition of the 32P-labeled NF-κB probe; lane 4, preincubation with antibodies against p50; lane 5, preincubation with control IgG; lanes 6 to 8 contain the NF-κB probe and the antibodies against the p65 subunit, p50 subunit, and control IgG, without nuclear extracts. The dark arrow shows the NF-κB–DNA complex. The open arrow represents the supershifted complex. NS = nonspecific protein-DNA binding. These results are representative of three different experiments.
Effect of IL-4 on the production of CRP by human primary hepatocytes.
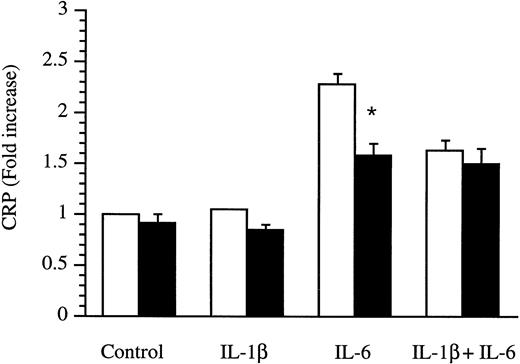
Because HepG2 cells do not produce CRP, we determined the effects of IL-4 on CRP production by human primary hepatocytes. Supernatants were obtained from hepatocytes cultured in the absence or presence of cytokines for 18 hours and were analyzed by ELISA. As shown in Fig 8, the regulation of CRP and IL-1Ra production was clearly different. CRP was only stimulated by IL-6, whereas IL-1β was devoid of any effect. Interestingly, IL-4 significantly downregulated the inducing effect of IL-6. Similar results were obtained with IL-13 (data not shown).
Production of CRP by human primary hepatocytes. Human primary hepatocytes were isolated from livers unsuitable for transplantation and cultured in the absence (control) or presence of IL-1β, IL-6, and a combination of IL-1β and IL-6 without (□) or with (▩) the addition of 10 ng/mL IL-4 for 18 hours. Levels of CRP were measured by ELISA in culture supernatants. Results are expressed as fold increase over unstimulated cells. Values represent the mean ± SEM of experiments performed with three donors. *P< .05 in comparison with cells cultured in the absence of IL-4.
Production of CRP by human primary hepatocytes. Human primary hepatocytes were isolated from livers unsuitable for transplantation and cultured in the absence (control) or presence of IL-1β, IL-6, and a combination of IL-1β and IL-6 without (□) or with (▩) the addition of 10 ng/mL IL-4 for 18 hours. Levels of CRP were measured by ELISA in culture supernatants. Results are expressed as fold increase over unstimulated cells. Values represent the mean ± SEM of experiments performed with three donors. *P< .05 in comparison with cells cultured in the absence of IL-4.
DISCUSSION
In this study, we confirmed that both sIL-1Ra and 16 kD icIL-1RaII, but not 18 kD icIL-1RaI, are produced by HepG2 and human primary hepatocytes. In addition, we showed that both IL-4 and IL-13 significantly amplified the stimulatory effects of IL-1β on production of IL-1Ra by both human hepatoma HepG2 cells and human primary hepatocytes. The effect of IL-4 was not mediated through the induction of another cytokine such as IL-1β or IL-6, because in contrast to IL-1Ra, IL-1β was not produced by either HepG2 cells or human primary hepatocytes (C. Gabay, unpublished results). In addition, we previously showed that human primary hepatocytes do not produce IL-6.11 The results of transfection studies with a construct containing the sIL-1Ra promoter as well as mRNA stability studies indicated that IL-4 primarily stimulated sIL-1Ra transcription. In addition, gel retardation assays showed that IL-4 induced the formation of a STAT6-DNA complex with a recently characterized STAT6 binding element within the sIL-1Ra promoter. We confirmed that IL-6 is the main inducer of CRP and showed that in contrast to IL-1Ra, IL-4 downregulated the effect of IL-6 on production of CRP by hepatocytes.
IL-4 and IL-13 are T-cell–derived cytokines that exibit a broad range of activities in the regulation of inflammatory and immune responses and are thought to play important roles during allergic responses.22 However, the effects of these cytokines are primarily antiinflammatory.23 Both cytokines reduce production of IL-1, TNF-α, and other proinflammatory mediators, whereas they increase production of IL-1Ra by monocytes and macrophages. In addition, IL-4 has been shown to downregulate the production of prostaglandin E2 by synovial macrophage-type cells through the inhibition of cyclooxygenase 2.24 Both IL-4 and IL-13 reduced inflammation in different animal models of arthritis.25-27 The observation that both IL-4 and IL-13 enhanced IL-1β–induced production of IL-1Ra by hepatocytes further emphasizes the antiinflammatory role of these cytokines during the acute-phase response. Interestingly, IL-4 has been shown to amplify the stimulatory effect of IL-1 on expression of monocyte chemotactic protein by endothelial cells28 and of IL-6 by endothelial cells and fibroblasts.21,28 IL-4 has also been reported to enhance the inducing effects of TNF-α on production of IL-1Ra by neutrophils29 and on VCAM-1 expression by endothelial cells.22 However, we did not observe any effect of the combination of TNF-α and IL-4 on production of IL-1Ra by HepG2 cells (C. Gabay, unpublished data).
Some of the antiinflammatory effects of IL-4 and IL-13 have been shown to be mediated through inhibition of NF-κB activation in macrophages.20,21 However, the effect of IL-4 on NF-κB activation has not been examined in hepatocytes. We recently showed that NF-κB plays an important role in the regulation of sIL-1Ra production in HepG2 cells after IL-1 stimulation.2 Our results showed that in HepG2 cells, IL-4 does not modify the IL-1–induced NF-κB binding activity. This observation is consistent with similar findings in dermal and synovial fibroblasts,21indicating that the effects of IL-4 on NF-κB activation varies in different cells.
The term IL-1Ra refers to three different proteins derived from the same gene. One isoform includes a leader sequence and is glycosylated and secreted, 17 kD sIL-1Ra, and two intracellular18kD icIL-1RaI and 16kD icIL-1RaII.14 Our results indicated that both HepG2 cells and primary hepatocytes produced only sIL-1Ra mRNA. In addition, the presence of IL-1Ra in the culture of both HepG2 cells and human primary hepatocytes is related to production and secretion of sIL-1Ra protein. By Western blot analysis, we observed that only icIL-1RaII was detected in the cell lysate of both cell types, whereas sIL-1Ra was present in the supernatant of HepG2 cells.2 These observations are consistent with the results obtained in mouse liver in vivo after the injection of LPS.13
Our results showed that CRP and IL-1Ra are regulated differently in human primary hepatocytes. IL-1β, which is the main inducer of IL-1Ra, exhibited no effect on the induction of CRP. Consistent with the results of previous studies in human primary hepatocytes, IL-1β reduced the stimulatory effects of IL-6.12 Another major finding of this study is that IL-4 significantly downregulated the effect of IL-6 in inducing CRP production by hepatocytes. Other investigators observed that IL-4 alone, but not in the presence of IL-6 significantly decreased the production of CRP.9 Differences between their results and ours may be due to variations in culture conditions.
The differential effects of IL-4 and IL-13 on regulation of CRP and IL-1Ra in vitro may have some clinical relevance. Although both CRP and IL-1Ra react as APP in some inflammatory conditions such as infectious diseases, several clinical observations indicate that CRP and IL-1Ra may be differently regulated in other inflammatory conditions. In systemic lupus erythematosus and dermatomyositis, circulating levels of IL-1Ra are generally elevated and correlate with disease activity whereas serum levels of CRP remain usually normal.30,31 In contrast, in patients with spondylarthrtopathies and rheumatoid arthritis, a reverse pattern has been observed.32 It can be hypothesized that these variations may reflect a difference in the balance of cytokines produced by T helper 1 (Th1) and Th2 lymphocytes. IL-4 and IL-13 are both considered as Th2 cytokines and evidence from experimental animal models suggests that the features of systemic lupus erythematosus and other connective tissue diseases are consistent with a prominent Th2 response.33-36 In addition, elevated circulating levels of IL-4 and IL-13 were observed in patients with scleroderma,37 38 another connective tissue disease generally associated with normal CRP levels. It is therefore possible that IL-4 and IL-13 contribute to the pattern of APP observed in these inflammatory diseases.
In conclusion, our results showed that both IL-4 and IL-13 amplified the inducing effect of IL-1β on production of IL-1Ra by hepatocytes, suggesting another possible mechanism through which IL-4 and IL-13 may exert their antiinflammatory effects in vivo. In addition, the observation that IL-4 and IL-13 downregulated the production of CRP by hepatocytes indicates that expression of different APP genes to common extracellular signals may vary.
Supported by National Institutes of Health grant AR40135 (W.P.A), by the Rocky Mountain Chapter of the Arthritis Foundation (W.P.A), and by a postdoctoral fellowship grant from the Swiss National Science Foundation and from la Fondation Suisse de Bourse de Medecine et Biologie (C.G.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal